Abstract
The vaccinia virus complement control protein (VCP) is secreted by infected cells and has been shown to inhibit complement activation through interactions with C3b/C4b. It contains four short consensus repeat (SCR) domains. It has been suggested that all four SCRs are required for VCP's activity. To elucidate which SCR domains are involved in abolishing complement-enhanced neutralization of vaccinia virus virions, we generated and characterized a panel of mouse monoclonal antibodies (MAbs) raised against VCP. Ten MAbs were isolated and all recognized VCP on Western blots under reducing conditions as well as native-bound VCP in a sandwich enzyme-linked immunosorbent assay. Three of the 10 MAbs (2E5, 3D1, and 3F11) inhibited VCP's abolition of complement-enhanced neutralization of vaccinia virus virions. These MAbs blocked the interaction of VCP with C3b/C4b. The seven remaining MAbs did not alter VCP function in the complement neutralization assay and recognized VCP bound to C3b/C4b. To understand MAb specificity and mode of interaction with VCP, we mapped the MAb binding regions on VCP. The seven nonblocking MAbs all bound to the first SCR of VCP. One of the blocking MAbs recognized SCR 2 while the other two recognized either SCR 4 or the junction between SCRs 3 and 4, indicating that structural elements involved in the interaction of VCP with C3b/C4b are located within SCR domains 2 and 3 and 4. These anti-VCP MAbs may have clinical significance as therapeutic inhibitors of VCP's complement control activity and may also offer a novel approach to managing vaccinia virus vaccine complications that occur from smallpox vaccination.
The vaccinia virus complement control protein (VCP) is a 35-kDa protein that is encoded by the C3L gene and secreted by cells infected with vaccinia virus (7, 8). VCP contains 4 short consensus repeat domains (SCR) that are also present in eukaryotic complement regulatory proteins (9). VCP inhibits activation of complement, binds C4b and C3b, acts as a cofactor in the enzymatic inactivation of C4b and C3b by factor I, prevents the formation of the classical and alternative pathway C3 convertases, and accelerates their decay (6, 12, 15, 18). The structure of VCP has been solved by nuclear magnetic resonance spectroscopy (1, 5) and crystallization (14). VCP is highly conserved in other members of the orthopoxvirus family (19, 20), most notably variola virus, the causative agent of smallpox. Interestingly, the variola virus homolog differs from VCP by only 11 amino acids, yet its human complement inhibitory activity is 100 times higher (16). Monkeypox has a functional VCP homolog; however, it lacks the majority of the fourth SCR (19, 20).
VCP inhibits the antibody-dependent, complement-enhanced neutralization of vaccinia virus virions (3). Animal studies comparing wild-type virus to a mutant vaccinia virus lacking VCP has shown that the mutant virus is attenuated (3, 6). This in vivo effect is likely due in part to enhanced complement-mediated neutralization of virus lacking expression of VCP (3) as well as the recruitment of inflammatory cells to areas of active infection (13). These results indicate that complement activation has a substantial antiviral role in vivo and that VCP helps protect the virus against this host defense mechanism. Thus, the poxvirus complement control proteins might be attractive therapeutic targets to treat poxvirus infections.
Because of concerns about the intentional or accidental release of smallpox (2), widespread smallpox vaccination may be needed. In past smallpox vaccination efforts, complications arising from the use of this live vaccine were treated with human vaccinia immune globulin (VIG) obtained from vaccinia virus-immunized people (17). Current supplies of VIG are low, and while new stocks are being generated, there are serious drawbacks to relying on a blood product. Consequently, there is a critical need to develop therapeutic interventions to counter complications from the current vaccine. We reasoned that inactivation of a viral immune evasion protein may be an approach to managing vaccinia virus vaccine complications. Antibodies with such activity may become part of a cocktail of human monoclonal antibodies (MAbs) directed at specific poxvirus proteins. Alternatively, such antibodies may be useful to augment the effectiveness of VIG.
Therefore, in the present study we raised MAbs against VCP in order to identify MAbs that abolish VCP's ability to inhibit complement-enhanced neutralization of vaccinia virus virions and to elucidate the critical regions on VCP responsible for its complement-inhibitory function.
MATERIALS AND METHODS
Expression and purification of rVCP in Pichia.
Recombinant VCP (rVCP) was expressed and purified as previously described (18) by using the Pichia pastoris yeast system. The recombinant yeast generated by recombination with the pHIL-S1-VCP vector, was placed in a 30°C shaker for 24 h in 10 ml of sterile buffered minimal glycerol complex (BMGY) medium. This small culture was used to inoculate 1 liter of sterile BMGY medium and then incubated at 30°C with vigorous shaking for 48 h. To induce rVCP expression, cells were centrifuged at ∼5,400 × g for 20 min, resuspended in 150 ml of BMMY medium (BMGY containing 0.5% methanol but without glycerol), and incubated with vigorous shaking at 30°C. After 48 h, the culture was centrifuged and supernatant containing rVCP was collected. The rVCP was purified by successively precipitating the culture supernatant with 20, 40, 60, and 80% ammonium sulfate. The resulting pellets were then dissolved in phosphate-buffered saline (PBS) and dialyzed overnight at 4°C. The fractions were run on a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE) gel, and those containing the most rVCP (40 and 60%) were then purified over a DEAE-Sephacel column preequilibrated with PBS. The protein was eluted with PBS and detected by the Bradford method (Bio-Rad), and fractions containing purified rVCP were identified by SDS-PAGE, pooled, and dialyzed against PBS.
Generation of mouse anti-rVCP hybridomas.
Mouse immunizations and hybridoma production were done at the Pocono Rabbit Farm and Laboratory, Inc. (Canadensis, Pa.) by using standard procedures. In brief, mice were immunized with rVCP, and the mouse that developed the highest titer of anti-VCP antibodies was sacrificed for fusion. The resulting hybridoma supernatants were screened by enzyme-linked immunosorbent assay (ELISA) with rVCP. Ten hybridomas were selected and subcloned. Selected hybridomas were injected intraperitoneally into pristane-primed BALB/c mice (Cocalico Biologicals). MAbs were purified from the ascitic fluid by sequential caprylic acid and ammonium sulfate precipitations. The use of animals in this research has complied with all relevant federal guidelines and institutional policies.
ELISA to assess the neutralization of VCP's complement inhibitory activity by MAbs.
Microtiter ELISA plates were coated with 50 μl of ovalbumin (10 mg/ml; Sigma). Rabbit anti-ovalbumin antiserum (ICN Cappel) was then added at a 1:100 dilution to form an immunocomplex on which complement could be activated. To determine whether our MAbs could neutralize VCP's complement inhibitory activity, 7 μM rVCP or bovine serum albumin (BSA) (as a control) was serially diluted in gelatin Veronal buffered saline with 0.5 mM MgCl2 and 0.15 mM CaCl2 (GVB++) (30 μl/well). Twenty microliters of an equimolar amount of MAb was added to each well, and then 10 μl of whole human plasma (collected in 50 μg of Refludan [Aventis]/ml and stored in aliquots at −70°C) diluted 1:25 in GVB++ was added. Bound C3b-iC3b was detected by using horseradish peroxidase (HRP)-conjugated goat anti-human C3 antibody (ICN Cappel). Color was developed by adding 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) peroxidase substrate. All steps after coating were performed at 25°C for 1 h, and unbound proteins were removed by washing with PBS containing 0.05% Tween 20.
Complement-mediated neutralization of vaccinia virus virions.
The in vitro assay of complement-mediated neutralization of vaccinia virus virions (intracellular mature virus [IMV]) was carried out as described previously (3) with the following modifications. Briefly, serum-free media containing VCP or no VCP was obtained from RK-13 cells infected with wild-type virus or vSIGK-1, a recombinant virus with the entire VCP open reading frame deleted (6), respectively. To remove virus that was released into the supernatants during the infection, the media were centrifuged at ∼28,000 × g for 1 h. Aliquots of the media were then incubated with 5 μg of MAbs for 1 h at room temperature, after which ∼5 × 103 PFU of IMV was added. One hundred-microliter samples were then incubated for 1.5 h at 37°C along with a 1:10 dilution of complement (guinea pig serum; Calbiochem) and/or a 1:100 dilution of hyperimmune anti-vaccinia virus rabbit serum. The resulting titer was obtained by counting plaques after serial dilution on BSC-1 cells.
Vaccinia virus and bacterial expression of VCP SCRs.
Vaccinia virus-produced VCP (vv-VCP) was obtained from wild-type vaccinia virus (strain WR) by infecting BSC-1 cells in serum-free medium (Opti-MEM; Life Technologies). The first SCR of VCP (vv-SCR1) was expressed and secreted from cells infected with vSI-13, a previously constructed recombinant vaccinia virus in which the VCP gene is disrupted just after the first SCR (4). The vaccinia virus negative control medium (vv-control) contained no VCP and was obtained from cells infected with recombinant vaccinia vSIGK-1, a recombinant virus from which the entire VCP gene had been deleted (6).
Since all the MAbs recognized denatured VCP on Western blots, we used an Escherichia coli expression system to express overlapping recombinant SCRs in order to further map the epitopes recognized by the MAbs. As detailed below, segments of the VCP gene were amplified by PCR and ligated into the E. coli expression vector Pin Point Xa-3 (Promega). The Pin Point system is designed to produce fusion proteins that are biotinylated in vivo. The biotinylation reaction in E. coli is catalyzed by the biotin ligase holoenzyme and results in the specific incorporation of a single biotin onto one lysine residue in the fusion tag. To generate SCRs 1 and 2, the oligonucleotide primers 5′-CAGGATCCTGCTGTACTATTCCGTCACGA-3′ (the BamHI site is underlined) and 5′-ATGCAACAGCGGCCGCTCAAACAGATTCACAAATAGGTGC-3′ (the NotI site is underlined and the stop codon is in boldface type) were used to amplify a 381-bp segment of the C3L gene of vaccinia virus (strain WR). To generate SCRs 2 and 3, the oligonucleotide primers 5′-ATGGATCCTGCCCATCGCCTCGAGATATC-3′ and 5′-ATGCAACAGCGGCCGCTCATTTAACAATCTGACACGTGGG-3′ were used to amplify a 360-bp DNA fragment. To generate SCRs 3 and 4, the oligonucleotide primers 5′-TGGGATCCTGCCAATCCCCTCCATCTATA-3′ and 5′-ATTAGGCAGCGGCCGCTCAGCGTACACATTTTGGAAGTTC-3′ were used to amplify a 348-bp DNA fragment. The PCR conditions were 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s (30 cycles). After amplification, both the PCR products and the vector PinPoint Xa-3 were digested with BamHI and NotI, gel purified, and ligated with T4 DNA ligase. The DNA constructs were used to transform JM109 E. coli cells.
For expression of recombinant proteins, 5 ml of Luria-Bertani containing 2 μM biotin and 100 μg of ampicillin/ml were inoculated with a single bacterial colony and incubated overnight at 37°C with shaking. The next day, the culture was diluted 1:100 in fresh Luria-Bertani medium and incubated for an additional 1 h at 37°C. Protein expression was induced by adding 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubating the mixture for an additional 5 h. After harvesting the cells by centrifugation at ∼5,400 × g for 15 min, the cells were lysed with 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5% glycerol, and 1 mg of lysozyme/ml. The crude lysates were centrifuged, and the supernatants were subsequently used for Western blot analysis.
Western blot analysis to determine the reactivity of MAbs to rVCP, vv-VCP, vv-SCR1, and overlapping SCRs.
Recombinant VCP (2 μg) or media (20 μl) from infected cells containing vv-VCP, vv-SCR1, or vv-control was run on an SDS (0.1%)-15% PAGE gel and transferred to a polyvinylidene difluoride (PVDF) membrane at 100 mA per gel for 1 h. Membranes were blocked with 10% milk in PBS for 1 h at 25°C. MAbs were diluted 1:20 in 10% milk-PBS containing 0.05% Tween 20 and incubated for 1 h at 25°C. After the membranes were washed two times for 15 min each in PBS containing 0.05% Tween 20, the MAbs bound to VCP were detected with HRP-conjugated goat anti-mouse antibody for 30 min at 25°C. Membranes were washed three times, and immunoreactive bands were visualized by chemiluminescence according to the manufacturer's instructions.
Sandwich ELISA to measure reactivity of MAbs with VCP.
Ninety-six-well microtiter ELISA plates were coated with 50 μl of 2-μg/ml goat anti-mouse immunoglobulin G (IgG) Fc (ICN Cappel) in coating buffer for 2 h at 25°C or overnight at 4°C. After coating, wells were blocked with 200 μl of 1% BSA (Sigma). Anti-VCP MAb supernatants were added at a 1:20 dilution in PBS. rVCP, vv-VCP, vv-SCR1 and vv-control were then serially diluted in PBS starting at 10 ng of rVCP/ml or a 1:5,000 dilution of vaccinia virus-produced supernatants. VCP bound to MAbs was detected by using rabbit anti-VCP polyclonal antiserum (generated by Cocalico Biologicals) at a 1:5,000 dilution, followed by HRP-conjugated goat anti-rabbit antibody diluted 1:1,000 in PBS (Bio-Rad). All steps following the blocking step involved incubation for 1 h at 25°C. Unbound proteins were washed with PBS containing 0.05% Tween 20. Color was developed by adding ABTS peroxidase substrate, and optical density (OD) was measured at 405 nm.
ELISA to measure the reactivity of MAbs with VCP bound to immunocomplex-fixed C3b-C4b.
Immune complexes were formed in microtiter wells as described above. A 1:80 dilution of whole human plasma (collected as described above) in GVB++ was added and incubated at 25°C for 1 h. Supernatant from vaccinia virus-infected cells (vv-VCP, vv-SCR1, or vv-control) was serially diluted starting at a 1:40 dilution in PBS and incubated as described above. To detect VCP bound to immunocomplex-fixed C3b-C4b, VCP MAb supernatants were then added at a 1:10 dilution in PBS for 1 h at 25°C. MAbs recognizing VCP bound to C3b-C4b were detected by using a 1:1,000 dilution of HRP-conjugated goat anti-mouse IgG antibody (Bio-Rad). Unbound proteins were removed by washing with PBS containing 0.05% Tween 20 between all steps after blocking.
ELISA to measure inhibition of VCP binding to immunocomplex-fixed C3b-C4b with MAbs.
Complement was activated by the immune complex and bound to microtiter ELISA plates as described above. Supernatants from vaccinia virus-infected cells (vv-VCP, vv-SCR1, or vv-control) were serially diluted starting at a 1:40 dilution in PBS in a separate nonadsorbable 96-well microtiter plate (Corning). Hybridoma culture supernatants were added to the vaccinia virus supernatants at a 1:10 dilution in PBS. This mixture was then transferred to the plate containing immunocomplex-fixed C3b-C4b and incubated at 25°C for 1 h. MAbs recognizing VCP bound to C3b-C4b were detected by using a 1:1,000 dilution of peroxidase-conjugated goat anti-mouse IgG antibody (Bio-Rad). Unbound proteins were washed between all steps after saturation by using PBS containing 0.05% Tween 20.
ELISA to assess the ability of MAb 3D1 to inhibit the binding of 3F11 to VCP.
Since two of the blocking MAbs reacted with the same SCR fragment, we used a competitive ELISA to assess the ability of MAbs 3D1 and 3F11 to compete for binding to the same epitope. MAb 3F11 was conjugated with HRP by following standard procedures. HRP (type VI-A; Sigma) was dissolved in double-distilled water and activated with 0.1 M sodium meta-periodate for 20 min at room temperature. The activated HRP and MAb were each dialyzed overnight against 0.005 M acetate buffer (pH 4.4) and 0.01 M carbonate-bicarbonate buffer (pH 9.5), respectively. The antibody and activated HRP were conjugated at pH 9.5 for 2 h at room temperature. After conjugation, 50 μl of sodium borohydride (4 mg/ml) was added and incubated for 2 to 3 h. The resulting conjugated antibody was dialyzed against 50 mM PBS for 2 days with five changes. The conjugated MAb (diluted with glycerol 1:1) was used in a competitive assay against MAb 3D1 and MAb 3B1 (as a negative control). Unconjugated MAbs were captured in a 96-well microtiter ELISA plate (Nunc) coated with 100 ng of goat anti-mouse Fc antibody (ICN Pharmaceuticals)/well. Then 200 ng of rVCP or BSA (as a control)/well was added to each well, and the mixtures were incubated at 37°C for 1 h followed by the addition of HRP-conjugated MAb 3F11 (1:12,000 dilution) for 1 h at 37°C. The bound HRP-conjugated antibody was detected as described above.
RESULTS
Generation of MAbs against rVCP.
We had previously constructed and isolated a recombinant Pichia that secreted functional rVCP into the medium after induction with methanol. Coomassie staining of a gel loaded with DEAE-Sephacel column-purified rVCP showed a single band, indicating the purity of the protein used to immunize mice (data not shown). Western blotting of this purified rVCP, together with medium from vaccinia virus-infected cells confirmed that the recombinant protein is identical in molecular size to the vaccinia virus-produced protein. The immunized mice developed high-titer polyclonal antibodies that reacted with rVCP in an ELISA and to both rVCP and vv-VCP on a Western blot (data not shown). Ten hybridoma clones secreting MAbs that recognized VCP were selected and subcloned.
Alteration of VCP's ability to inhibit complement activation by anti-VCP MAbs.
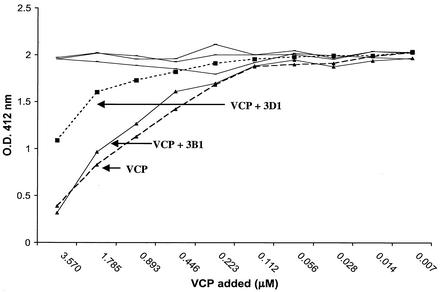
To determine if any of the MAbs interfered with VCP's ability to inhibit complement activation, we developed an ELISA-based assay in which complement was activated with ovalbumin-antiovalbumin immune complexes. We used an anti-C3 antibody to measure the amount of C3b deposited on a plate. VCP, MAb, and human complement were premixed before being added to the preformed immune complexes in a 96-well plate (Fig. 1). If no VCP or MAb was added, a set amount of C3b was deposited (OD, ∼2.0), representing full activation of complement on the immune complexes. Since VCP is known to inhibit complement activation, one would expect less C3b to be deposited in the presence of VCP. As expected, less C3b was deposited as the concentration of rVCP was increased (Fig. 1). When MAbs were incubated along with VCP, we found two types of activity. One set of MAbs (represented in Fig. 1 by 3B1) did not inhibit VCP's activity, and thus, less C3b was deposited, indicating no alteration in the inhibitory effect of VCP on complement activation. We called this group of MAbs nonblocking MAbs. Another group of MAbs (represented in Fig. 1 by 3D1) resulted in the deposition of more C3b, demonstrating that this type of blocking MAb neutralized the ability of VCP to inhibit complement activation.
FIG. 1.
Alteration by MAbs of VCP's complement inhibitory activity. Serial dilutions of rVCP were incubated with human serum in the presence or absence of MAb and then added to the wells containing immune complexes. The amount of C3b deposited was detected by using anti-C3 antibody. Since the reactivity of all of the MAbs within each of the two groups of MAbs was similar, representative data are shown for MAbs 3B1 (nonblocking) and 3D1 (blocking). O.D., optical density.
Abolition of VCP's inhibition of complement-enhanced neutralization of vaccinia virus virions by anti-VCP MAbs.
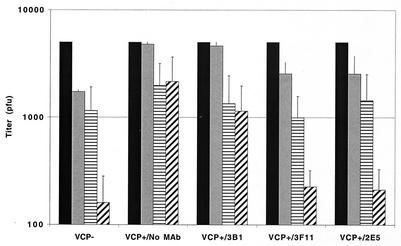
Since one of VCP's immune evasion activities is to protect infected cells and released virus from the host complement-mediated attack (3), we determined whether the MAbs could abolish VCP's inhibition of complement-enhanced neutralization of vaccinia virus virions. Similar to published results (3), we again found that in the absence of VCP, IMV is neutralized by complement (Fig. 2, VCP−). When VCP is present, complement-enhanced neutralization of IMV is inhibited (Fig. 2, VCP+/no MAb). A similar result was obtained when a nonblocking MAb was included (Fig. 3, VCP+/3B1), indicating that antibody binding to VCP alone did not affect the protein's function. However, when blocking MAbs were incubated, complement was once again able to neutralize virions (Fig. 2, VCP+/2E5 or 3F11) and antibody dependent, complement-enhanced neutralization was restored (Fig. 2, VCP+/2E5 or 3F11).
FIG. 2.
Complement-mediated neutralization of vaccinia virus virions. Vaccinia virus virions (∼5 × 103 PFU) were incubated in 100-μl aliquots of media from cells infected with a recombinant virus that does not express VCP (VCP−) or from cells infected with vaccinia virus (VCP+) in the absence of MAb (VCP+/No MAb) or in the presence of 5 μg of nonblocking MAb (VCP+/3B1) or blocking MAbs (VCP+/2E5 or 3F11). Bars represent input virus without complement or anti-vaccinia virus antibody (solid bars), complement alone (at 1:10 final dilution) (gray bars), anti-vaccinia virus antibody alone (at 1:100 final dilution) (horizontally striped bars), or both complement and anti-vaccinia virus antibody (diagonally striped bars). The data represent the means (± standard deviations) of 3 experiments. The data were standardized to the titer of 5 × 103 PFU.
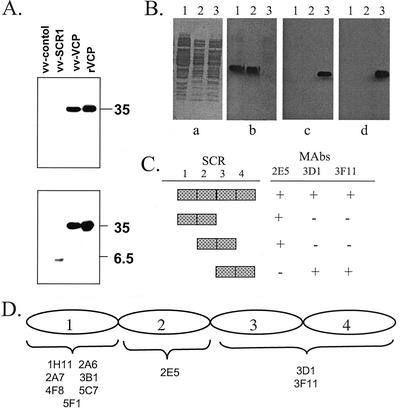
FIG. 3.
Mapping of SCR domains of VCP recognized by MAbs. (A) rVCP, along with medium from recombinant vaccinia virus that did not express VCP (vv-control) or only expressed the first SCR (vv-SCR1) or full-length VCP (vv-VCP), was resolved on SDS (0.1%)-15% PAGE, blotted onto a PVDF membrane, and incubated with the panel of anti-VCP MAbs. The upper panel shows the reactivity of MAb 3F11. The lower panel shows the reactivity of MAb 3B1. Since all of the MAbs within each group of MAbs had identical reactivities, only representative Western blots are shown. (B) Epitope mapping of MAbs against VCP by using overlapping recombinant SCRs. Culture lysates were resolved on SDS (0.1%)-10% PAGE, blotted onto a PVDF membrane, and incubated with the panel of anti-VCP MAbs. (a) Coomassie blue staining of whole lysates (lane 1, SCRs 1 and 2; lane 2, SCRs 2 and 3; lane 3, SCRs 3 and 4); (b) reactivity with MAb 2E5; (c) reactivity with MAb 3D1; (d) reactivity with MAb 3F11. (C) Table summarizing the selective reactivity of MAbs against overlapping SCRs of VCP. +, band seen on Western blots; −, no band seen on Western blots. (D) Schematic diagram summarizing the SCR domains of VCP recognized by the 10 MAbs in Western blots and ELISA.
Mapping of the SCR domains of VCP recognized by MAbs.
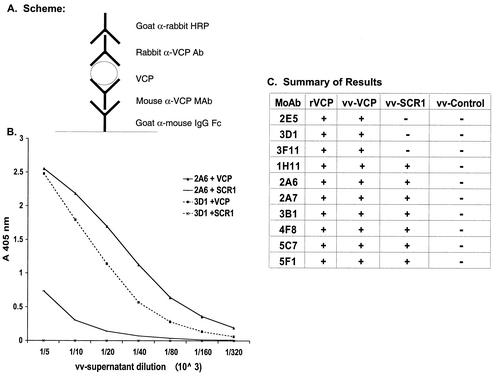
To begin to understand the specificity and mode of interaction of the blocking and nonblocking MAbs, we mapped the regions on VCP to which the MAbs bound. All 10 MAbs recognized full-length VCP produced by Pichia or vaccinia virus on Western blots under reducing conditions (Fig. 3A). Using a previously constructed recombinant vaccinia virus (4), in which the VCP gene was disrupted after SCR 1 and the first SCR was therefore expressed and secreted from vaccinia virus-infected cells, we found that seven of the MAbs (1H11, 2A6, 2A7, 3B1, 4F8, 5C7, and 5F1) reacted with this first SCR and three of the MAbs (2E5, 3D1, and 3F11) did not (Fig. 3A). All of the MAbs reacted with native bound VCP in a sandwich ELISA. Here, the MAbs were captured by using a goat anti-mouse antibody followed by incubation with either rVCP or medium from vaccinia virus-infected cells (Fig. 4). Bound VCP was detected by using a rabbit polyclonal anti-VCP antibody. This ELISA assay also confirmed the results of the Western blot experiment, in that seven MAbs (1H11, 2A6, 2A7, 3B1, 4F8, 5C7, and 5F1) recognized the first SCR and three of the MAbs (2E5, 3D1, and 3F11) did not.
FIG. 4.
Sandwich ELISA showing reactivity of representative MAbs with VCP. (A) Schematic diagram summarizing the experimental design. MAbs were captured, and serial dilutions of medium from vaccinia virus-infected cells were added. VCP or VCP fragments were detected by using an anti-VCP polyclonal rabbit antibody. (B) Since the reactivity of all the MAbs within each of the two groups was similar, representative data are shown for MAbs 2A6 (solid line, binds to VCP and SCR1) and 3D1 (dashed lines, binds only VCP). (C) Summary data showing the reactivity of all of the MAbs with serial dilutions of vaccinia virus-produced VCP, SCR 1, control, and recombinant Pichia-produced VCP. +, reactivity in ELISA; −, no reactivity in ELISA.
Since we found that all of the MAbs recognized denatured VCP on Western blots, we used a bacterial expression system to express SCRs 1 and 2, SCRs 2 and 3, or SCRs 3 and 4 in order to allow us to determine which SCR domains were recognized by the blocking MAbs (Fig. 3B and C). Using Western blots (Fig. 3B) and ELISA (data not shown), we found that MAb 2E5 recognized bacterially expressed SCRs 1 and 2 and SCRs 2 and 3 but did not recognize SCRs 3 and 4. This finding suggests that MAb 2E5 recognizes an epitope in SCR 2. The other two MAbs, 3D1 and 3F11, only recognized bacterially expressed SCRs 3 and 4 but not SCRs 2 and 3 (Fig. 3B), indicating that the epitopes for these MAbs were on SCR 4 or the junction between SCR 3 and SCR 4. A competition assay in which one of these two antibodies was labeled revealed that 3D1 and 3F11 recognized the same epitope on VCP (data not shown). The mapping data for the overlapping SCRs are summarized in Fig. 3C, and all of the mapping results are schematically summarized in Fig. 3D.
MAb inhibition of VCP's interaction with C3b-C4b.
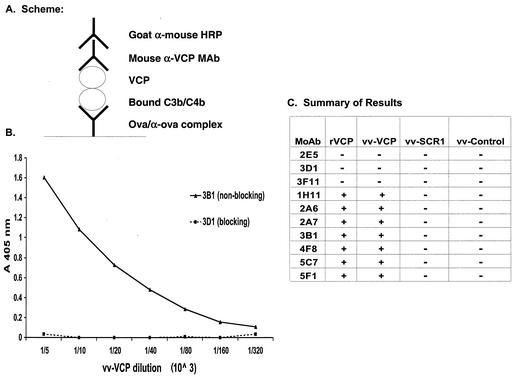
To begin to understand the mechanism by which the MAbs that bound to SCR 1 did not interfere with VCP's in vitro activity while MAbs that bound SCR 2 and SCRs 3 and 4 inhibited VCP's activity, we asked whether any of the MAbs would recognize VCP bound to C3b-C4b. Since VCP is known to bind C3b and C4b (12), we hypothesized that MAbs that did not interfere with VCP's function would not block VCP interacting with C3b and C4b while MAbs that inhibited VCP's function would also inhibit its interactions with C3b-C4b. We used the ELISA-based assay in which complement was activated with ovalbumin-antiovalbumin immune complexes, onto which C3b-C4b was deposited (Fig. 5). We then added VCP to the wells and detected VCP that bound to deposited C3b-C4b with either the polyclonal rabbit antibody against VCP (in this case we used a goat anti-ovalbumin antibody [ICN Cappel] to generate immune complexes) or the MAbs. Figures 5B and C depict a representative result and a summary of reactivity for all the MAbs, respectively. The seven MAbs that reacted with SCR 1 all recognized VCP bound to C3b-C4b. The remaining three MAbs (2E5, 3D1, and 3F11) that recognized SCRs other than SCR 1 and reversed VCP's abolition of complement-enhanced neutralization of IMV did not recognize bound C3b-C4b. This assay also revealed that soluble VCP-SCR 1 did not bind to deposited C3b-C4b.
FIG. 5.
Reactivity of MAbs with VCP bound to fixed C3b-C4b. (A) Schematic diagram summarizing the experimental design. As described in Materials and Methods, C3b-C4b was deposited by immune complex-mediated complement activation. Serial dilutions of medium from vaccinia virus-infected cells were added and followed by the addition of the MAb. (B) Since the reactivity of all of the MAbs within each of the two groups was similar, representative data are shown for MAbs 3B1 (solid line, nonblocking) and 3D1 (dashed line, blocking). (C) Summary data showing reactivity of all the MAbs with serial dilutions of vaccinia virus-produced VCP, SCR 1, control, and recombinant Pichia-produced VCP. +, reactivity in ELISA; −, no reactivity in ELISA.
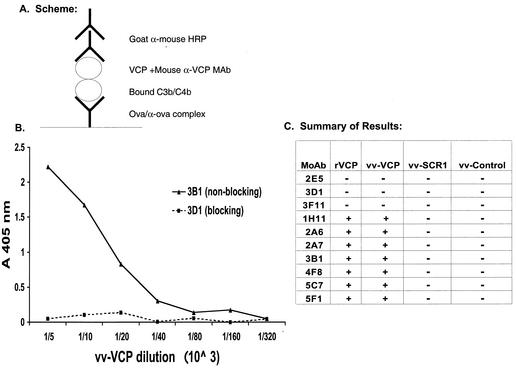
Since three of the 10 MAbs (2E5, 3D1, and 3F11) interfered with VCP function in complement-mediated neutralization of IMV and did not bind to VCP bound to deposited C3b-C4b, we hypothesized that these MAbs might block the interaction of VCP with C3b-C4b. To test this hypothesis, we modified the ELISA-based assay described above. We incubated VCP with the various MAbs before adding it to the wells containing immune complex-activated complement (Fig. 6). Figures 6B and C illustrate a representative result and a summary of reactivity for all the MAbs, respectively. None of the seven MAbs that reacted with SCR 1 interfered with VCP's interaction with C3b-C4b. In contrast, all three MAbs (2E5, 3D1, and 3F11) that did not recognize VCP bound to C3b-C4b blocked the interaction between VCP and C3b-C4b.
FIG. 6.
MAb inhibition of VCP binding to fixed C3b-C4b. (A) Schematic diagram summarizing the experimental design. As described in Materials and Methods, C3b-C4b was deposited by immune complex-mediated complement activation. Serial dilutions of medium from vaccinia virus-infected cells were incubated with MAbs and then added to the wells. (B) Since the reactivity of all of the MAbs within each of the two groups was similar, representative data are shown for MAbs 3B1 (solid line, nonblocking) and 3D1 (dashed line, blocking). (C) Summary data showing the reactivity of all the MAbs with serial dilutions of vaccinia virus-produced VCP, SCR 1, control, and recombinant Pichia-produced VCP. +, reactivity in ELISA; −, no reactivity in ELISA.
DISCUSSION
In the present study we have generated and characterized MAbs raised against VCP, with the goal of identifying site-specific MAbs that blocked VCP's ability to inhibit complement-mediated neutralization of vaccinia virus virions. Screening of hybridomas generated to a recombinant VCP resulted in 10 MAbs that recognized VCP. Seven of the 10 MAbs recognized VCP bound to C3b-C4b and did not inhibit VCP's ability to inhibit complement activation. These seven nonblocking MAbs all interacted with SCR 1 and did not interfere with VCP's abolition of complement-enhanced neutralization of IMV. Three of the 10 MAbs did not recognize VCP when bound to C3b-C4b. When preincubated with VCP, they blocked VCP binding to C3b-C4b. These blocking antibodies all prevented VCP's inhibition of complement-enhanced neutralization of vaccinia virus virions. One of the blocking MAbs (2E5) recognized an epitope on SCR 2. The other two blocking MAbs (3D1 and 3F11) recognized an epitope on SCRs 3 and 4. Since these MAbs did not recognize SCRs 2 and 3, they are either reacting to an epitope on SCR4 or the hinge region between SCR 3 and SCR 4. Furthermore, a competition assay revealed that they recognized the same or overlapping epitopes.
Our observation that the blocking MAbs mapped to three of the four SCRs may not be that surprising. It has previously been shown that all four SCRs were required for C3b binding to VCP (15). These data were generated by using a membrane-bound chimeric protein in which VCP SCRs replaced SCRs 1 and 2 of complement receptor 2. The requirement of all four SCRs for inhibition of complement activation was supported by the expression of various soluble truncations and mutations (19), although an SCR 1-to-3 construct was not reported. While VCP is highly conserved among members of the orthopoxvirus family (19, 20), it is intriguing that the absence of most of the SCR 4 in monkeypox may point to the functional components of the soluble complement inhibitor being contained within SCRs 1 to 3. Furthermore, the 11 amino acids that differ between VCP and the variola virus homolog (10, 11) are all in the same SCRs where the blocking MAbs map. These amino acid differences confer enhanced activity of the variola virus protein in humans (16).
In light of the fact that the structural elements of VCP involved in the interaction with C4b-C3b have not yet been determined, the mapping of these MAbs along with the known structure of VCP (1, 5, 14) will permit us to determine critical regions in VCP that allow interactions with C3b-C4b. Although it is possible that the antibodies are inhibiting VCP complement-modulating activity by steric hindrance or by induction of conformational changes, since the MAbs recognize linear epitopes on VCP, the mapping of these epitopes will allow identification of smaller areas on an SCR needed for C3b-C4b binding and might help future studies that seek to design chemical compounds with similar specificities and functions to those of the blocking antibodies.
The strategy of targeting VCP as a potential treatment of complications from vaccinia virus vaccination is supported by the data presented for in vitro complement-mediated neutralization. The ability of blocking MAbs to abolish VCP's ability to inhibit complement-mediated neutralization of vaccinia virus virions may result in the complement system helping the host regain control of a vaccinia virus infection. The attenuated phenotype that resulted from a genetic knockout of VCP indicates an in vivo role of VCP during viral infection. The MAbs that block VCP's ability to inhibit complement activation will be useful in future studies addressing the in vivo role of VCP during viral infection. If such future studies are successful, the variola virus homolog of VCP may also be a promising target for altering the disease caused by smallpox. Furthermore, the MAbs may be useful for determining specific SCR reactivities of VIG preparations.
Acknowledgments
S.N.I. and E.A. contributed equally to this work.
This work was supported by Public Health Service grants AI-48487 and AI-47237 from the National Institutes of Health.
REFERENCES
- 1.Henderson, C. E., K. Bromek, N. P. Mullin, B. O. Smith, D. Uhrin, and P. N. Barlow. 2001. Solution structure and dynamics of the central CCP module pair of a poxvirus complement control protein. J. Mol. Biol. 307:323-339. [DOI] [PubMed] [Google Scholar]
- 2.Henderson, D. A. 1999. The looming threat of bioterrorism. Science 283:1279-1282. [DOI] [PubMed] [Google Scholar]
- 3.Isaacs, S. N., G. J. Kotwal, and B. Moss. 1992. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc. Natl. Acad. Sci. USA 89:628-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isaacs, S. N., E. J. Wolffe, L. G. Payne, and B. Moss. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 66:7217-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirkitadze, M. D., C. Henderson, N. C. Price, S. M. Kelly, N. P. Mullin, J. Parkinson, D. T. Dryden, and P. N. Barlow. 1999. Central modules of the vaccinia virus complement control protein are not in extensive contact. Biochem. J. 344:167-175. [PMC free article] [PubMed] [Google Scholar]
- 6.Kotwal, G. J., S. N. Isaacs, R. McKenzie, M. M. Frank, and B. Moss. 1990. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science 250:827-830. [DOI] [PubMed] [Google Scholar]
- 7.Kotwal, G. J., and B. Moss. 1988. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology 167:524-537. [PubMed] [Google Scholar]
- 8.Kotwal, G. J., and B. Moss. 1988. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature 335:176-178. [DOI] [PubMed] [Google Scholar]
- 9.Liszewski, M. K., and J. P. Atkinson. 1998. Regulatory proteins of complement, p. 149-166. In J. E. Volanakis and M. M. Frank (ed.), The human complement system in health and disease. Decker, New York, N.Y.
- 10.Massung, R. F., J. J. Esposito, L.-L. Liu, J. Qi, T. R. Utterback, J. C. Knioght, L. Aubom, T. E. Yuran, J. M. Parsons, V. N. Loparev, N. A. Selivanov, K. F. Cavallaro, A. R. Kerlavate, B. W. J. Mahy, and J. C. Venter. 1993. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature 366:748-751. [DOI] [PubMed] [Google Scholar]
- 11.Massung, R. F., L. I. Liu, J. Qi, J. C. Knight, T. E. Yuran, A. R. Kerlavage, J. M. Parsons, J. C. Venter, and J. J. Esposito. 1994. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology 201:215-240. [DOI] [PubMed] [Google Scholar]
- 12.McKenzie, R., G. J. Kotwal, B. Moss, C. H. Hammer, and M. M. Frank. 1992. Regulation of complement activity by vaccinia virus complement-control protein. J. Infect. Dis. 166:1245-1250. [DOI] [PubMed] [Google Scholar]
- 13.Miller, C. G., D. E. Justus, S. Jayaraman, and G. J. Kotwal. 1995. Severe and prolonged inflammatory response to localized cowpox virus infection in footpads of C5-deficient mice: investigation of the role of host complement in poxvirus pathogenesis. Cell. Immunol. 162:326-332. [DOI] [PubMed] [Google Scholar]
- 14.Murthy, K. H., S. A. Smith, V. K. Ganesh, K. W. Judge, N. Mullin, P. N. Barlow, C. M. Ogata, and G. J. Kotwal. 2001. Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell 104:301-311. [DOI] [PubMed] [Google Scholar]
- 15.Rosengard, A. M., L. C. Alonso, L. C. Korb, W. M. Baldwin, III, F. Sanfilippo, L. A. Turka, and J. M. Ahearn. 1999. Functional characterization of soluble and membrane-bound forms of vaccinia virus complement control protein (VCP). Mol. Immunol. 36:685-697. [DOI] [PubMed] [Google Scholar]
- 16.Rosengard, A. M., Y. Liu, Z. Nie, and R. Jimenez. 2002. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc. Natl. Acad. Sci. USA 99:8808-8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenthal, S. R., M. Merchlinsky, C. Kleppinger, and K. L. Goldenthal. 2001. Developing new smallpox vaccines. Emerg. Infect. Dis. 7:920-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahu, A., S. N. Isaacs, A. M. Soulika, and J. D. Lambris. 1998. Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J. Immunol. 160:5596-5604. [PubMed] [Google Scholar]
- 19.Smith, S. A., N. P. Mullin, J. Parkinson, S. N. Shchelkunov, A. V. Totmenin, V. N. Loparev, R. Srisatjaluk, D. N. Reynolds, K. L. Keeling, D. E. Justus, P. N. Barlow, and G. J. Kotwal. 2000. Conserved surface-exposed K/R-X-K/R motifs and net positive charge on poxvirus complement control proteins serve as putative heparin binding sites and contribute to inhibition of molecular interactions with human endothelial cells: a novel mechanism for evasion of host defense. J. Virol. 74:5659-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uvarova, E. A., and S. N. Shchelkunov. 2001. Species-specific differences in the structure of orthopoxvirus complement-binding protein. Virus Res. 81:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]