Abstract
Murine gammaherpesvirus 68 (γHV68; also known as MHV-68) can establish a latent infection in both inbred and outbred strains of mice and, as such, provides a tractable small-animal model to address mechanisms and cell types involved in the establishment and maintenance of chronic gammaherpesvirus infection. Latency can be established at multiple anatomic sites, including the spleen and peritoneum; however, the contribution of distinct cell types to the maintenance of latency within these reservoirs remains poorly characterized. B cells are the major hematopoietic cell type harboring latent γHV68. We have analyzed various splenic B-cell subsets at early, intermediate, and late times postinfection and determined the frequency of cells either (i) capable of spontaneously reactivating latent γHV68 or (ii) harboring latent viral genome. These analyses demonstrated that latency is established in a variety of cell populations but that long-term latency (6 months postinfection) in the spleen after intranasal inoculation predominantly occurs in B cells. Furthermore, at late times postinfection latent γHV68 is largely confined to the surface immunoglobulin D-negative subset of B cells.
Members of the gammaherpesvirus subfamily represent important pathogens that infect a wide variety of mammalian species, including humans. Representative biological characteristics include the ability to establish a latent infection within lymphocytes and association with lymphomas and lymphoproliferative diseases. γHV68 is closely related to the human gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (also known as human herpesvirus 8) (11, 12, 44). Infection of mice with γHV68 provides a tractable model system for characterizing fundamental aspects of gammaherpesvirus infection, pathobiology, host control, and tumor induction (for reviews, see references 9, 10, 25, 26, 30, 31, 34, and 43). As such, it is a potentially valuable tool for the development of strategies to disrupt or prevent the establishment of latency by gammaherpesviruses.
γHV68 was originally isolated from a bank vole (Clathriomys glariolum), is a natural pathogen of free-living murid rodents (6), and is able to infect both inbred and outbred mice (6, 23, 28, 36). Inbred mice inoculated with γHV68 establish productive virus replication in the lung (36) and establish a latent infection in the spleen (36, 37, 45). Establishment of splenic latency is characterized by a transient phase of CD4+-T-cell-dependent splenomegaly involving proliferation of both B and T lymphocytes (13, 33, 36, 41) and by lymph node enlargement, which peaks 2 to 3 weeks postinoculation. The virus is also able to establish a latent infection in the thymus (28) and bone marrow (47, 48) and may also establish a persistent infection in the lungs (35).
Analogous to EBV infection, B lymphocytes appear to be the major reservoir harboring latent γHV68 (14, 37), although macrophages (14, 48), splenic dendritic cells (14), and lung epithelial cells (35) have been shown to harbor latent virus. In peripheral blood, EBV appears to reside exclusively within memory B cells, providing the virus with access to a long-lived and immunologically privileged site (reviewed in references 38 and 39). There is a growing body of evidence to support the notion that EBV has evolved mechanisms to take advantage of the natural process of B-cell differentiation in order to gain access to the memory B-cell compartment. EBV infection of a resting B cell results in expression of a subset of viral gene products that are able to provide the requisite signals leading to activation of the B cell. It is hypothesized that these latently infected B cells, once activated, migrate to follicles and form germinal centers. A subset of viral genes provide surrogate signals that allow the latently infected B cells to follow the normal B-cell differentiation pathway, ultimately exiting the cell cycle by differentiating into memory cells. Once the virus has gained access to the memory compartment, EBV latency-associated gene expression is downregulated, allowing the virus to avoid detection by the host immune system.
It is currently unclear what cell types are responsible for maintaining long-term γHV68 latency and whether viral infection follows a route similar to that of EBV. The purpose of the studies described here was to address the relative distribution of long-term latent γHV68 infection in the spleen among non-B-cell and various B-cell populations after intranasal challenge. At early and intermediate times postinfection the virus was predominantly found within the B-cell compartment and resides within both naive and more mature B cells lacking surface immunoglobulin D (sIgD), which exhibit features representative of memory B cells. Moreover, germinal-center cells appear to harbor very high frequencies of latent virus at these times. At 6 months postinfection, we show that latent γHV68 infection is largely confined to the sIgD− B-cell compartment. The present study complements a recently published report that demonstrated that at 3 months postinfection, γHV68 latently infected cell types included germinal-center B cells and isotype-switched memory B cells (15). In contrast to that work, we did not detect significant numbers of latently infected naive B cells by 6 months postinfection. These findings suggest that EBV and γHV68 share a common strategy for maintaining latency by accessing a pool of long-lived, immunologically privileged cells, namely, memory B cells.
MATERIALS AND METHODS
Cells, viruses, and virus culture.
γHV68 strain WUMS (ATCC VR-1465) was used for all virus infections. Virus passage, maintenance, and titers were performed as described previously (8). NIH 3T12 cells and mouse embryonic fibroblast (MEF) cells were maintained at 37°C in a 5% CO2 environment in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS; HyClone Laboratories, Inc., Logan, Utah), 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 2 mM l-glutamine (cMEM). MEF cells were obtained as described previously (27). Briefly, BALB/c mouse embryos were homogenized, the resulting homogenate was plated in tissue culture flasks for 1 to 2 weeks, and the adherent cells were harvested after trypsinization.
Mice, infections, and organ harvests.
Female C57BL/6J mice (catalog no. 000664; The Jackson Laboratory, Bar Harbor, Maine) between 8 and 12 weeks of age were housed at the Yerkes vivarium in accordance with all federal, university, and facility regulations. Mice were placed under isofluorane anesthesia prior to intranasal inoculation with 1,000 PFU in 20 μl of cMEM. Mice were sacrificed by cervical dislocation under isofluorane anesthesia, and spleens were harvested and treated as described below. Sentinel mice were assayed every 3 months and were negative for adventitious mouse pathogens as determined by parasitology and serology.
Cell preparation.
Spleens were disrupted in a TenBroek homogenizer and filtered through a 100-μm-pore-size nylon cell strainer (Becton Dickinson, Franklin Lakes, N.J.). After removal of erythrocytes with red blood cell lysing buffer (Sigma, St. Louis, Mo.), cells were collected by centrifugation, washed, refiltered, and counted on a hemocytometer. Mononuclear cells were obtained according to the manufacturer's directions by layering 2 × 107 cells/ml onto Lympholyte-M lymphocyte separation medium (Cedarlane, Hornby, Ontario, Canada). The resulting mononuclear cell preparation was enumerated and assayed for viability with trypan blue (Sigma). Control experiments in the absence of Lympholyte-M gradient separation on bulk unsorted splenocytes and on sorted B-cell (CD19+) and non-B-cell (CD19−) populations demonstrated no significant difference in the frequency of cells harboring or reactivating viral genome (data not shown). Pooled splenocytes from 5 to 10 mice were used in all experiments.
Antibodies for flow cytometry.
Rat anti-mouse CD16/CD32 (Fc block) was used to block Fc receptors prior to staining in some cases. Cells were stained for fluorescence-activated cell sorting (FACS) analysis by using combinations of the following antibodies and lectins: phycoerythrin (PE)-conjugated antibodies directed against CD19, surface immunoglobulin, and IgD (Southern Biotech, Birmingham, Ala.); fluorescein isothiocyanate (FITC)-conjugated antibodies to CD19, IgD, IgG1, IgG2a, IgG2b, IgG3, IgA, and IgE; allophycocyanin-conjugated CD19, and FITC-conjugated peanut agglutinin (PNA; Vector Laboratories, Burlingame, Calif.). FITC-Ig2a and PE-Ig2a (Southern Biotech) were used as isotype controls. All reagents were purchased from Pharmingen (San Diego, Calif.) except as noted.
Flow cytometry.
Cells were prepared as described above and resuspended at 5 × 107 cells/ml in FACS blocking buffer (8% FCS, 10% normal rabbit serum, 10% normal goat serum, 5% bovine serum albumin, and 0.5 mg of mouse immunoglobulin/ml in 1× Hanks balanced salt solution [0.15 M NaCl, 0.015 M sodium citrate]) or in 1× phosphate-buffered saline containing 1% FCS and Fc block (0.125 μg/106 cells) and then maintained on ice for 30 min. Cells were stained with PE conjugates at 0.05 μg/106 cells, FITC conjugates at 0.25 μg/106 cells, and allophycocyanin conjugates at 0.1 μg/106 cells by incubation on ice for 1 h in the dark. FITC-PNA was used at a 1:250 dilution (20 μg/ml). Cells were washed twice with phosphate-buffered saline containing 1% FCS, counted, and resuspended to 5 × 107 cells/ml. Stained cell populations were immediately acquired by using a FACSVantage SE (Becton Dickinson, Mountian View, Calif.) or a MoFlo (Cytomation, Fort Collins, Colo.) flow cytometer. Data were analyzed by using FloJo software (TreeStar, Inc., San Carlos, Calif.). Sorted populations were resuspended at 106 to 107 cells/ml in cMEM supplemented with 10% dimethyl sulfoxide and stored at −80°C for limiting-dilution PCR (LD-PCR) assays or at 4°C in cMEM for limited dilution ex vivo reactivation assays as described below.
Limiting-dilution ex vivo reactivation analyses.
Limiting-dilution analysis to detect reactivation from latency was performed as previously described (45, 48). Briefly, bulk splenocyte or FACS-sorted populations were obtained from mice at days 16 and 42 postinfection as described above. Cells were resuspended in cMEM and plated in 12 serial twofold dilutions (starting with 5 × 104 or 5 × 105 cells) onto MEF indicator cell monolayers in 96-well tissue culture plates. Twelve to twenty-four replicate wells were plated per dilution. The wells were scored microscopically for cytopathic effect (CPE) 21 and 28 days postplating. To detect preformed infectious virus, duplicate cell samples were mechanically disrupted (latent virus cannot reactivate from killed cells) and plated in parallel with live cells. This procedure kills >99% of the cells with, at most, a twofold effect on the titer of preformed infectious virus (45, 47, 48). This assay has a sensitivity of 0.2 PFU (45), which represents an approximate 5- to 10-fold increase in sensitivity for detection of preformed infectious virus compared to traditional plaque assay (45, 47, 48).
LD-PCR determination of the frequency of cells harboring viral genome.
The frequency of cells containing the γHV68 genome was determined by using a single-copy sensitivity nested PCR assay as described previously, with the following modifications (47, 48). Briefly, splenocytes harvested at days 16, 42, and 182 postinfection and stored at −80°C in cMEM supplemented with 10% dimethyl sulfoxide were thawed, counted, and resuspended in isotonic buffer. A series of six threefold serial dilutions, starting with 104 cells per well, were plated in a background of 104 uninfected NIH 3T12 cells in 96-well PCR plates (MWG Biotech, High Point, N.C.) containing 5 μl of lysis solution. Twelve replicate PCRs were performed for each cell dilution per sample per experiment. Within each set of PCR samples, negative controls (water) and positive controls consisting of 10 copies, 1 copy, or 0.1 copy of pBamHI-N plasmid (12) diluted into a background of 104 uninfected NIH 3T12 cells (48) were used as sensitivity controls. Plates were sealed with PCR foil (Eppendorf Scientific, Westbury, N.Y.), and the cells subjected to lysis by proteinase K at 56°C for 6 h, followed by enzyme inactivation at 95°C for 20 min. A total of 10 μl of round 1 PCR cocktail was added directly to each well by foil puncture, and the first round of PCR was performed. Subsequently, 10 μl of round 2 PCR cocktail was added directly to the wells by foil puncture and subjected to a second round of PCR. All cell lysis and PCR protocols were performed as described previously (47, 48) on a PrimusHT thermal cycler (MWG Biotech). Reaction products were separated on 1.5% agarose gels in 1× TAE buffer containing 0.5 mg of ethidium bromide/ml. All of the assays demonstrated near-single-copy sensitivity with no false positives. All PCR setup and manipulations were conducted in a PCR-dedicated room by using either positive-displacement pipettors or traditional pipettors with aerosol-resistant barrier tips.
Statistical analyses.
All data analysis was conducted by using GraphPad Prism software (GraphPad Software, San Diego, Calif.). For limiting-dilution analysis of latency and reactivation, data were subjected to nonlinear regression analysis (with a sigmoidal dose-response algorithm for best fit). Frequencies of reactivation and viral-genome-positive cells were obtained from the nonlinear regression fit of the data where the regression line intersected 63.2%. Based on the Poisson distribution, this is the frequency at which there is at least one event present in a population.
RESULTS
Reactivation of latent γHV68 from splenic B-cell and non-B-cell populations at day 16 postinfection.
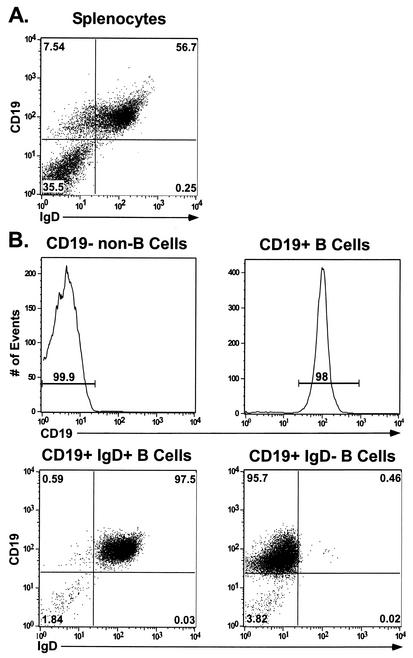
Mononuclear single-cell preparations were generated from spleens harvested from γHV68-infected mice at 16 days postinfection (dpi). Splenocytes were stained with antibody directed against the pan-B-cell marker CD19 (a transmembrane glycoprotein that is selectively expressed on the surface of B lymphocytes) (Fig. 1A) (49). CD19+ (B-cell) and CD19− (non-B-cell) fractions were purified from bulk splenocytes by FACS (Fig. 1). After this approach we were able to obtain significant enrichment of each cell population, as illustrated in Fig. 1B. We determined the frequency of cells reactivating virus from unfractioned splenocytes and from the CD19+ and CD19− splenic cell subsets by using a limiting-dilution ex vivo reactivation assay (see Materials and Methods) (45, 47). This assay involves serially diluting harvested cells and plating them on MEF indicator monolayers (which are permissive for γHV68 replication). Spontaneous virus reactivation from latency is scored as viral CPE on the MEF monolayers at days 21 and 28 postplating of the latently infected cells. Since reactivation from latency requires plating live cells, the relative contribution of preformed infectious virus can be assessed by plating mechanically disrupted cells in parallel with live cells.
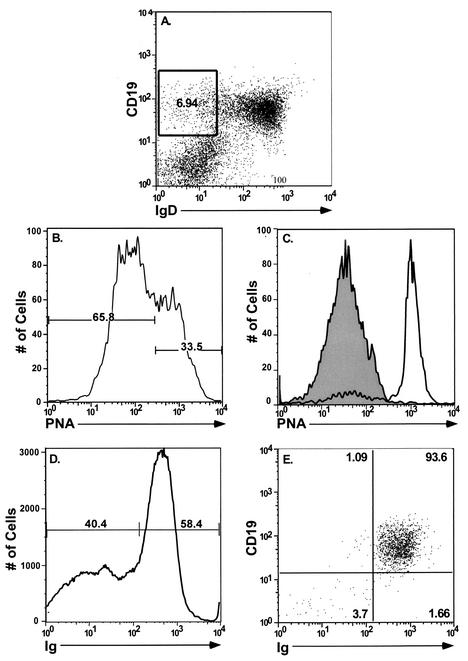
FIG. 1.
FACS analysis of purified splenic lymphocyte populations. Splenocytes collected from C57BL/6J mice were isolated and prepared as described in Materials and Methods. Cells were fractionated into B-cell and non-B-cell populations by staining with a PE-conjugated antibody to the pan-B-cell marker CD19. CD19+ cells were further fractionated into IgD+ and IgD− B-cell subsets by using an FITC-conjugated antibody to IgD. Flow cytometric dot plots from one representative experiment are shown. (A) Unfractionated total splenocytes stained with a pan-B-cell marker (CD19-PE) and FITC-conjugated IgD. (B) Postsort FACS analysis indicating purity of sorted lymphocyte subsets. The mean purities over four replicate experiments were as follows: CD19+ (96.5% ± 2.82%), CD19− (97.6% ± 4.72%), CD19+ IgD+ (95.7% ± 2.4%), and CD19+ IgD− (92% ± 3.7%). The extent of contamination of sorted populations was limited to 2.72% ± 0.6 and 0.93% ± 0.46 for the CD19+ and CD19− fractions, respectively. Purified IgD+ and IgD− B-cell subsets demonstrated only minimal contamination of 0.63% ± 0.08% and 0.75% ± 0.18%, respectively, with the unwanted cell type.
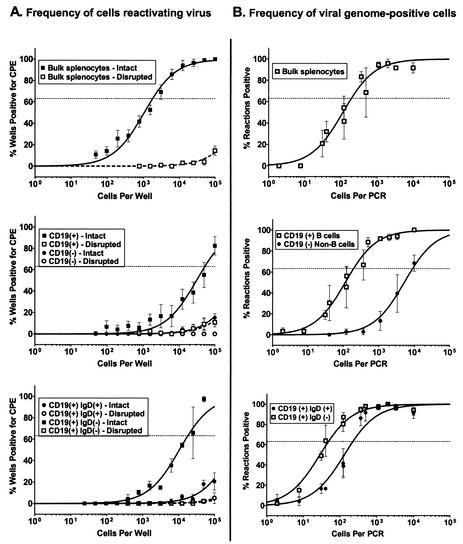
At 16 dpi, unsorted splenocytes demonstrated high levels of reactivation (ca. 1 in 1,900 cells) (Fig. 2A and Table 1), whereas there is a marked difference in the ability of virus to reactivate virus from the purified B-cell (CD19+) and non-B-cell (CD19−) fractions. The B-lymphocyte fraction showed detectable levels of reactivation (ca. 1 in 53,000 cells) but, in contrast, reactivation from the non-B-cell fraction was below the limit of detection of this assay (Fig. 2A and Table 1). In addition, these results showed very low or undetectable levels of preformed infectious virus (dashed lines) in the bulk-unsorted and CD19+ samples (Fig. 2A), which is consistent with previous studies demonstrating the clearance of lytic virus by this time postinfection (36, 46). Thus, the majority of detectable CPE in this assay can be attributed to reactivation of latent virus.
FIG. 2.
Survey of splenic latency 16 days after intranasal infection. Bulk splenocytes or FACS-sorted splenic cell populations were obtained from γHV68-infected C57BL/6J mice at 16 dpi and analyzed by limiting-dilution ex vivo reactivation and limiting-dilution viral-genome PCR assays. (A) Frequency of cells reactivating virus. For the ex vivo reactivation assay, serial dilutions of live, intact cells (solid symbols and lines) were plated on MEF indicator monolayers in parallel with samples that had been mechanically disrupted in order to distinguish between virus reactivation from latency and virus replication resulting from preformed infectious virus (open symbols and dashed lines) (see Materials and Methods). (B) Frequency of cells harboring viral genome. The frequency of viral-genome-positive cells was determined by using an LD-PCR assay. Serial dilutions of cells were plated into a background of 104 uninfected cells, lysed, and analyzed by a nested PCR to detect viral genome (see Materials and Methods). For both assays, curve fit lines were derived from nonlinear regression analysis, and symbols represent mean percentages of wells positive for virus (viral DNA or CPE) ± the standard error of the mean (SEM). The dashed line represents 63.2%, from which the frequency of viral-genome-positive cells or the frequency of cells reactivating virus was calculated based on a Poisson distribution. The data shown represent at least three independent experiments, with cells pooled from 10 mice per experimental group.
TABLE 1.
Frequency of cells reactivating γHV68: explant cultures of unsorted and sorted splenocytes at 16 dpi
| Cell fraction | Frequency of reactivationa
|
||
|---|---|---|---|
| Frequencyb | Total no. of cellsc | Total no. of cells reactivatingd | |
| Unsorted | 1/1,850 | 8.1 × 107 | 43,784 |
| CD19+ | 1/53,000 | 5.2 × 107 | 981 |
| CD19− | ND | 2.9 × 107 | NA |
| CD19+ IgD+ | ND | 4.6 × 107 | NA |
| CD19+ IgD− | 1/18,000 | 6.2 × 106 | 344 |
Reactivation frequencies were calculated from splenocyte subsets obtained 16 dpi.
Frequencies of reactivating cells were derived from the mean of at least three independent experiments with splenocytes pooled from 10 mice per experimental group. ND, not determined because the values were below the limit of detection of the assay.
Cell numbers were derived from total cells per spleen (8.1 × 107) and the percentage of the total spleen cells that each subset represents, as calculated from FACS gating (CD19+, 64%; CD19−, 36%; CD19+ IgD+, 56.3%; CD19+ IgD−; 7.7%).
The total number of reactivating cells was determined by using the experimental frequency data and the estimated total cell numbers per subset. NA, not applicable due to the inability to accurately estimate the frequency of reactivating cells.
The FACS-sorted populations showed a significant decrease in the frequency of cells spontaneously reactivating virus compared to bulk unsorted splenocyte population. As has been reported previously, the physical process of cell sorting can have dramatic effects on reactivation frequencies (14). Consistent with these findings, we routinely observed a 1- to 1.5-log decrease in reactivation efficiency of mock-sorted cells compared to bulk unsorted samples (data not shown). Importantly, this decrease in reactivation efficiency was a not the result of antibody staining or due to prolonged incubations at 4°C. Moreover, the viability of cells postsort was comparable to that of presort samples, as judged by trypan blue staining (data not shown), although the extended viability of these sorted cells has not been addressed.
Frequency of splenic B-cell and non-B cells harboring latent γHV68 at day 16 postinfection.
Having established that the bulk of CPE detected by the ex vivo reactivation analyses arises from reactivation of latent virus, purified B-cell and non-B-cell populations were assayed for the frequency of cells harboring viral genome by using a LD-PCR assay (see Materials and Methods). Serial dilutions of these purified cell populations were plated into a background of 104 uninfected cells and then replicates of each dilution were assayed for the presence of viral DNA by nested PCR. By measuring the frequency of positive PCRs through a dilution series and by using Poisson distribution statistics, we were able to calculate the frequency of cells harboring virus for any given cell population. Viral-genome-positive cells were easily detected from within the unsorted spleen preparation (ca. 1 in 200 cells) (Fig. 2B and Table 2). An examination of the frequency of viral-genome-positive cells within the sorted populations revealed that both B-cell and non-B-cell fractions harbor virus at day 16 postinfection. The frequency of viral-genome-positive cells in the purified B-cell fraction (CD19+) was equivalent to that observed in the bulk, unsorted splenocyte population (1 in 200 B cells harbored viral genome) (Fig. 2B and Table 2). Notably, there was a significantly lower frequency of viral-genome-positive cells in the non-B-cell population (CD19−) (1 in 9,000 cells harbored viral genome).
TABLE 2.
Frequency of cells harboring γHV68: analysis of splenocyte populations
| Cell fraction | Frequency of viral-genome-positive cells
|
|||
|---|---|---|---|---|
| dpi | Frequencya | Total no. of cellsb | No. of latently infected cellsc | |
| Unsorted | 16 | 1/200 | 8.1 × 107 | 405,000 |
| 42 | 1/3,500 | 5.9 × 107 | 16,857 | |
| 182 | 1/18,500 | 3.8 × 107 | 2,054 | |
| CD19+ | 16 | 1/200 | 5.2 × 107 | 259,000 |
| 42 | 1/4500 | 3.8 × 107 | 8,400 | |
| 182 | 1/28,000 | 2.4 × 107 | 868 | |
| CD19− | 16 | 1/9000 | 2.9 × 107 | 3,244 |
| 42 | ND | 2.1 × 107 | NA | |
| 182 | ND | 1.4 × 107 | NA | |
| CD19+ IgD+ | 16 | 1/230 | 4.6 × 107 | 200,000 |
| 42 | 1/12250 | 3.3 × 107 | 2,710 | |
| 182 | ND | 2.1 × 107 | NA | |
| CD19+ IgD− | 16 | 1/50 | 6.2 × 106 | 124,740 |
| 42 | 1/700 | 4.5 × 106 | 6,586 | |
| 182 | 1/2000 | 2.9 × 106 | 1,465 | |
| Isotype-switched CD19+ IgD−d | 84 | 1/1500 | 2.5 × 106 | 1692 |
Frequencies of viral-genome-positive cells represent the mean of at least three independent experiments with splenocytes pooled from 10 mice per experimental group. ND, not determined because values were below the limit of detection of the assay.
Cell numbers were derived from the calculated total number of spleen cells at days 16, 42, 84, and 182 postinfection (8.1 × 107, 5.9 × 107, 5.4 × 107, and 3.8 × 107, respectively) and from the percentage of total spleen cells that each subset represents, as calculated from FACS gating (CD19+, 64%; CD19−; 36%; CD19+ IgD+, 56.3%; CD19+ IgD−, 7.7%, isotype-switched IgD− B cells, 4.7%).
The total number of latently infected cells was determined by using the experimental frequency data and the estimated total cell numbers per subset. NA, not applicable due to the inability to accurately estimate the frequency of cells harboring viral genome.
Isotype-switched memory B cells were stained and sorted by using a cocktail of antibodies directed against IgG1, IgG2a/b, IgG3, IgA, and IgE.
Both naive and sIgD− splenic B cells are latently infected at day 16 postinfection, but virus reactivates preferentially from the sIgD− B-cell population.
It is clear from the analyses described above that B cells harbor a significant fraction of the latent virus present in the spleen at 16 dpi. However, it is unknown whether specific subsets of B cells are preferential targets for latent virus infection. Recent studies have shown that within the peripheral blood of EBV-infected individuals virus is restricted to the sIgD− population of B cells, but within the tonsils EBV is found within both sIgD+ and sIgD− B cells (for a review, see references 38 and 39). sIgD is a marker for mature, naive B cells but is downregulated sometime during the germinal-center reaction as naive B cells differentiate into either plasma or memory B cells. The resulting sIgD− B cells represent memory B cells that either maintain surface IgM expression or express a switched immunogloblulin isotype on their surfaces (IgG, IgA, or IgE). Although it has been demonstrated that activated B cells expressing markers representative of germinal-center B cells preferentially harbor γHV68 infection (14), it is currently not known whether γHV68 has any bias for B cells with respect to sIgD expression. Spleen cells isolated at 16 dpi were simultaneously stained with antibodies directed against CD19 and IgD and sorted into CD19+ IgD+ (naive) and CD19+ IgD− fractions by FACS (Fig. 1). These populations were subsequently assayed for their ability to reactivate virus upon coculture, as described above.
Ex vivo reactivation assays demonstrated that the naive B-cell fraction showed a very low but detectable frequency of virus reactivation (Fig. 2A), although the frequency was not much greater than that seen for preformed infectious virus (Fig. 2A, dashed line). Although the level of reactivation seen in this fraction is not statistically significant, we could approximate a reactivation frequency of 1 in 2 × 105 cells by extrapolation. In contrast, the IgD− B-cell subset demonstrated significant reactivation (ca. 1 in 18,000) while exhibiting no detectable presence of preformed infectious virus in the disrupted cell samples (Fig. 2A and Table 1). Based on this analysis, reactivation of latent virus from purified B cells appears to arise almost exclusively from the sIgD− splenic B-cell population. Thus, although we cannot directly compare the reactivation frequencies of unsorted versus sorted cells populations due to the effects of cell sorting on the efficiency of virus reactivation, it appears that we can compare reactivation between sorted populations since the reactivation frequency observed in the CD19+ fraction can largely be accounted for by virus reactivation from the sIgD− fraction (Fig. 2A and Table 1). However, these results must be interpreted with caution since it is possible that the ability to reactivate virus from specific B-cell and non-B-cell populations is differentially affected by the cell-sorting process. In addition, the ex vivo reactivation assay may not detect reactivation from latently infected cell populations whose survival in culture is very limited.
To extend the above analyses, the frequency of sIgD+ and sIgD− B cells that harbor latent viral genome was determined. As expected, these analyses demonstrated that the sIgD− subset of B cells contained a higher frequency of viral-genome-positive cells (1 in 50) compared to the sIgD+ (naive) B-cell fraction (1 in 230) (Fig. 2B and Table 2). By comparing the frequency of cells harboring virus within these two B-cell subsets to the frequency of virus present within the total B-cell populations, it is clear that they can account for all of the virus within the total splenic B-cell population (Table 2), and thus the process of fractionating B-cell subsets does not appear to have led to a selective loss of a virus-infected B-cell population. By comparing the frequency of cells in each population that (i) reactivate latent virus and (ii) harbor viral genome, we determined that the efficiencies of virus reactivation from total splenic B cells, sIgD− B cells, and sIgD+ B cells at day 16 postinfection were ca. 0.38, 0.28, and 0.1%, respectively. Thus, based on these analyses it appears that sIgD− B cells reactivate γHV68 with a slightly greater efficiency than γHV68-infected naive B cells, although the physiological relevance of this difference is unclear at this time.
sIgD− B cells are the predominant latently infected cell population in the spleen at intermediate and late times postinfection.
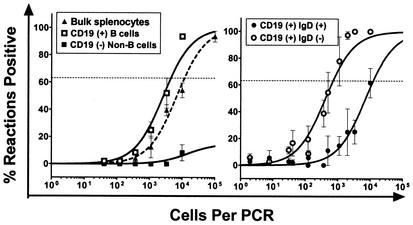
To extend the analyses described above, we examined viral latency in the spleen 6 weeks and 6 months postinfection. At 6 weeks postinfection, spleen cells were again sorted into B-cell and non-B-cell populations, with the B cells being further sorted based on sIgD expression. LD-PCR assays and limiting-dilution ex vivo reactivation assays were conducted on unsorted and sorted cell populations to determine the frequency of cells harboring virus, and the frequency of cell reactivating from latency, respectively (see Fig. 3 and Tables 1 and 2). We observed a drop of ca. 1.5 logs from day 16 to day 42 postinfection in the frequency of cells harboring viral genome in bulk unsorted splenocytes and an equivalent drop in the frequency of viral-genome-positive cells in the purified B-cell fraction. Analysis of the non-B-cell splenocyte population at 6 weeks postinfection revealed a frequency of viral-genome-positive cells below the limit of detection of the assay. Thus, by 6 weeks postinfection virtually all of the latent virus appears to be present within the B-cell fraction (compare the frequencies of viral-genome-positive cells in B cells versus the bulk unsorted sample; Fig. 3 and Table 2). Examination of the sIgD+ and sIgD− B-cell subsets revealed a further increase in the bias toward infection of the sIgD− fraction (Fig. 3). Notably, from day 16 to 6 weeks postinfection there was a substantial decrease in the frequency of naive B cells harboring viral genome (barely detectable), whereas there was only a modest decrease in the frequency of viral-genome-positive cells in the CD19+ sIgD− fraction (Fig. 3 and Table 2).
FIG. 3.
Analysis of splenic latency 42 dpi after intranasal infection. Bulk splenocytes or specific FACS-sorted splenic cell subsets were obtained at 42 dpi. Cell populations were analyzed for the frequency of viral-genome-positive cells by LD-PCR as described in Materials and Methods. The data shown represent at least three independent experiments, with cells pooled from 10 mice per experimental group. Curve fit lines were derived from nonlinear regression analysis, and symbols represent mean percentages of wells positive for viral genome. The error bars represent the SEM. Frequency analysis was based on the Poisson distribution with the horizontal dashed line at 63.2%. Symbols: ▴, unsorted splenocytes; □, CD19+ (B cells); ▪, CD19− (non-B cells); ○, CD19+ IgD−; •, CD19+ IgD+ (naive B cells).
Reactivation frequencies at 42 dpi were also assessed for the unsorted and sorted splenocyte populations. Notably, the frequency of splenocytes reactivating virus at this time postinfection was extremely low and precluded any comparison of reactivation frequencies (data not shown). This finding is consistent with previous observations showing a marked lack of reactivation in the spleen after intranasal infection at 6 weeks postinfection (16, 48). As such, monitoring spontaneous virus reactivation from the spleen at late times postinfection is not informative.
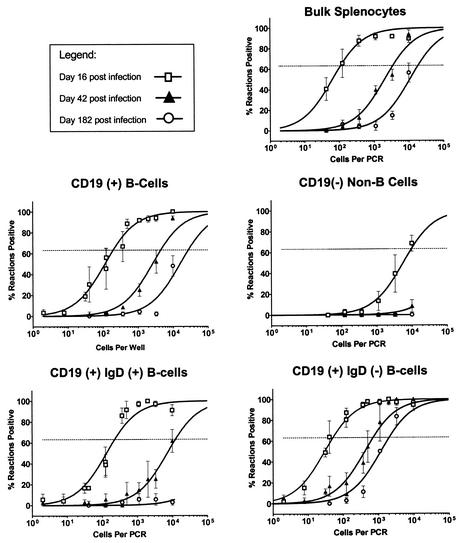
To complete the time course of γHV68 latency in the spleen of C57BL/6 mice, we examined the frequency of cells harboring virus in various splenocyte subsets at 6 months postinfection. Figure 4 illustrates the presence of viral-genome-positive cells within the bulk unsorted fraction, the B-cell fraction, and the IgD− B-cell subset. By 6 months postinfection we could not detect any viral-genome-positive cells in the non-B-cell fraction. In addition, we could not detect virus in the naive B-cell population (CD19+, IgD+), indicating that infection of naive B cells in the spleen occurs transiently during the early stages of chronic infection. In contrast to the disappearance of virus from non-B-cell and naive B-cell reservoirs, we readily detected virus in the CD19+, sIgD− B cells, which continue to maintain moderate levels of latent γHV68 (ca. 1 in 2,000) out to 182 dpi (Fig. 4 and Table 2). Throughout our analyses, the CD19+ IgD− fraction of cells comprised 7.7% ± 2.2% of total splenocytes, as judged by presort analysis, with very little variation between the time points examined. As a result of selective enrichment for this cell population by FACS, we are roughly able to account for the bulk of viral-genome-positive cells seen in the unsorted bulk fraction (ca. 1 in 18,500) (Fig. 4 and Table 2). These analyses demonstrate that at late times postinfection latent virus is mostly excluded from the CD19− and naive B-cell populations and preferentially associated with a subset of B cells lacking sIgD.
FIG. 4.
Long-term splenic latency is maintained within the IgD− subset of B cells in vivo. Bulk splenocytes and FACS-sorted splenic subsets were obtained at days 16 (□), 42 (▴), and 182 (○) postinfection after infection of C57BL/6J mice with γHV68. Cells were analyzed for the frequency of genome-positive cells by LD-PCR as described in Materials and Methods. The data demonstrate a decrease in overall frequency of viral-genome-positive cells from all compartments over the time course analyzed. The data shown represent three to four independent experiments, with cells pooled from 10 mice per experiment. Curve fit lines were derived from nonlinear regression analysis, and symbols represent mean percentages of wells positive for viral genome ± the SEM. Frequency analysis was based on the Poisson distribution with the horizontal dashed line at 63.2%. Symbols: □, 16 dpi (early); ▴, 42 dpi (intermediate); ○, 182 dpi (late).
Germinal-center B cells and surface immunoglobulin-expressing CD19+ IgD− cells are reservoirs of latent virus at early and intermediate times postinfection.
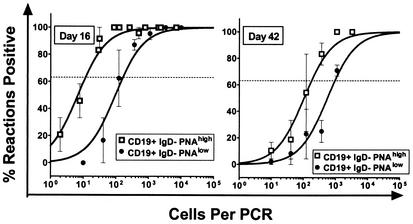
To further characterize the phenotypes of sIgD− B cells harboring latent γHV68 at early and intermediate times postinfection, we assessed whether virus was present in sIgD− germinal-center B cells. Germinal-center B cells avidly bind the lectin PNA (29). We sorted CD19+ IgD− cells into PNAhigh (germinal-center B cells) and PNAlow B cells (Fig. 5). At both early and intermediate times postinfection, a higher frequency of germinal-center B cells harbored viral genome than the PNAlow B-cell population (Fig. 6). This difference was greater at day 16 postinfection (1 in 12 versus 1 in 170) than at day 42 postinfection (1 in 180 versus 1 in 960). Notably, at day 16 postinfection the total number of PNAhigh B cells harboring viral genome was nearly 10-fold higher than the total number of PNAlow B cells (204,261 versus 21,738; see Table 3). However, by day 42 postinfection the total number of PNAhigh and PNAlow were roughly equivalent (5,822 and 3,562, respectively). Thus, γHV68 latency persists in germinal-center B cells over an extended period of time. These results are consistent with a recent report from Flano et al. (15) demonstrating that germinal-center B cells are a major reservoir of γHV68 latency at 16 days and 3 months postinfection.
FIG. 5.
Analysis of PNA and surface immunoglobulin staining of the sIgD− B-cell population. Bulk splenocytes were harvested at 16 and 42 dpi and stained with antibodies directed to CD19, sIgD, and PNA or surface immunoglobulin. (A) Cells phenotypically CD19+ IgD− were gated and used for subsequent fractionation with additional cell surface markers. This population represented on average ca. 7.7% of the total splenocyte population. (B) Profile of PNA staining for cells within the gated area at 16 dpi. At this point, 39.3% ± 8.2% of CD19+ IgD− cells were PNAhigh, and 59.25% ± 9.3% were PNAlow. At 42 dpi these numbers were 23.3% ± 1.3% and 76.0% ± 2.3%, respectively. (C) Postsort analysis showing an overlay of CD19+ IgD− PNAlow (shaded area, 98.9% pure) and CD19+ IgD−PNAhigh cells (open area, 82% pure). (D) Staining profile of total splenocytes with a pan-immunoglobulin antibody. (E) Postsort fraction of CD19+ IgD− immunoglobulin-positive cells (93.6% pure).
FIG. 6.
Latent virus is preferentially found within PNAhigh IgD− B cells at 16 and 42 dpi. Splenocytes were harvested at day 16 (left panel) and day 42 (right panel) and FACS sorted into CD19+ IgD− PNAhigh and CD19+ IgD− PNAlow cell subsets. The purified fractions were analyzed for the frequency of cells harboring viral genome by using the LD-PCR assay as described in Materials and Methods. Viral genome was found preferentially within the PNAhigh subset of CD19+ IgD− at both early and intermediate times postinfection. The data shown represent two independent experiments, with cells pooled from 10 mice per experiment. Curve fit lines were derived from nonlinear regression analysis, and symbols represent mean percentages of wells positive for viral DNA ± the SEM. Frequency analysis was based on the Poisson distribution with the horizontal dashed line at 63.2%. Symbols: □, CD19+ IgD− PNAhigh; •, CD19+ IgD− PNAlow.
TABLE 3.
γHV68 latency at early and intermediate times postinfection can be found within germinal-center cells and surface immunoglobulin-positive, sIgD− B cells
| Cell fractiona | Frequency of viral-genome-positive cells
|
|||
|---|---|---|---|---|
| dpi | Frequencyb | Total no. of cellsc | No. of latently infected cellsd | |
| PNAhigh | 16 | 1/12 | 2.4 × 106 | 204,261 |
| 42 | 1/180 | 1.0 × 106 | 5,822 | |
| PNAlow | 16 | 1/170 | 3.7 × 106 | 21,738 |
| 42 | 1/960 | 3.4 × 106 | 3,562 | |
All cell fractions were derived from initial gating on CD19+ IgD− cells and then further fractionated into the indicated subsets.
The data shown are the mean values of at least two replicate experiments, with cell populations derived from the pooled spleens recovered from 5 to 10 mice per experiment.
Cell numbers were derived from the calculated total number of spleen cells at days 16 and 42 postinfection (8.1 × 107 and 5.9 × 107, respectively) and the percentage of total spleen cells that each subset represents, as calculated from FACS gating.
The total number of latently infected cells was determined by using the experimental frequency data and the estimated total cell numbers per subset.
Having examined the sIgD− B-cell fraction with respect to PNA binding, we also examined this specific subset of B cells for expression of surface immunoglobulin, since the loss of sIgD from B cells does not identify them conclusively as memory B cells. Memory B cells arising from GC reactions can either maintain surface IgM expression or alternatively express an isotype-switched phenotype (IgG, IgA, and IgE) on their cell surface. To control for the possibility that γHV68 may reside in an uncharacterized population of CD19+ cells lacking surface Ig, we examined purified IgD− B cells with respect to the expression of surface immunoglobulin by using a pan-immunoglobulin antibody and FACS-sorted IgD− surface immunoglobulin-positive B cells at day 16 and day 42 postinfection. Importantly, virtually all CD19+ IgD− cells were surface immunoglobulin positive (data not shown). Furthermore, this purified B-cell subset harbored viral genome at levels comparable to that seen in the IgD− B-cell fraction (1 in 205 at 16 dpi and 1 in 1,700 at 42 dpi). We also examined the presence of latent γHV68 in isotype-switched memory B cells by using a cocktail of antibodies (IgG1, IgG2a, IgG2b, IgG3, IgA, and IgE) at 3 months postinfection. At this late time point there was a significant frequency of viral genome present in the isotype-switched IgD− B-cell fraction (1 in 1,500; see Table 2). Taken together, these results indicate that γHV68 latency is preferentially found in both germinal-center and memory B cells by day 42 postinfection and is maintained at a relatively high frequency in isotype-switched memory B cells at least out to 3 months postinfection.
DISCUSSION
Analogous to the findings with EBV, there appears to be a unique role for B cells in the establishment of long-term latency in γHV68-infected mice. B cells represent the principal reservoir of splenic latency after intranasal inoculation (14, 37, 47), and there is a failure to establish latency in the spleens of B-cell-deficient mice after intranasal inoculation (42, 47). These results suggest a role for B cells in trafficking virus to the spleen. Consistent with this hypothesis, adoptive transfer of B cells into latently infected B-cell-deficient mice is able to restore splenic latency (35).
Although B cells have been implicated as a major reservoir of splenic latency, up until now it has been unclear what populations of B cells are integral for maintenance of life-long latency within the host. Studies on EBV infection of humans suggest that gammaherpesviruses may have evolved a clever strategy to usurp control of normal B-cell differentiation pathways to gain access to the memory B-cell compartment, where they are able to hide from immune surveillance (for reviews, see references 38 and 39). EBV is able to infect any resting B cell (24) and, through the coordinated expression of a subset of viral genes (3), is able to activate infected B cells and drive them to proliferate (40). These proliferating B lymphoblasts are thought to migrate to peripheral lymphoid tissues, where they form germinal centers. During normal B-cell differentiation, B cells entering germinal centers, provided they are positively selected for during the process of affinity maturation, leave the germinal center as either antibody-producing plasma cells or long-lived memory B cells. Expression of the EBV latency-associated membrane antigens LMP1 and LMP2A in infected germinal-center B cells is thought to provide signals that promote survival and differentiation of these germinal-center B cells into memory B cells.
In the peripheral blood, EBV latency is tightly restricted to resting memory B cells (2, 16, 24), whereas in sites of active virus replication, such as the tonsils, EBV infection of naive, germinal-center, and memory B cells populations can be detected (4, 19). Additionally, EBV resides within both sIgD+ and sIgD− B cells in the spleen (19). Memory B cells are able to circulate throughout the bloodstream and are also known to take up residency within the bone marrow, spleen, and lymph nodes. The longevity of memory B cells provides a stable reservoir for EBV latency while still providing the potential for reactivation, infection of naive cells, and shedding of virus at specific anatomic sites.
We have shown here that during the initial stages of infection γHV68 latently infects both B cells and some non-B cells, but in our analyses only the B-cell fraction exhibited appreciable reactivation from latency. This is in agreement with previously published reports demonstrating reactivation of virus from B cells (37, 45). Reactivation from the B-cell compartment was derived almost exclusively from the sIgD− subset of B cells, with naive B cells contributing only minimally to the overall B-cell reactivation frequency. Latent virus within the non-B-cell fraction presumably represents infection of both macrophages and dendritic cells, which are known to be targets of γHV68 infection (14, 48). Although reactivation from both macrophages (14, 48) and dendritic cells (14) has been reported previously, we consistently observed only a low level of reactivation from the non-B-cell subset. This may, at least in part, be due in our analyses to the lack of specific enrichment for these cell types, which account for only a small fraction of total spleen cells at this time. Macrophages and dendritic cells account for 5.6 and 6.7%, respectively, of total cells in the spleen of γHV68-infected mice at 14 dpi (14). In addition, we cannot rule out that the physical rigors of cell sorting may also have contributed to the low levels of reactivation observed within the non-B-cell fraction. Throughout our analyses, little or no preformed infectious virus was present, indicating that all of the virus growth detected in the ex vivo reactivation analyses arose from latently infected cells reactivating virus.
Having established acute viral infection had been largely cleared by this time, we extended the above analysis to determine the frequency of cells harboring viral genome from within our sorted cell populations. Viral genome is present within the non-B-cell fraction, although the frequency of viral-genome-positive cells within the B-cell fraction is much greater and accounts for the vast majority of latently infected cells at this time. We examined various subsets of B cells and showed that naive, IgD−, and germinal-center B cells all harbor significant levels of latent virus. In agreement with previously published reports (14, 15), we saw a very high frequency of virus in both IgD− (1 in 50 cells) and germinal-center B cells (1 in 12) which cannot be accounted for by contaminating cell populations. We know from these analyses that the CD19+ IgD− fraction of cells are additionally surface immunoglobulin positive (many of which have undergone isotype switching). Based on these analyses, we conclude that the majority of latent γHV68 in the spleen at late times postinfection is harbored in germinal-center and memory B cells.
By 42 d.p.i. there is little or no detectable virus within the non-B cell reservoir. The lack of dectectable virus in the non-B cell reservoir may be due to a more rapid turnover of these cells types without reseeding of these reservoirs, and/or may be due to an effective CTL response directed against these virally infected cells. Moreover, infection of macrophage and dendritic cells may lead to dead-end infections, although these cell types may play pivotal roles during the early stages of infection and establishment of latency. In contrast, high levels of virus within the B-cell population are maintained at 42 d.p.i, which can account for much of the detectable virus seen in the unsorted splenocyte population. Although the absolute frequencies of viral-genome-positive cells decreased from two to six-weeks postinfection, an even more pronounced bias for virus within the sIgD− B-cell population was apparent. Within this population, germinal-center B cells still maintain significant numbers of latently infected cells. Germinal-center reactions are thought to reach peak development by 10 to 12 postexposure to primary antigen, after which they begin to dissociate by around 21 to 28 days after initial antigen encounter (reviewed in reference 21). Although germinal centers have been visualized as late as 100 dpi (5), it is unclear whether the germinal-center cells seen at this time represent germinal centers found in response to the initial infection which are beginning to dissipate or if there is ongoing germinal-center formation during the chronic phase of γHV68 infection.
Extending the analysis of splenic latency afforded an examination of what is likely to reflect the steady-state maintenance of γHV68 latency. Strikingly, at late times postinfection, only sIgD− B cells harbor significant levels of latent virus; many of these cells have undergone isotype switching by 3 months postinfection. It is possible that we have unknowingly selected against a specific cell population(s) that contains latent virus through our choice of FACS gates. However, it should be noted that the sIgD− B-cell population, which on average represents ca. 7% of the total splenocytes, is able to account for nearly all of the viral-genome-positive cells detected in the bulk unsorted fraction at 6 months postinfection and thus suggests that IgD− B cells are the primary reservoir involved in maintaining long-term latency in the spleen. In contrast to a recently published report demonstrating that naive B cells harbor viral genome (15), we were unable to detect any viral-genome-positive cells within the naive B-cell population by 6 months postinfection. Flano et al. (15) assessed infection at 3 months postinoculation, indicating that at this time point latency is still a dynamic process and has not yet reached steady state. How the pool of latently infected memory B cells is maintained is unknown. Long-term latency could be maintained as follows: (i) low-level reactivation and reseeding of latency reservoirs or (ii) antigen-independent, or possibly antigen-dependent, turnover of existing γHV68 latently infected memory B-cell pools. It is unclear whether latently infected memory B cells traveling in the peripheral blood are activated upon entry into secondary lymphoid tissues, resulting in terminal differentiation into either plasma cells or memory cells once again. Survival of circulating memory B cells reentering secondary lymphoid tissues requires two signals: (i) antigen-specific T-cell help and (ii) B-cell antigen receptor signaling (20, 22). EBV encodes two gene products—LMP1 and LMP2a—that are capable of supplying these survival signals (1, 7, 18, 50). γHV68 does not appear to encode homologs of either of these EBV gene products (44). Further studies will identify whether any of the genes unique to γHV68 can provide surrogate signals analogous to those provided by LMP1 and LMP2A of EBV.
A major question raised by the observation of γHV68 in memory B cells, shown here and by the studies of Flano et al. (15), is whether γHV68 infection of B cells drives their differentiation into memory B cells or, alternatively, whether the bias for infection of the memory B-cell reservoir at late times postinfection simply reflects the slow turnover of this cell population. Clearly in the case of EBV infection, as discussed above, the known activities of the EBV latency-associated antigens supports the former model, i.e., EBV driving terminal B-cell differentiation. γHV68 infection of mice provides an ideal small-animal model with which to directly address this issue. Since γHV68 latency-associated genes are identified, their impact on the establishment of long-term latency in the memory B-cell reservoir can be directly assessed. Such analyses may lead to the identification of specific γHV68 gene products required for driving B-cell differentiation.
A final issue to consider is how γHV68 might persist in the memory B-cell reservoir without alerting the host immune system. Recent data from the analysis of EBV latency in cells exhibiting a resting memory B-cell phenotype suggest that expression of immunogenic viral proteins may be tolerated within this reservoir, at least with respect to detection by CD8+ T cells (17, 32). It is also clear from studies on EBV that, during the transition from virus-driven B-cell activation and proliferation to long-term latency in resting memory B cells, there is an associated downregulation of EBV latency-associated gene expression (3, 4). Thus, we can speculate that in the case of γHV68 infection, memory B cells may provide an immunoprivileged site that, in conjunction with limited viral gene expression, affords the virus a safe haven from the host T-cell response.
Acknowledgments
S.H.S. was supported by NIH grants CA43143, CA52004, CA58524, and CA87650. D.O.W. is a Research Fellow of the National Cancer Institute of Canada supported with funds provided by the Terry Fox Run.
We thank Robert E. Karaffa II and Mike Hulsey for their expertise and assistance with the FACS analyses and members of the Speck lab for comments and advice on this research.
REFERENCES
- 1.Adler, B., E. Schaadt, B. Kempkes, U. Zimber-Strobl, B. Baier, and G. W. Bornkamm. 2002. Control of Epstein-Barr virus reactivation by activated CD40 and viral latent membrane protein 1. Proc. Natl. Acad. Sci. USA 99:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 3.Babcock, G. J., D. Hochberg, and A. D. Thorley-Lawson. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13:497-506. [DOI] [PubMed] [Google Scholar]
- 4.Babcock, G. J., and D. A. Thorley-Lawson. 2000. Tonsillar memory B cells, latently infected with Epstein-Barr virus, express the restricted pattern of latent genes previously found only in Epstein-Barr virus-associated tumors. Proc. Natl. Acad. Sci. USA 97:12250-12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachmann, M. F., B. Odermatt, H. Hengartner, and R. M. Zinkernagel. 1996. Induction of long-lived germinal centers associated with persisting antigen after viral infection. J. Exp. Med. 183:2259-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaskovic, D., M. Stancekova, J. Svobodova, and J. Mistrikova. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 24:468.. [PubMed] [Google Scholar]
- 7.Caldwell, R. G., J. B. Wilson, S. J. Anderson, and R. Longnecker. 1998. Epstein-Barr virus LMP2A drives B-cell development and survival in the absence of normal B-cell receptor signals. Immunity 9:405-411. [DOI] [PubMed] [Google Scholar]
- 8.Clambey, E. T., H. W. t. Virgin, and S. H. Speck. 2000. Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency. J. Virol. 74:1973-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doherty, P. C., J. P. Christensen, G. T. Belz, P. G. Stevenson, and M. Y. Sangster. 2001. Dissecting the host response to a gamma-herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty, P. C., R. A. Tripp, A. M. Hamilton-Easton, R. D. Cardin, D. L. Woodland, and M. A. Blackman. 1997. Tuning into immunological dissonance: an experimental model for infectious mononucleosis. Curr. Opin. Immunol. 9:477-483. [DOI] [PubMed] [Google Scholar]
- 11.Efstathiou, S., Y. M. Ho, S. Hall, C. J. Styles, S. D. Scott, and U. A. Gompels. 1990. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J. Gen. Virol. 71:1365-1372. [DOI] [PubMed] [Google Scholar]
- 12.Efstathiou, S., Y. M. Ho, and A. C. Minson. 1990. Cloning and molecular characterization of the murine herpesvirus 68 genome. J. Gen. Virol. 71:1355-1364. [DOI] [PubMed] [Google Scholar]
- 13.Ehtisham, S., N. P. Sunil-Chandra, and A. A. Nash. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J. Virol. 67:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 15.Flano, E., I. J. Kim, D. L. Woodland, and M. A. Blackman. 2002. γ-Herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J. Exp. Med. 196:1363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joseph, A. M., G. J. Babcock, and D. A. Thorley-Lawson. 2000. EBV persistence involves strict selection of latently infected B cells. J. Immunol. 165:2975-2981. [DOI] [PubMed] [Google Scholar]
- 17.Kelly, G., A. Bell, and A. Rickinson. 2002. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat. Med. 8:1098-1104. [DOI] [PubMed]
- 18.Kilger, E., A. Kieser, M. Baumann, and W. Hammerschmidt. 1998. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 17:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laichalk, L. L., D. Hochberg, G. J. Babcock, R. B. Freeman, and D. A. Thorley-Lawson. 2002. The dispersal of mucosal memory B cells: evidence from persistent EBV infection. Immunity 16:745-754. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Y. J., and C. Arpin. 1997. Germinal center development. Immunol. Rev. 156:111-126. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y. J., J. Zhang, P. J. Lane, E. Y. Chan, and I. C. MacLennan. 1991. Sites of specific B-cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur. J. Immunol. 21:2951-2962. [DOI] [PubMed] [Google Scholar]
- 22.MacLennan, I. C. 1994. Germinal centers. Annu. Rev. Immunol. 12:117-139. [DOI] [PubMed] [Google Scholar]
- 23.Mistrikova, J., and D. Blaskovic. 1985. Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol. 29:312-317. [PubMed] [Google Scholar]
- 24.Miyashita, E. M., B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1997. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 71:4882-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nash, A. A., and N. P. Sunil-Chandra. 1994. Interactions of the murine gammaherpesvirus with the immune system. Curr. Opin. Immunol. 6:560-563. [DOI] [PubMed] [Google Scholar]
- 27.Pollock, J. L., and H. W. t. Virgin. 1995. Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J. Virol. 69:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajcani, J., D. Blaskovic, J. Svobodova, F. Ciampor, D. Huckova, and D. Stanekova. 1985. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 29:51-60. [PubMed] [Google Scholar]
- 29.Rose, M. L., M. S. Birbeck, V. J. Wallis, J. A. Forrester, and A. J. Davies. 1980. Peanut lectin binding properties of germinal centres of mouse lymphoid tissue. Nature 284:364-366. [DOI] [PubMed] [Google Scholar]
- 30.Simas, J. P., and S. Efstathiou. 1998. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 6:276-282. [DOI] [PubMed] [Google Scholar]
- 31.Speck, S. H., and H. W. Virgin. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2:403-409. [DOI] [PubMed] [Google Scholar]
- 32.Staege, M. S., S. P. Lee, T. Frisan, J. Mautner, S. Scholz, A. Pajic, A. B. Rickinson, M. G. Masucci, A. Polack, and G. W. Bornkamm. 2002. MYC overexpression imposes a nonimmunogenic phenotype on Epstein-Barr virus-infected B cells. Proc. Natl. Acad. Sci. USA 99:4550-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevenson, P. G., and P. C. Doherty. 1999. Non-antigen-specific B-cell activation following murine gammaherpesvirus infection is CD4 independent in vitro but CD4 dependent in vivo. J. Virol. 73:1075-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart, J. P. 1999. Of mice and men: murine gammaherpesvirus 68 as a model. Epstein-Barr Virus Rep. 6:31-34. [Google Scholar]
- 35.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 37.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 38.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 1:75-82. [DOI] [PubMed] [Google Scholar]
- 39.Thorley-Lawson, D. A., and G. J. Babcock. 1999. A model for persistent infection with Epstein-Barr virus: the stealth virus of human B cells. Life Sci. 65:1433-1453. [DOI] [PubMed] [Google Scholar]
- 40.Thorley-Lawson, D. A., and K. P. Mann. 1985. Early events in Epstein-Barr virus infection provide a model for B-cell activation. J. Exp. Med. 162:45-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Usherwood, E. J., A. J. Ross, D. J. Allen, and A. A. Nash. 1996. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J. Gen. Virol. 77:627-630. [DOI] [PubMed] [Google Scholar]
- 42.Usherwood, E. J., J. P. Stewart, K. Robertson, D. J. Allen, and A. A. Nash. 1996. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J. Gen. Virol. 77:2819-2825. [DOI] [PubMed] [Google Scholar]
- 43.Virgin, H. W., and S. H. Speck. 1999. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 44.Virgin, H. W. t., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed]
- 45.Weck, K. E., M. L. Barkon, L. I. Yoo, S. H. Speck, and H. I. Virgin. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 70:6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weck, K. E., A. J. Dal Canto, J. D. Gould, A. K. O'Guin, K. A. Roth, J. E. Saffitz, S. H. Speck, and H. W. Virgin. 1997. Murine gamma-herpesvirus 68 causes severe large-vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus-induced vascular disease. Nat. Med. 3:1346-1353. [DOI] [PubMed] [Google Scholar]
- 47.Weck, K. E., S. S. Kim, H. I. Virgin, and S. H. Speck. 1999. B cells regulate murine gammaherpesvirus 68 latency. J. Virol. 73:4651-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weck, K. E., S. S. Kim, H. I. Virgin, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, L. J., D. C. Ord, A. L. Hughes, and T. F. Tedder. 1991. Structure and domain organization of the CD19 antigen of human, mouse, and guinea pig B lymphocytes: conservation of the extensive cytoplasmic domain. J. Immunol. 147:1424-1432. [PubMed] [Google Scholar]
- 50.Zimber-Strobl, U., B. Kempkes, G. Marschall, R. Zeidler, C. Van Kooten, J. Banchereau, G. W. Bornkamm, and W. Hammerschmidt. 1996. Epstein-Barr virus latent membrane protein 1 (LMP1) is not sufficient to maintain proliferation of B cells but both it and activated CD40 can prolong their survival. EMBO J. 15:7070-7078. [PMC free article] [PubMed] [Google Scholar]