Abstract
From 1988 to 1993, 30 cases of poliomyelitis associated with poliovirus type 2 were found in seven governorates of Egypt. Because many of the cases were geographically and temporally clustered and because the case isolates differed antigenically from the vaccine strain, it was initially assumed that the cases signaled the continued circulation of wild type 2 poliovirus. However, comparison of sequences encoding the major capsid protein, VP1 (903 nucleotides), revealed that the isolates were related (93 to 97% nucleotide sequence identity) to the Sabin type 2 oral poliovirus vaccine (OPV) strain and unrelated (<82% nucleotide sequence identity) to the wild type 2 polioviruses previously indigenous to Egypt (last known isolate: 1979) or to any contemporary wild type 2 polioviruses found elsewhere. The rate and pattern of VP1 divergence among the circulating vaccine-derived poliovirus (cVDPV) isolates suggested that all lineages were derived from a single OPV infection that occurred around 1983 and that progeny from the initiating infection circulated for approximately a decade within Egypt along several independent chains of transmission. Complete genomic sequences of an early (1988) and a late (1993) cVDPV isolate revealed that their 5′ untranslated region (5′ UTR) and noncapsid- 3′ UTR sequences were derived from other species C enteroviruses. Circulation of type 2 cVDPVs occurred at a time of low OPV coverage in the affected communities and ceased when OPV coverage rates increased. The potential for cVDPVs to circulate in populations with low immunity to poliovirus has important implications for current and future strategies to eradicate polio worldwide.
Evidence is growing that wild type 2 poliovirus has already been eradicated (7, 72, 73). The last reported human infection with indigenous wild type 2 poliovirus was associated with a poliomyelitis (polio) case in Aligarh, Uttar Pradesh, India, in October 1999 (73). Apart from a recent localized breach in containment of type 2 poliovirus (MEF-1 strain) in northern India (74), the only known current type 2 poliovirus infections are from the oral poliovirus vaccine (OPV) itself. All type 2 poliovirus isolates obtained since 1999, either from clinical specimens or from environmental samples, have been derived from OPV (72). The large majority of these isolates are closely related (>99% VP1 nucleotide sequence identity) to the Sabin type 2 OPV strain (Sabin 2) and are described as OPV-like by this genetic criterion (69). Close sequence matches to the vaccine are an indication of limited virus replication since administration of the initiating OPV dose, consistent with the rapid evolution rate of poliovirus genomes (23, 25, 32, 35), the short duration of OPV infections (typically 3 to 4 weeks) in immunologically normal individuals (1, 60), and the normally limited spread of vaccine virus to close contacts of OPV recipients (10).
Some OPV-derived isolates, however, are more divergent (≤99% VP1 sequence identity to the corresponding Sabin strain), indicative of prolonged replication of the vaccine virus. These isolates are described as vaccine-derived polioviruses (VDPVs). Two categories of VDPV isolates have been identified. First to be recognized and extensively characterized were the immunodeficient VDPV (iVDPV) isolates, obtained from patients with defects in antibody production (generally common variable immunodeficiency or X-linked agammaglobulinemia) (4, 25, 60, 68, 79). Some iVDPV isolates are highly divergent (∼90% VP1 sequence identity to the parental OPV strain), suggesting that the chronic poliovirus infections had persisted for 10 years or more (25, 68). So far, all reports of persistent iVDPV infections have been from countries with high or intermediate levels of development, where the rates of OPV coverage are high and where the survival times of immunodeficient patients may be extended by their access to appropriate clinical management (N. A. Halsey, personal communication). Chronic iVDPV infections are independent events (57, 60), and the isolates obtained from such infections trace separate pathways of divergence from the original OPV strains (25, 35; R. Park, unpublished results). Currently, there is no clear evidence of spread of iVDPVs from immunodeficient patients to the wider community (57, 63, 68).
The second, more recently recognized category of divergent OPV-derived isolates includes circulating VDPVs (cVDPVs). The immediate public health importance of cVDPVs was underscored by the occurrence of an outbreak of 21 confirmed polio cases (including 2 fatal cases) associated with type 1 cVDPV on the Caribbean island of Hispaniola in 2000 to 2001 (22). Person-to-person transmission of VDPVs was initially suspected when it was found that the first two outbreak isolates were 2 to 3% divergent in VP1 sequence from the parental Sabin 1 OPV strain but shared numerous nucleotide differences from Sabin 1. Phylogenetic analysis of VP1 sequences of the 31 outbreak isolates suggested that the outbreak originated from a single OPV dose given in 1998 to 1999 (22). A separate, apparently much more limited outbreak of polio in the Philippines in 2001 was also found to be associated with type 1 cVDPV (8, 64). A third outbreak, in Madagascar, involving type 2 cVDPV, is currently under investigation (9). The critical risk factors for all three outbreaks were declining rates of OPV coverage and the prior eradication of the corresponding serotype of indigenous wild poliovirus (22).
In this report, we describe the widespread circulation of type 2 VDPV in Egypt in the 1980s and early 1990s. Like the outbreaks in Hispaniola, the Philippines, and Madagascar, VDPV circulation occurred after the apparent elimination of the corresponding serotype of indigenous wild poliovirus. However, unlike the recent outbreaks, where VDPV circulation was estimated to have occurred for less than 3 years (8, 22), type 2 VDPV circulation probably continued in Egypt for approximately a decade. Circulation occurred at a time when the rates of OPV coverage in Egypt were insufficient to eliminate the indigenous wild poliovirus types 1 and 3 (70, 71). VDPV circulation ceased when OPV coverage rates increased. Here we describe key genetic and biological properties of the Egyptian type 2 cVDPV isolates and discuss the implications of our findings to the overall strategy for global polio eradication.
MATERIALS AND METHODS
Viruses.
Poliovirus isolates were contributed by several laboratories (Table 1). The 1988 to 1993 cVDPVs from Egypt were originally isolated at The Egyptian Organization for Biological Products and Vaccine Production (VACSERA) from specimens taken at the Poliomyelitis Institute in Cairo, the main rehabilitation hospital in Egypt for polio patients. The VACSERA studies were initiated in 1971 to monitor poliovirus circulation in Egypt, 2 decades before the establishment of the national polio surveillance system. Unfortunately, poliovirus isolates obtained before 1988 were discarded, as were vaccinelike isolates (see below) obtained before 2001. All isolates in the present study had been previously characterized by neutralization with hyperimmune equine sera, diagnostic reverse transcription-PCR (75), probe hybridization (11), and partial genomic sequencing (50). The Sabin Original + 2 (52) master seed strain of Sabin 2 (strain P712 ch 2ab) was kindly provided by Carolyn Weeks-Levy of Lederle Laboratories, Pearl River, N.Y. Viruses were propagated in monolayers of L20B cells (mouse L cells expressing the human receptor for poliovirus) (45) to select for poliovirus and in RD cells (human rhabdomyosarcoma cell line; ATCC CCL 136) to produce high-titer cultures for extraction of poliovirus RNA.
TABLE 1.
Type 2 poliovirus isolates
| Isolate | Categorya | Location | Type of case or contact | Date of specimen | Contributorb | GenBank access no. |
|---|---|---|---|---|---|---|
| MEF-1/EGY42 | Wild | Egypt | Polio case | 1942 | JHN | AY082677 |
| 0317/EGY55 | Wild | Egypt | Polio case | 1955 | JHN | AF551795 |
| 2996/SWE77 | Wild | Sweden | Polio case | 1977 | JHN | AF551796 |
| 0295/ISR78 | Wild | Israel | Polio case | 1978 | AvL | AF551797 |
| 0297/EGY79 | Wild | Egypt | Polio case | 1979 | AvL | AF551798 |
| 0301/CAE80 | Wild | Cameroon | Polio case | 1980 | JHN | AF551799 |
| 2203/BAN82 | Wild | Bangladesh | Contactc | 1982 | RG | AF551800 |
| 7079/IND86 | Wild | India | Polio case | 1986 | KD | AF551801 |
| 0864/GEO87 | Wild | Georgia | Polio case | 1987 | GYL | AF551802 |
| 0176/PER89 | Wild | Peru | Polio case | 1989 | JB | AF551803 |
| 5890/VTN88 | Wild | Vietnam | Polio case | 1988 | PVT | AF551804 |
| 3825/PAK91 | Wild | Pakistan | Polio case | 1991 | HA | AF551805 |
| Giza/88 | VDPV | Egypt | Polio case | 7 September 1988 | TN | AF551806 |
| Qalyubiya/88-1 | VDPV | Egypt | Polio case | 22 September 1988 | TN | AF551807 |
| Qalyubiya/88-2 | VDPV | Egypt | Polio case | 29 October 1988 | TN | AF551808 |
| Menufiya/89-1 | VDPV | Egypt | Polio case | 16 April 1989 | TN | AF551809 |
| Cairo/89 | VDPV | Egypt | Polio case | 8 June 1989 | TN | AF551810 |
| Giza/89 | VDPV | Egypt | Polio case | 14 September 1989 | TN | AF551811 |
| Menufiya/89-2 | VDPV | Egypt | Polio case | 28 September 1989 | TN | AF551812 |
| Qalyubiya/89 | VDPV | Egypt | Polio case | 20 November 1989 | TN | AF551813 |
| Fayyum/90-1 | VDPV | Egypt | Polio case | 6 January 1990 | TN | AF551814 |
| Qalyubiya/90-1 | VDPV | Egypt | Polio case | 5 February 1990 | TN | AF551815 |
| Qalyubiya/90-2 | VDPV | Egypt | Polio case | 26 June 1990 | TN | AF551816 |
| Giza/90-1 | VDPV | Egypt | Polio case | 26 June 1990 | TN | AF551817 |
| Giza/90-2 | VDPV | Egypt | Polio case | 8 August 1990 | TN | AF551818 |
| Giza/90-3 | VDPV | Egypt | Polio case | 30 August 1990 | TN | AF551822 |
| Giza/90-4 | VDPV | Egypt | Polio case | 12 September 1990 | TN | AF551819 |
| Qalyubiya/90-3 | VDPV | Egypt | Polio case | 11 November 1990 | TN | AF551820 |
| Qalyubiya/90-4 | VDPV | Egypt | Polio case | 24 November 1990 | TN | AF551821 |
| EGY/91-1 | VDPV | Egypt | Polio case | 4 April 1991 | TN | AF551823 |
| EGY/91-2 | VDPV | Egypt | Polio case | 13 June 1991 | TN | AF551824 |
| Giza/91-1 | VDPV | Egypt | Polio case | 18 October 1991 | TN | AF551825 |
| Beni Suef/91 | VDPV | Egypt | Polio case | 30 October 1991 | TN | AF551826 |
| Giza/91-2 | VDPV | Egypt | Polio case | 27 November 1991 | TN | AF551827 |
| Sharqiya/92 | VDPV | Egypt | Polio case | 1992 | TN | AF551828 |
| Minya/92 | VDPV | Egypt | Polio case | 1992 | TN | AF551829 |
| Beni Suef/92-1 | VDPV | Egypt | Polio case | 8 August 1992 | TN | AF551830 |
| Beni Suef/92-2 | VDPV | Egypt | Polio case | 10 October 1992 | TN | AF551831 |
| Beni Suef/92-3 | VDPV | Egypt | Polio case | 2 November 1992 | TN | AF551832 |
| Beni Suef/92-4 | VDPV | Egypt | Polio case | 10 November 1992 | TN | AF551833 |
| Beni Suef/92-5 | VDPV | Egypt | Polio case | 19 November 1992 | TN | AF551834 |
| Beni Suef/93 | VDPV | Egypt | Polio case | 9 February 1993 | TN | AF551835 |
| Aswan/95 | OPV-L | Egypt | AFP case | 2 July 1995 | TN | AF551836 |
| Behaira/97 | OPV-L | Egypt | AFP case | 7 November 1997 | TN | AF551837 |
| Beni Suef/98 | OPV-L | Egypt | AFP case | 22 January 1998 | TN | AF551838 |
| Giza/98 | OPV-L | Egypt | AFP case | 27 January 1998 | TN | AF551839 |
| Alexandria/98 | OPV-L | Egypt | AFP case | 2 July 1998 | TN | AF551840 |
| Sohag/00 | OPV-L | Egypt | AFP case | 24 July 2000 | LEB | AF551841 |
OPV-L, OPV-like.
Abbreviation of donor names: HA, Humayun Asghar; LEB, Laila El-Bassioni; JB, Jorge Boshell; KD, Kanu Dave; RG, Roger Glass; GYL, Galina Y. Lipskaya; AvL, Anton van Loon; TN, Tary Naguib; JHN, James H. Nakano; PVT, Phan Van Tu.
Isolate obtained from patient with diarrhea but with no signs of AFP.
Antigenic characterization.
Initial intratypic differentiation of cVDPV isolates used highly specific cross-absorbed antisera in an enzyme-linked immunosorbent assay (ELISA) format (65). Briefly, clinical isolates were tested with two different antisera preparations, one of which reacts with Sabin 2 and another which reacts with wild type 2 polioviruses. In this assay, isolates can have one of four different antigenic properties: (i) vaccinelike, typical of Sabin 2-related isolates, (ii) non-vaccinelike, typical of wild isolates, (iii) double-reactive (DR), and (iv) nonreactive (NR). Isolates with DR or NR properties are either antigenic variants of Sabin 2 or wild type 2 polioviruses of unusual antigenicity. Reactivity as DR is also observed when an isolate is a mixture of vaccine-related and wild polioviruses of the same serotype (65). Further genetic and antigenic analyses have excluded the presence of such mixtures in all isolates described here.
Nucleic acid sequencing.
Conditions for reverse transcription-PCR amplification and cycle sequencing were as described previously (32), using the primers listed in Table 2. Sequencing was performed in both directions, and every nucleotide position was sequenced at least once from each strand. Terminal sequences were determined by using the 5′ and 3′ rapid amplification of cDNA ends system kits (Life Technologies, Gaithersburg, Md.) according to the manufacturer's instructions.
TABLE 2.
PCR and sequencing primers
| Primera | Target sequence (nucleotides) | Primer sequenceb |
|---|---|---|
| 1S | 1-19 | TTAAAACAGCTCTGGGGTT |
| 446Sc | 446-470 | TCCGGCCCCTGAATGCGGCTAATCC |
| 559Ac | 533-559 | ACACGGACACCCAAAGTAGTCGGTTCC |
| 636A | 613-636 | GGATGGCCAATCCAAYTCGCTTTA |
| 780A | 761-780 | TCCAACTTTTTGAGATGAAA |
| 823S | 823-846 | AATTACACTACAATCAATTACTAT |
| 1018A | 995-1018 | TTGAATTGCCCAGAGTTAGCTGCA |
| 1135S | 1135-1156 | TGCAGGTTCTACACATTAGATA |
| 1493A | 1472-1493 | AAGAGGTAATCAACTGGGCAGA |
| 1681S | 1681-1700 | TCCACTGAGATACCCATTAC |
| 1773A | 1752-1773 | TAATCCTTGGGTTCTTGGCACA |
| 1951S | 1951-1974 | AAGAACACAATGGACATGTATAGG |
| 1973A | 1954-1973 | CTATACATGTCCATTGTGTT |
| 2031A | 2011-2031 | GGACAACGAGAGACACAAGAT |
| 2401Sd | 2401-2423 | GGTTTTGTGTCAGCGTGTAATGA |
| 2507Se | 2507-2526 | CCGTTGAAGGGATTACTAAA |
| 2577Ae | 2562-2577 | CGGCTTTGTGTCAGGC |
| 2858Sd,f | 2858-2877 | CITAITCIMGITTYGAYATG |
| 2899S | 2899-2922 | TCAAACTATACTGATGCAAACAAC |
| 2936Ad,f | 2917-2936 | TTIAIIGCRTGICCRTTRTT |
| 3008A | 2986-3008 | TGCCACGTATAGTCATTCCATTT |
| 3356S | 3356-3375 | CACTACCAGAAAAGGGATTA |
| 3418A | 3397-3418 | CTGTATACACAGCTTTGTTTTG |
| 3503Ad | 3484-3503 | AAGAGGTCTCTRTTCCACAT |
| 3838S | 3838-3858 | TCCAACTACATTGAGTCCCTT |
| 4129S | 4129-4149 | AGTTGGTTGAAGAAATTCACA |
| 4411S | 4411-4431 | GAAGCAAAAAGGATTCAGAAG |
| 4496A | 4475-4496 | AAACATACTGGCTCAATACGGT |
| 4957S | 4957-4977 | AAGAGATGTTGCCCTTTAGTG |
| 5036A | 5017-5036 | TGATCAATGCTGTATCTAAC |
| 5456S | 5456-5479 | CAGTGGCCATGGCTAAGAGAAACA |
| 5555A | 5533-5555 | TGGGTTGGTAGAATAGCCACGTT |
| 5953S | 5953-5974 | GCCCTGAAGCGATCATACTTCA |
| 6104A | 6084-6104 | GGTTCCTTAACTCCTTCAAAC |
| 6539S | 6539-6560 | TGGCCATGAGAATGGCATTTGG |
| 6656A | 6636-6656 | AGCACTGGGATTTTGCTCCAA |
| 7078S | 7078-7101 | ACAGTCACATGGGAGAATGTAACA |
| 7195A | 7175-7195 | TCCATCTAATTGATTCATGAA |
| 7241S | 7241-7263 | TATTGGCCTGGCACAATGGTGAA |
A, antisense polarity; S, sense polarity. Numbering of nucleotide positions follows that for the Sabin 2 sequence (49).
Bases in inosine-containing and degenerate primers: I, inosine; R, A + G; Y, C + T.
Primers previously described as EV/PCR-2 (446S) and EV/PCR-1 (559A) (76).
Primers used for VP1 sequencing.
Primers previously described as Sabin 2/PCR-2 (2507S) and Sabin 2/PCR-1 (2577A) (75).
Primers previously described as panPV/PCR-2 (2858S) and panPV/PCR-1 (2936A) (27).
Phylogenetic analysis.
Evolutionary relationships among diverse type 2 poliovirus genotypes were estimated from VP1 sequences (all codon positions) by using the K2P method (28) to correct for multiple substitutions at a site (the transition/transversion [Ts/Tv] ratio was set at 10). Genetic distances were calculated using the program DNADIST and summarized in a tree constructed by the neighbor-joining method (53) using the program NEIGHBOR; both programs are part of the PHYLIP 3.572c program package (14).
Trees of VP1 sequence relationships among Egyptian type 2 cVDPV isolates were constructed by using the maximum likelihood method implemented in the fastDNAml program (40). A heuristic search was performed with 1,000 bootstrap replicates, with the input order of the sequences randomized for each replicate. The Ts/Tv ratio was initially set to 1, and the base frequencies were determined from the input data to simulate the F81 proportional model (13). The candidate tree was further analyzed using the program PAML (77) to compare other models of evolution in likelihood ratio tests (19). The TN93 model of nucleotide substitution (62) gave a significantly higher likelihood score than the F81 model; more complex models did not significantly improve the likelihood score. Parameters in TN93 were set to a Ts/Tv ratio of 10, and the shape parameter of the gamma distribution was calculated by PAML as 0.5 with eight categories of discrete gamma rates. The tree with the highest log likelihood score was rooted to the VP1 sequence of Sabin 2 and displayed using the program TreeView (41).
Estimation of the time of the initiating OPV dose.
The time of the initiating OPV dose was estimated from the rate of fixation into VP1 of synonymous nucleotide substitutions among the 28 cVDPV isolates for which the dates of sample collection were known (Minya/92 and Sharqiya/92 were accordingly excluded). The number of VP1 substitutions at synonymous sites (Ks) that accumulated from the Sabin 2 sequence were computed according to the method of Yang and Nielsen (78) implemented in the program YN00 included in the PAML package (77). This method corrects for the Ts/Tv rate bias, the codon frequency bias, and multiple substitutions at a site. Corrected Ks values relative to the Sabin 2 root sequence (for which Ks is set to zero) for each cVDPV isolate were plotted as a function of the date of sample collection. The rate of accumulation of synonymous substitutions was estimated by weighted linear regression (where the weight for each data point was proportional to the reciprocal of the error variance for the corrected Ks value) using statistical applications within the SAS system, version 8 (SAS Institute, Inc., Cary, N.C.). The date of the initiating OPV dose was estimated from the intercept on the abscissa at Ks = 0, and the 95% confidence interval (CI) for the estimated date was calculated following procedures described by Sokal and Rohlf (56).
Estimation of nucleotide diversity.
Nucleotide diversity was calculated from VP1 sequence differences and corrected for superimposed substitutions by use of the K2P algorithm (Ts/Tv ratio set at 10) by using the MEGA program, version 2 (30).
Comparison of complete genomic sequences of with those of Sabin 2.
Complete genomic sequences of the recombinant cVDPV isolates, Qalyubiya/88-1 and Beni Suef/93, were compared with those of the Sabin 2 master seed stock. Sequence relationships for each viral gene (i.e., the 5′ untranslated region [5′ UTR] and each interval encoding a major cleavage product from the poliovirus polyprotein) were summarized in neighbor-joining trees constructed using the parameters described above for diverse wild type 2 poliovirus genotypes. Insertion-deletion 5′ UTR differences were treated as single nucleotide substitutions.
Numbering of nucleotide and amino acid positions.
VP1 sequences of all type 2 isolates were colinear. The coding sequences of Qalyubiya/88-1 and Beni Suef/93 were colinear with those of Sabin 2, but their 5′ UTRs were not. To facilitate comparisons, numbering of nucleotide positions of all three isolates followed that described for Sabin 2 (49), with serial letters assigned to insertions.
Neurovirulence testing in PVR-Tg21 mice.
Neurovirulence tests on cVDPV isolates were carried out on PVR-Tg21 mice as previously described (29, 31). The mice were purchased from the Central Laboratories for Experimental Animals (Kanagawa, Japan). The type 2 reference strains were MEF-1/EGY42 (neurovirulent) and Sabin 2 (attenuated). Six mice (equal numbers of males and females) were inoculated (30 μl/mouse) intracerebrally for each virus dilution (in 10-fold increments; range: 1.5 to 7.5 log 50% tissue culture infective doses/mouse). Mice were examined daily for 14 days postinoculation, and the times of paralysis or death were recorded. The virus titer that induced paralysis or death in 50% of inoculated mice was calculated by the method of Kärber (20) and expressed as 50% tissue culture infective doses/mouse.
Single-step growth curves.
The growth rates and plaque yields at 39.5°C of Qalyubiya/88-1 and Beni Suef/93 were compared with those of Sabin 2, Beni Suef/98 (OPV-like), and MEF-1/EGY42 in single-step growth experiments (input multiplicity, 10 PFU/cell) in S3 HeLa cell suspension cultures (human cervical carcinoma, ATCC CCL-2.2). Virus titers were determined in plaque assays on HeLa cell monolayer cultures (ATCC CCL-2) incubated at 37°C essentially as described previously (51).
Nucleotide sequence accession numbers.
VP1 sequences of the type 2 polioviruses described in this study were submitted to the GenBank library under accession numbers AF551795 to AF551841 (Table 1). Complete genomic sequences of isolates Qalyubiya/88-1 and Beni Suef/93 were submitted under accession numbers AF448782 and AF448783.
RESULTS
Epidemiologic and virologic background.
Archaeological evidence suggests that polio was present in Egypt at the dawn of civilization (44). As recently as the late 1970s, all three serotypes of indigenous wild poliovirus were circulating in Egypt (23). The last wild type 2 poliovirus from Egypt to be characterized was isolated from a 1979 case. However, the last year of circulation of the indigenous wild type 2 poliovirus in Egypt is unknown, because sensitive surveillance for cases of acute flaccid paralysis (AFP) and wild poliovirus was not initiated in the country until the 1990s (70, 71). Wild poliovirus type 3 was last detected in December 2000; wild poliovirus type 1 was still repeatedly isolated in Egypt (primarily in the southern governorates of Minya, Asyut, Sohag, and Qina) in 2001 and 2002 from both clinical specimens from AFP cases and environmental samples (70, 71).
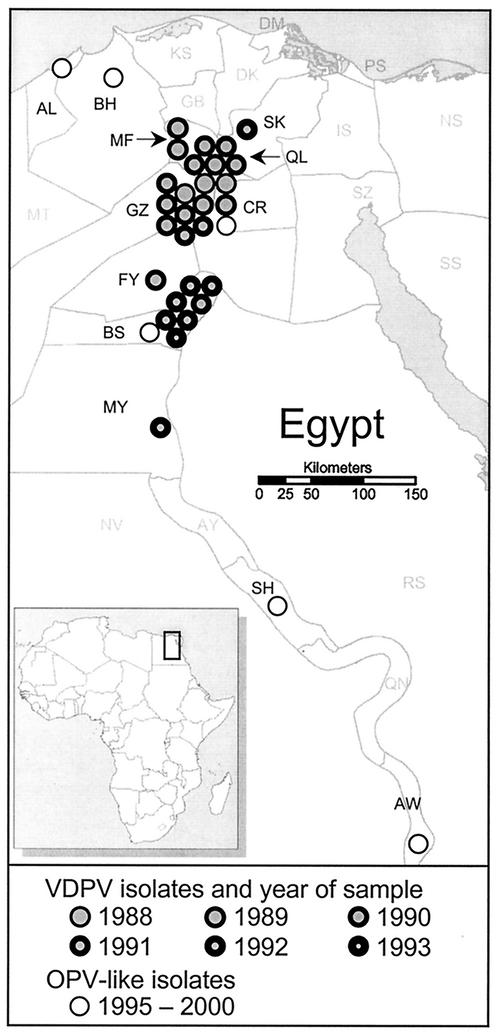
From 1988 to 1993, type 2 poliovirus was frequently isolated from patients with clinically confirmed poliomyelitis in Egypt (Fig. 1; Table 1). Because many of the patients were unimmunized and had no history of recent exposure to OPV, it was assumed at the time that these cases signaled the continued circulation of the indigenous wild poliovirus type 2. This view was reinforced by the finding that 30 of the type 2 isolates had DR antigenic properties when tested in the standard ELISA intratypic differentiation test using cross-absorbed antisera (65). The 30 DR case isolates were selected for further characterization as part of a broader study monitoring the eradication of wild poliovirus in Egypt.
FIG. 1.
Geographic distribution of polio cases in Egypt associated with type 2 cVDPV from 1988 to 1993. Individual cVDPV cases, represented by filled circles, are mapped by governorate (2 of the 30 cVDPV isolates were not mapped because their source governorates were not recorded). cVDPV cases occurring in different years are differentiated by increasing thickness of circle rims from 1988 to 1993. OPV-like isolates (>99% VP1 sequence identity to Sabin 2) from AFP cases with onset from 1995 to 2000 are identified by open circles. Governorate abbreviations: AL, Alexandria; AW, Aswan; AY, Asyut; BH, Behaira; BS, Beni Suef; CR, Cairo; DQ, Daqahliya; DM, Damietta; FY, Fayyum; GB, Gharbiya; GZ, Giza; IS, Ismailiya; KS, Kafr El-Sheikh; MF, Menufiya; MT, Matrouh; MY, Minya; NS, North Sinai; NV, New Valley; PS, Port Said; QL, Qalyubiya; QN, Qina; RS, Red Sea; SH, Sohag; SQ, Sharqiya; SS, South Sinai; SZ, Suez.
Vaccine derivation of type 2 isolates from polio cases from 1988 to 1993.
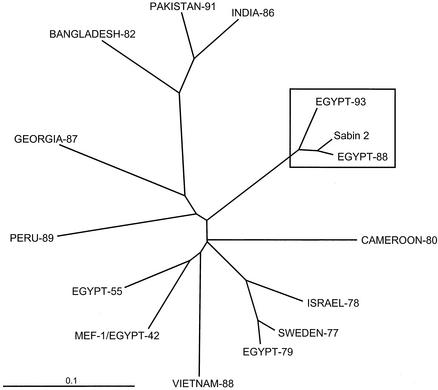
When the 30 DR isolates were tested by nucleic acid probe hybridization (11) and diagnostic PCR (75), they were found to be genetically related to Sabin 2 (data not shown). To confirm this identification, VP1 sequences (903 nucleotides) of an early (Qalyubiya/88-1) and a late (Beni Suef/93) DR isolate were compared with those of Sabin 2, three Egyptian wild type 2 poliovirus isolates (the reference strain, MEF-1/EGY42; 0317/EGY55; and 0297/EGY79), and nine wild type 2 isolates from cases occurring in Asia, Africa, Europe, and South America from 1977 to 1991 (Fig. 2). The DR isolates Qalyubiya/88-1 and Beni Suef/93 were clearly VDPVs, as they were more closely related to Sabin 2 (97.2 and 93.3% VP1 sequence identity, respectively) than they were to any of the wild polioviruses (76.6 to 81.1% VP1 sequence identity). However, direct pairwise comparisons of VP1 sequence differences severely underestimate the true genetic distances separating the type 2 VDPVs from wild polioviruses. The underestimate occurs because sequence differences do not increase linearly with increasing genetic distance but instead increase asymptotically as superimposed base substitutions are fixed into variable sites (18, 61). In the rapidly evolving poliovirus genome (16, 25, 32, 35), variable sites (which represent ∼33% of total VP1 nucleotides, primarily at positions of codon degeneracy) saturate within ∼30 years. However, synonymous transversions are fixed into variable poliovirus sites at an approximately 10-fold lower rate than synonymous transitions (J. Jorba, unpublished results). Therefore, closely related isolates can be better resolved from more divergent viruses by comparing only transversional differences at fourfold degenerate sites. By this method of comparison, Sabin 2, Qalyubiya/88-1, and Beni Suef/93 are indeed very closely related (0.2 to 0.4% transversional differences in VP1) and are only distantly related to any of the wild type 2 polioviruses (3.7 to 10.6% transversional differences in VP1).
FIG. 2.
Unrooted neighbor-joining tree summarizing VP1 (nucleotides 2482 to 3384) sequence relationships among the Sabin 2 OPV strain, the Egyptian cVDPV isolates Qalyubiya/88-1 and Beni Suef/93, three Egyptian wild type 2 isolates (MEF-1/EGY42, 0317/EGY55, and 0297/EGY79), and nine wild type 2 poliovirus isolates from cases occurring in Africa, Asia, Europe, and South America from 1977 to 1991.
VP1 sequence relationships among Egyptian VDPV isolates.
The VDPV isolates Qalyubiya/88-1 and Beni Suef/93 shared 15 VP1 nucleotide substitutions distinguishing them from Sabin 2, consistent with their derivation from a common ancestral infection. Such sequence relationships are typical among isolates of locally circulating wild polioviruses (32, 55) and cVDPVs (22) but not among OPV-like or iVDPV isolates from different patients, which generally follow independent pathways of divergence from the vaccine strain (22, 25, 35; R. Park, unpublished results). To visualize the pathways of divergence among the Egyptian cVDPV isolates, their VP1 sequence relationships were summarized in a phylogenetic tree rooted to the VP1 sequence of Sabin 2 (Fig. 3).
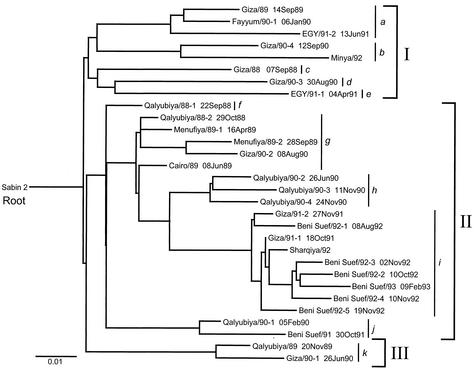
FIG. 3.
Maximum likelihood tree of VP1 sequence relationships among the Sabin 2 OPV strain (root of tree) and 30 type 2 cVDPV isolates from Egypt (1988 to 1993). Brackets enclose separate genetic groups (I to III) and their component clusters (a to k).
The tree resolved 11 main clusters of genetic lineages (a to k), which correspond to independent pathways of evolutionary divergence from Sabin 2. The clusters distributed into three broad groups (group I, clusters a to e; group II, clusters f to j; and group III, cluster k) (Fig. 3). Several individual clusters (a, b, g, h, i, j, and k) had one or more branches representing additional diverging lineages established by separate chains of transmission. Four clusters (c, d, e, and f) were represented by a single isolate. The most closely related clusters were f and g, with an average of 26 VP1 nucleotide differences between them. The most distantly related clusters were b and d, with an average of 73 VP1 nucleotide differences between them.
A single node connected the sequences of the 30 cVDPV isolates to the root Sabin 2 sequence. This node represents the VP1 sequence of the inferred most recent common ancestor to all 30 cVDPV isolates. Although the bootstrap values (14) supporting the node shown in Fig. 3 were only moderate (∼44%), replicate trees constructed from the same data set by using the neighbor-joining (53) and quartet puzzling (58) algorithms had similarly placed ancestral nodes. The location of the ancestral node on the tree and the short length of the branch joining it to the Sabin 2 root sequence suggest that the most recent ancestral infection common to all subsequent lineages occurred about a year after the initiating OPV dose (see next section) and well before 1988, the year of the cases yielding the earliest available cVDPV isolates (Giza/88, Qalyubiya/88-1, and Qalyubiya/88-2) (Fig. 3). Soon after the ancestral infection, the virus radiated along separate chains of transmission, observed as separate branches on the tree. We have no further information about the early events in the spread of cVDPVs in Egypt because OPV-derived isolates dating prior to 1988 are not available.
The isolates within group I had the most diverse VP1 sequences (0.071 nucleotide differences per site), with the eight isolates distributing into five clusters (a to e). Most group I sequences were connected to each other by long branches. Long branches indicate missing sequences, as many polio cases were likely missed and specimens were not available for virologic testing. At least half (four of eight) of the group I isolates (the source governorates for two of the isolates were not recorded) were from Giza. Isolates from Giza were found in all three groups and in seven different clusters (a, b, c, d, g, i, and k). The high sequence diversity of the Giza isolates (0.070 nucleotide differences per site) does not necessarily indicate that multiple cVDPV lineages originated from that governorate. It is more likely that different cVDPV lineages were introduced into Giza from other communities in Egypt, as Giza is host to migrants from other parts of the country. Only one group I isolate (Minya/92, cluster b) was from the southern governorates, where OPV coverage and AFP surveillance has been lower than in the rest of Egypt. It is possible that several of the Giza cVDPV isolates (such as those comprising clusters b, c, and d) were derived from viruses circulating in areas remote from the Poliomyelitis Institute in Cairo.
Group II isolates were more closely related to each other (0.046 nucleotide differences per site). Most of the isolates were from the adjoining governorates of Qalyubiya, Menufiya, Giza, and Cairo (Fig. 1 and 3). The sequence of Qalyubiya/88-1 is close to the ancestral node connecting all group II isolate sequences. Some of the genetic clusters correlated with the geographic source of the virus (Fig. 1 and 3). For example, all three isolates of cluster h were from Qalyubiya, and most (six of nine) isolates of cluster i were from Beni Suef. Cluster g represents a lineage that circulated over four governorates and then apparently faded out after 1990. Cluster h, apparently localized to Qalyubiya, was also not detected after 1990. Isolates of cluster i were the last type 2 cVDPVs to be found in Egypt (last isolate: Beni Suef/93, 9 February 1993). There is clear evidence of sustained localized cVDPV transmission during a single polio season (which in Egypt runs from about the beginning of April to the end of March of the following calendar year). For example, isolates of cluster h were obtained from polio patients in Qalyubiya from 26 June 1990 to 24 November 1990, and isolates of cluster i were obtained from polio patients in Beni Suef from 8 August 1992 to 9 February 1993. There is also evidence of continued transmission of cVDPV lineages through one or more winter low-transmission seasons for poliovirus, as is seen in clusters g (Qalyubiya/88-2 [29 October 1988] to Giza/90-2 [8 August 1990]) and i (Giza/91-1 [18 October 1991] to Beni Suef/93 [9 February 1993]).
The two isolates forming group III (cluster k), Qalyubiya/89 and Giza/90-1, represent a lineage that diverged from the others well before 1988 and may have established a pocket of localized circulation in 1989 to 1990 (Fig. 3).
Genetic distance from the Sabin 2 VP1 sequence correlated with time of specimen collection (Fig. 3). Sequences of the earliest (1988 to 1989) isolates were located at the tips of branches closest to the root (left side) of the tree, sequences of isolates from 1990 to 1991 were located centrally, and sequences of isolates from 1992 to 1993 were located at the tips of branches at the right side of the tree.
Estimated time of occurrence of initiating OPV dose.
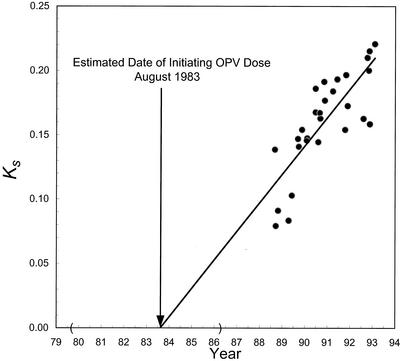
The approximate time of the initiating OPV infection was estimated from the VP1 sequence differences from Sabin 2 among the cVDPV isolates, the dates of specimen collection, and the rate of VP1 evolution (22). The rate of substitution into synonymous sites of VP1 was approximately linear from 1988 to 1993 (R2 = 0.670) (Fig. 4). When the regression line was extrapolated to zero VP1 substitutions, the intercept was August 1983 (95% CI: July 1979 to March 1986). The CI was wide because of the stochastic nature of base substitution, because the earliest available cVDPV isolates were obtained from specimens taken about 5 years after the initiating OPV dose and because of the bias in the sources of isolates available for analysis. The VP1 evolution rate estimated from the slope of the regression line was (2.47 ± 0.69) × 10−2 substitutions per synonymous site per year, approximately 80% of the rate observed for circulating wild type 1 polioviruses (16, 25, 32).
FIG. 4.
Estimate of the date of initiating OPV dose from the rate of accumulation of synonymous substitutions into VP1 among the 28 type 2 cVDPV isolates for which the dates of sample collection are known. Abscissa: date of sample collection for each isolate. Ordinate: Ks, the number of substitutions (Sabin 2 sequence set to zero substitutions) at synonymous sites in VP1. The evolution rate was estimated by weighted linear regression (R2 = 0.670) as described in the Materials and Methods. The 95% CI for the estimated date of the initiating OPV dose, July 1979 to March 1986, is bounded by parentheses along the abscissa.
Disappearance of type 2 cVDPV after 1993.
The last type 2 cVDPV isolate from Egypt, Beni Suef/93, was isolated from a polio case with onset in February 1993, near the end of the 1992 polio season (39). After 1993, all six DR type 2 polioviruses isolated from patients with AFP in Egypt were OPV-like (Table 1), having only 0 to 1 VP1 nucleotide sequence differences from Sabin 2. In addition, 29 other Sabin 2-derived polioviruses were isolated from sewage samples taken in 2001 to 2002 in urban areas of Gharbiya, Menufiya, Sharqiya, Cairo, Fayyum, Beni Suef, Minya, Asyut, and Sohag. These isolates had 0 to 4 (mean, 1.4) VP1 nucleotide sequence differences from Sabin 2 (L. El-Bassioni, unpublished results). All OPV-like type 2 polioviruses isolated after 1993 reacted as vaccinelike in the ELISA with cross-absorbed antisera. Because surveillance improved throughout the 1990s, the inability to detect any VDPVs in clinical specimens from AFP patients or in environmental samples during the 9 years after 1993 strongly suggests that VDPV circulation had ceased. We believe that the most important factor for the disappearance of cVDPV was the rising rate of OPV coverage.
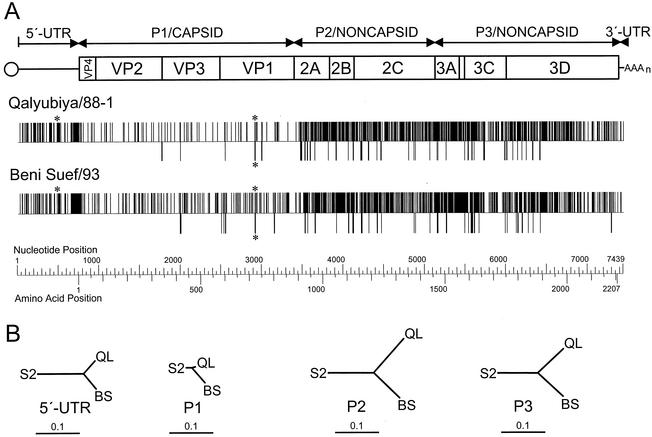
Tripartite recombinant structure of Qalyubiya/88-1 and Beni Suef/93 genomes.
The complete genomic sequences of an early type 2 cVDPV isolate, Qalyubiya/88-1, and the last isolate, Beni Suef/93 (Table 1), were compared with that of the Sabin type 2 OPV strain (Fig. 5). Qalyubiya/88-1 was selected as the early cVDPV isolate for comparison because it appeared to be closely related to the ancestor of clusters f to j (which includes Beni Suef/93, cluster i) (Fig. 3). While the capsid sequences were homologous to those of Sabin 2 (97.4% [Qalyubiya/88-1] and 94.4% [Beni Suef/93] nucleotide sequence identity) and to each other (95.1% nucleotide sequence identity), the flanking sequences were dissimilar. The 5′ UTR sequences of both Qalyubiya/88-1 and Beni Suef/93 were apparently derived from viruses other than Sabin 2 (having <85% nucleotide sequence identity to the 5′ UTR of Sabin 2). The conserved 5′ UTR sequences at positions 1 to 646 (maximum observed divergence among poliovirus isolates: ∼89%) of both cVDPV isolates were not closely related for that interval (93.1% nucleotide sequence identity). By contrast, the highly variable and rapidly evolving (H.-M. Liu, unpublished results) 5′ UTR sequences at positions 647 to 747 (maximum observed divergence among poliovirus isolates: ∼46%) appeared to be relatively more closely related (90.1% nucleotide sequence identity). Similarly, the noncapsid region and 3′ UTR sequences of the two cVDPV isolates differed from Sabin 2 (82.7% [Qalyubiya/88-1] and 83.2% [Beni Suef/93] nucleotide sequence identity) and from each other (86.4% nucleotide sequence identity) (Fig. 5). The sequence relationships among the genomes of Sabin 2, Qalyubiya/88-1, and Beni Suef/93 suggest that at least two rounds of recombination occurred in both the 5′ UTR and noncapsid region during cVDPV evolution. The heterologous sequences were not derived from the Sabin 1 or Sabin 3 OPV strains or any other characterized poliovirus and were most likely derived from unidentified species C enteroviruses. A type 2 OPV-like isolate having a similar tripartite recombinant structure has been described previously (17).
FIG. 5.
(A) Nucleotide (upper bars) and amino acid (lower bars) substitutions into the genomes of cVDPV isolates Qalyubiya/88-1 (above) and Beni Suef/93 (below). The reference sequence is that of Sabin 2. The substitution maps are aligned with a schematic of the poliovirus genome; the single open reading frame is indicated by a rectangle, flanked by the 5′ and 3′ UTRs. Asterisks indicate attenuating mutations replaced by recombination (5′ UTR) or nucleotide substitution (VP1). (B) Neighbor-joining trees summarizing sequence relationships in the 5′ UTR and in each interval encoding a major cleavage product from the poliovirus polyprotein; all trees are drawn to the same scale. Isolate sequence abbreviations: S2, Sabin 2; QL, Qalyubia/88-1; BS, Beni Suef/93.
Replacement of attenuating mutations by recombination and back mutation.
Two substitutions have been identified as principal determinants of the attenuated phenotype in the Sabin 2 genome: an A at nucleotide position 481 in the 5′ UTR and an Ile residue at amino acid position 143 in VP1 (34, 48). Both of these substitutions are under strong selective pressure for reversion when OPV replicates in the human intestine. A large proportion (>90%) of type 2 OPV-like isolates from healthy and paralyzed patients (34, 36) and from the environment (80) are 481 A→G revertants, restoring the consensus nucleotide at that position (47). Both Qalyubiya/88-1 and Beni Suef/93 had the consensus G at position 481, but unlike in most OPV-like isolates, the surrounding 5′ UTR sequences were probably derived from other enteroviruses. Isolates Qalyubiya/88-1 and Beni Suef/93 shared the same VP1-143 Ile→Thr amino acid replacement (restoring the consensus residue for wild type 2 polioviruses) encoded by a U→C substitution at nucleotide position 2909.
Neurovirulence of Qalyubiya/88-1 and Beni Suef/93 to PVR-Tg21 transgenic mice.
All 30 of the cVDPVs were isolated from patients with paralytic polio (Table 1), demonstrating their neurovirulence for humans under natural conditions. Moreover, the sequence properties of isolates Qalyubiya/88-1 and Beni Suef/93 suggested that they would be neurovirulent when introduced into the central nervous systems of experimental animals (34, 48). When the neurovirulence of these isolates was measured quantitatively in PVR-Tg21 transgenic mice expressing the human receptor for poliovirus, they were found to be as neurovirulent as the prototype wild type 2 poliovirus strain, MEF-1/EGY42 (Table 3), isolated in 1942 from patients with paralytic polio.
TABLE 3.
Neurovirulence of Qalyubiya/88-1 and Beni Suef/93 in PVR-Tg21 transgenic mice
| Isolate | No. of mice killed/total no. inoculated with a dose (TCID50/mouse) of:
|
PD50b | ||||||
|---|---|---|---|---|---|---|---|---|
| 7.5 | 6.5 | 5.5 | 4.5 | 3.5 | 2.5 | 1.5 | ||
| Qalyubiya/88-1 | —a | — | — | — | 5/6 | 3/6 | 1/6 | 2.5 |
| Beni Suef/93 | — | — | — | — | 6/6 | 2/6 | 0/6 | 2.7 |
| MEF-1/EGY42 | — | — | — | — | 6/6 | 3/6 | 0/6 | 2.5 |
| Sabin 2 | 0/6 | — | — | — | — | — | — | >8.0 |
—, not done.
Values represent the virus titer that induced paralysis or death in 50% of inoculated mice (PD50) and are expressed as a multiple of the TCID50.
Single-step growth curves of Qalyubiya/88-1 and Beni Suef/93.
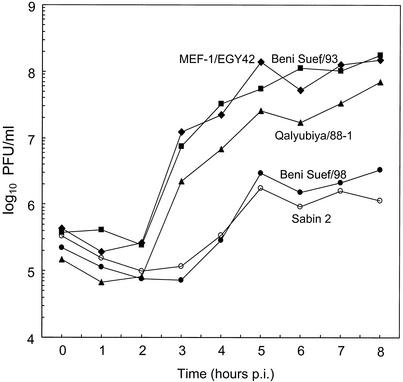
All three Sabin strains are temperature sensitive, producing lower virus yields at supraoptimal temperatures than wild polioviruses (37). Temperature sensitivity may contribute to the attenuation of OPV by reducing virus yields at the early stages of replication in the human intestine. However, the temperature-sensitive phenotype is under intense negative selection during natural replication, and variants with reduced temperature sensitivity are frequently excreted by OPV recipients (37). Reversion of temperature sensitivity appears to occur most rapidly with Sabin 3 and more slowly with Sabin types 1 and 2 (37).
The growth rates and virus yields at 39.5°C of Qalyubiya/88-1 and Beni Suef/93 were compared with those of Sabin 2, Beni Suef/98, and MEF-1/EGY42 in single-step growth experiments in S3 HeLa cells (Fig. 6). Beni Suef/98 is an OPV-like isolate, having vaccinelike antigenic properties, one nucleotide substitution in VP1 (a U→A substitution at nucleotide position 2909 encoding a VP1-143 Ile→Asn replacement), and the consensus G nucleotide at position 481. The wild isolate, MEF-1/EGY42, grows to high yields in cell culture, a property that favored its selection as the type 2 component of the inactivated poliovirus vaccine (IPV) (46). MEF-1/EGY42 grew more rapidly and produced ∼90-fold higher titers than Sabin 2 during infection of S3 HeLa cells incubated at 39.5°C (Fig. 6). Interestingly, the late cVDPV isolate, Beni Suef/93, had growth properties at 39.5°C similar to those of MEF-1/EGY42. By contrast, the OPV-like isolate, Beni Suef/98, grew to only slightly higher titers (∼twofold) than Sabin 2. The early cVDPV isolate, Qalyubiya/88-1, grew more rapidly and to much higher titers than Sabin 2 and Beni Suef/98, but the virus yield at 8 h was about one-third of the yields for MEF-1/EGY42 and Beni Suef/93 (Fig. 6).
FIG. 6.
Single-step growth curves at 39.5°C in S3 HeLa cells of MEF-1/EGY42 (wild), Qalyubiya/88-1 (cVDPV), Beni Suef/93 (cVDPV), Beni Suef/98 (OPV-like), and Sabin 2.
DISCUSSION
The key finding from this study is that derivatives of the OPV strain had restored poliovirus type 2 endemicity to Egypt within a few years of the presumed eradication of the indigenous wild type 2 poliovirus. The type 2 cVDPV had recovered the two most important biological properties of wild polioviruses, namely, (i) the capacity to cause paralytic disease in humans and (ii) the capacity for continuous person-to-person transmission. The most critical risk factor for the spread of type 2 cVDPV in Egypt was low population immunity to poliovirus type 2, a result of the combination of low OPV coverage in many communities and the prior elimination of the indigenous wild poliovirus type 2. The other risk factors appear to be the same as for wild poliovirus circulation and include crowding, high birth rates, poor hygiene and sanitation, and tropical climate (15, 39). Under such conditions, the cVDPV established several independent foci of endemicity in separate communities. The total number of human infections associated with the type 2 cVDPV probably was very large, most likely in the hundreds of thousands. This view is consistent with the prolonged duration of cVDPV endemicity (estimated at ∼10 years), the high nucleotide diversity of the cVDPV isolates (consistent with multiple chains of transmission), the generally very low paralytic attack rate for type 2 poliovirus infections (39), and the likelihood that the large majority of polio cases that had occurred during the period of cVDPV endemicity in Egypt had not been investigated.
VDPV circulation in Egypt apparently ceased after 1993 as the rates of OPV coverage increased (70, 71). We do not know what levels of OPV coverage actually achieved cessation of VDPV circulation in Egypt because of the absence of complete national coverage data for the early 1990s. It is likely that the OPV coverage rates necessary to interrupt virus transmission would be comparatively high in Egypt because of the prevalence of conditions favorable to poliovirus circulation (15, 39, 42). Nonetheless, the threshold rates of coverage necessary to stop poliovirus transmission are lower for type 2 than for types 1 and 3, probably because seroconversion to type 2 antigen is more efficient (43). The lower OPV coverage threshold required to control type 2 poliovirus may explain why circulation of the indigenous type 2 wild poliovirus and the subsequent type 2 cVDPV ceased in Egypt several years before elimination of the indigenous wild poliovirus types 3 (in 2000) and 1 (circulation continued throughout 2002) (71). We also do not know when the last case associated with type 2 cVDPV occurred in the country because surveillance gaps persisted into the 1990s, especially in the southern governorates where the rates of OPV coverage were lowest. However, as the sensitivity of AFP surveillance improved, no evidence has been found for VDPV circulation after 1993.
The endemic circulation of VDPV in Egypt differs in several important respects from the recent cVDPV outbreaks in Hispaniola (Dominican Republic and Haiti) (22), the Philippines (8, 64), and Madagascar (9). The most significant difference was the much longer period of VDPV circulation in Egypt than in the three recent outbreaks (estimated at ∼3 years). Moreover, the outbreaks in Egypt and Madagascar were associated with type 2 cVDPV, whereas the outbreaks in Hispaniola and the Philippines were associated with type 1 cVDPV. Genetic evidence suggests that cVDPV spread along multiple independent chains of transmission in both Egypt and Haiti (22). Two separate lineages of type 2 cVDPV have been found in Madagascar, with the first lineage arising about a year before the second (9). By contrast, the more localized outbreak in the Dominican Republic appears to have resulted from the point source introduction of cVDPV from neighboring Haiti (22). cVDPV spread appears to have been even more limited in the Philippines, as the genetic properties of the outbreak isolates suggest that transmission occurred along a single, minimally branched chain (8, 64).
The early virologic events leading to cVDPV outbreaks have not been observed directly. Such events would be difficult to observe even under ideal conditions because of the inherently low paralytic attack rates of poliovirus infections (39). However, the prospects for early detection of cVDPV outbreaks are further reduced in the areas of highest risk, because gaps in surveillance usually accompany gaps in OPV coverage. Nonetheless, a reasonable model for the genesis of cVDPV outbreaks may be proposed from the known properties of the Sabin OPV strains. Because the base substitutions contributing to the attenuated phenotype of the Sabin strains (48) readily revert during replication in the human intestine (34, 36), the virus excreted by healthy vaccine recipients is frequently less attenuated than the original OPV strains. Reversion is very rapid for the Sabin 2 strain because attenuation is primarily determined by only two highly unstable substitutions (34, 36, 48, 80). The intense selection against the attenuating mutations suggests that the revertants replicate more efficiently in human intestine (36). This may be expressed as an overall increase in the titers of excreted virus, an extended period of virus excretion, and possibly an increased potential for person-to-person transmission. Introduction of OPV at low rates of coverage could trigger the transmission of excreted vaccine virus. The extent of spread would depend on several factors, including the number and density of nonimmune susceptible persons in the population. It therefore appears likely that Sabin 2 revertants with increased potentials for neurovirulence and transmissibility are constantly selected in all communities where OPV is used, but their spread is normally restricted by high population immunity.
The observation that all cVDPV isolates were derived from a single initiating OPV dose does not necessarily indicate that only one cVDPV lineage emerged in Egypt during the 1980s. The conditions leading to the observed lineages may have also favored the independent generation of other cVDPV lineages, as has recently been found in Madagascar (9). However, the other lineages might have escaped detection in several different ways, namely, (i) by circulating in communities outside of those sampled, (ii) by spontaneously fading out before 1988, or (iii) by being displaced before 1988 by the observed lineages.
All outbreak-associated cVDPV isolates described thus far have been recombinants with other species C enteroviruses (8, 22, 64). However, this observation does not necessarily imply that recombination plays an obligatory mechanistic role in the phenotypic reversion of OPV, because the main determinants of attenuation of all three Sabin strains map to 5′ UTR and capsid region sites (21, 34, 48, 66), and most of the observed recombination sites map to the noncapsid region (9, 22, 64; J. Iber, unpublished results). Although we observed that the interval containing the critical attenuating sites in the 5′ UTR had been exchanged out by recombination in the Egyptian cVDPV isolates, it is likely that back mutation preceded recombination, because selection of 5′ UTR revertants is very rapid during replication of OPV in the human intestine (33, 36, 80). Moreover, the successive recombination events involving noncapsid region (22) and 5′ UTR sequences may not confer any progressive increase in replicative fitness. Rather, poliovirus recombination with other species C enteroviruses may at times occur during mixed infection, with the frequency of recombination being a function of the enterovirus carriage rate and the total number of mixed infections. Thus, if a vaccine-related isolate has significant divergence in its capsid nucleotide sequences (>1% from the parental OPV strains) and has noncapsid or 5′ UTR sequences of non-Sabin strain origin, it should be considered a possible cVDPV isolate and the associated case should be investigated further.
The episodes of VDPV circulation in Egypt and other developing countries challenge the assumption that poliovirus endemicity can be restored only by the reintroduction of wild poliovirus and underscore the urgency of reaching the goal of global polio eradication as quickly as possible. The first priority is to eliminate the remaining reservoirs of wild poliovirus endemicity in South Asia and Sub-Saharan Africa (24, 63, 72). At the same time, it is essential to maintain high levels of poliovaccine coverage in all countries, both to prevent the spread of imported wild polioviruses and to suppress the emergence of cVDPVs. Areas at highest risk for emergence of cVDPVs are those where poliovaccine coverage rates have declined, the competing wild polioviruses have been eliminated, and epidemiologic conditions had previously favored wild poliovirus transmission. In response to the recent cVDPV outbreaks, the World Health Organization (WHO) has recommended reinstatement of mass immunization campaigns to close the immunity gap in areas where the rates of routine OPV coverage have been insufficient to prevent a growing susceptibility to polio (63). The optimal frequency of the mass campaigns follows from the rate of accumulation of susceptible persons in the population and the basic reproduction number for poliovirus in the highest risk populations in each area (15, 42). No additional measures have been recommended for the remaining countries of polio endemicity, because activities currently planned for elimination of the last pockets of types 1 and 3 poliovirus circulation will also effectively prevent dissemination of cVDPVs.
The occurrence of cVDPV cases also highlights the need to maintain sensitive poliovirus surveillance into the foreseeable future. Intensive screening of recent poliovirus isolates for cVDPVs has been initiated by laboratories within the WHO Global Polio Laboratory Network (5). Beginning in 2001, all vaccine-related poliovirus isolates from AFP cases have been screened for evidence of prolonged replication or circulation (6, 69). Poliovirus isolates are identified according to their genetic properties by probe hybridization (11), diagnostic PCR assays (26, 75), or PCR-restriction fragment polymorphism analysis (3). All isolates are also tested for antigenic change by using specific cross-absorbed sera in an ELISA format (65) or panels of monoclonal antibodies in neutralization tests (65). Alternatively, isolates have been screened for recombinant noncapsid sequences using PCR primers targeting P2 and P3 region sequences characteristic for each Sabin strain (D. R. Kilpatrick, unpublished results). Any isolate having non-vaccinelike, DR, or NR antigenic properties or having a recombinant genome is characterized further by VP1 sequencing (6). The combined strategy of molecular and antigenic characterization of poliovirus isolates led to the recognition of the cVDPV outbreak in the Philippines (8, 64) and to the identification of several candidate iVDPV isolates from developing countries (6).
Several observations suggest that the risk for emergence of cVDPVs may be highest for poliovirus type 2 (15). The type 2 OPV strain appears to spread more readily to unimmunized people than the other Sabin strains, as shown by the more frequent association of contact cases of vaccine-associated paralytic poliomyelitis with poliovirus type 2 (57) and by the much higher seroprevalence to poliovirus type 2 (relative to types 1 and 3) found among unvaccinated individuals in the United States (10, 15, 59) and Europe (2). Type 2 VDPVs resembling the cVDPVs from Egypt and Madagascar had been isolated from polio cases in other developing countries during times when OPV coverage rates were low (R. Park, unpublished results). However, because of the low paralytic attack rates for type 2 infections, circulation of type 2 VDPVs is the most difficult to detect by AFP surveillance. Consequently, early detection of any future type 2 cVDPV outbreaks will require maintenance, and possible augmentation in some countries (54), of the present very high global standard for AFP and poliovirus surveillance (63, 72).
The recognition that cVDPVs can cause polio outbreaks and potentially reestablish polio endemicity to polio-free areas has prompted a reassessment of global strategies for maintaining polio-free status after wild poliovirus circulation has ceased (12, 15, 38, 63, 68). The number of viable options for the endgame strategy using the existing poliovirus vaccines now appears to be quite limited (12, 38, 63). OPV use should continue in countries of polio endemicity and in developing countries where polio is not endemic but which are at high risk for outbreaks caused by imported wild polioviruses or emerging cVDPVs. Transition from OPV to IPV is likely to continue in polio-free countries, especially in developed countries in temperate zones where IPV efficacy is highest (67) and where high rates of IPV coverage can be maintained through routine immunization (63). Soon after certification of global eradication, countries still using OPV may discontinue its use immediately after completion of the last round of synchronous mass OPV campaigns (12, 63). In addition, IPV use might be implemented in all countries for an interim period until evidence is compelling that the risk of recurrence of poliovirus circulation is very low (12, 63). Whatever the strategy, implementation will be most challenging in low-income, tropical developing countries where the risks of cVDPV emergence are highest. In the meantime, universally high rates of polio vaccine coverage must be maintained at least until implementation of the WHO endgame strategy (63).
Acknowledgments
We thank A. J. Williams, Naomi Dybdahl-Sissoko, Nada Mishrik, Deborah Moore, and Michelle Staples (CDC, Atlanta) for preparation of poliovirus isolates for molecular analyses; Laila El-Bassioni for contributing the 2000 to 2002 type 2 isolates from Egypt; Hongmei Liu for sequencing many of those isolates; Carolyn Weeks-Levy for contributing the reference Sabin 2 strain; Paul Chenoweth for mapping the Egyptian cVDPV isolates; and Larry Anderson, Jorge Boshell, Victor Cáceres, Steve Cochi, Esther de Gourville, Francis Delpeyroux, Walter Dowdle, Howard Gary, Hamid Jafari, Bob Keegan, David Kilpatrick, Chris Maher, Steve Oberste, Ray Sanders, and Roland Sutter for helpful discussions. We thank the virologists from the World Health Organization Global Polio Laboratory Network for contributing the wild poliovirus isolates used in our comparisons.
Hiroyuki Shimizu, Tetsuo Yoneyama, and Tatsuo Miyamura were supported by a grant in aid for Promotion of Polio Eradication from the Ministry of Health, Welfare and Labor, Japan.
REFERENCES
- 1.Alexander, J. P., Jr., H. E. Gary, Jr., and M. A. Pallansch. 1997. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J. Infect. Dis. 175(Suppl. 1):S176-S182. [DOI] [PubMed] [Google Scholar]
- 2.Aylward, R. B., D. Porta, L. Fiore, B. Ridolfi, P. Chierchini, and F. Forastiere. 1997. Unimmunized Gypsy populations and implications for the eradication of poliomyelitis in Europe. J. Infect. Dis. 175(Suppl. 1):S86-S88. [DOI] [PubMed] [Google Scholar]
- 3.Balanant, J., S. Guillot, A. Candréa, F. Delpeyroux, and R. Crainic. 1991. The natural genomic variability of poliovirus analyzed by a restriction fragment polymorphism assay. Virology 184:645-654. [DOI] [PubMed] [Google Scholar]
- 4.Bellmunt, A., G. May, R. Zell, P. Pring-Akerblom, W. Verhagen, and A. Heim. 1999. Evolution of poliovirus type I during 5.5 years of prolonged enteral replication in an immunodeficient patient. Virology 265:178-184. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2000. Developing and expanding contributions of the Global Laboratory Network for Poliomyelitis Eradication, 1997-1999. Morb. Mortal. Wkly. Rep. 49:156-160. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Laboratory surveillance for wild poliovirus and vaccine-derived poliovirus, 2000-2001. Morb. Mortal. Wkly. Rep. 51:369-371. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1999. Progress toward the global interruption of wild poliovirus type 2 transmission, 1999. Morb. Mortal. Wkly. Rep. 48:736-738. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2001. Public health dispatch: acute flaccid paralysis associated with circulating vaccine-derived poliovirus—Philippines, 2001. Morb. Mortal. Wkly. Rep. 50:874-875. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2002. Public health dispatch: poliomyelitis—Madagascar, 2002. Morb. Mortal. Wkly. Rep. 51:622. [Google Scholar]
- 10.Chen, R. T., S. Hausinger, A. S. Dajani, M. Hanfling, A. L. Baughman, M. A. Pallansch, and P. A. Patriarca. 1996. Seroprevalence of antibody against poliovirus in inner-city preschool children. Implications for vaccination policy in the United States. JAMA 275:1639-1645. [PubMed] [Google Scholar]
- 11.De, L., B. K. Nottay, C.-F. Yang, B. P. Holloway, M. A. Pallansch, and O. Kew. 1995. Identification of vaccine-related polioviruses by hybridization with specific RNA probes. J. Clin. Microbiol. 33:562-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowdle, W. R., E. de Gourville, O. M. Kew, M. A. Pallansch, and D. J. Wood. Polio eradication: the OPV paradox. Rev. Med. Virol., in press. [DOI] [PubMed]
- 13.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. University of Washington, Department of Genetics.
- 15.Fine, P. E. M., and I. A. M. Carneiro. 1999. Transmissibility and persistence of oral polio vaccine viruses: implications for the global poliomyelitis eradication initiative. Am. J. Epidemiol. 150:1001-1021. [DOI] [PubMed] [Google Scholar]
- 16.Gavrilin, G. V., E. A. Cherkasova, G. Y. Lipskaya, O. M. Kew, and V. I. Agol. 2000. Evolution of circulating wild poliovirus and of vaccine-derived poliovirus in an immunodeficient patient: a unifying model. J. Virol. 74:7381-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgescu, M.-M., F. Delpeyroux, and R. Crainic. 1995. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J. Gen. Virol. 76:2343-2348. [DOI] [PubMed] [Google Scholar]
- 18.Graur, D., and W.-H. Li. 2000. Fundamentals of molecular evolution, 2nd ed. Sinauer Associates, Sunderland, Mass.
- 19.Huelsenbeck, J. P., and B. Rannala. 1997. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science 276:227-232. [DOI] [PubMed] [Google Scholar]
- 20.Kärber, G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 162:480-483. [Google Scholar]
- 21.Kawamura, N., M. Kohara, S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1989. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kew, O. M., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. André, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 23.Kew, O. M., M. N. Mulders, G. Y. Lipskaya, E. E. da Silva, and M. A. Pallansch. 1995. Molecular epidemiology of polioviruses. Semin. Virol. 6:401-414. [Google Scholar]
- 24.Kew, O. M., and M. A. Pallansch. 2002. The mechanism of polio eradication, p. 481-491. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 25.Kew, O. M., R. W. Sutter, B. Nottay, M. McDonough, D. R. Prevots, L. Quick, and M. Pallansch. 1998. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J. Clin. Microbiol. 36:2893-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilpatrick, D. R., B. Nottay, C.-F. Yang, S.-J. Yang, E. da Silva, S. Peñaranda, M. Pallansch, and O. Kew. 1998. Serotype-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J. Clin. Microbiol. 36:352-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilpatrick, D. R., B. Nottay, C.-F. Yang, S.-J. Yang, M. N. Mulders, B. P. Holloway, M. A. Pallansch, and O. M. Kew. 1996. Group-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J. Clin. Microbiol. 34:2990-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 29.Koike, S., C. Taya, T. Kurata, S. Abe, I. Ise, H. Yonekawa, and A. Nomoto. 1991. Transgenic mice susceptible to poliovirus. Proc. Natl. Acad. Sci. USA 88:951-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetic Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 31.Li, J., L.-B. Zhang, T. Yoneyama, H. Yoshida, H. Shimizu, K. Yoshii, M. Hara, T. Nomura, H. Yoshikura, T. Miyamura, and A. Hagiwara. 1996. Genetic basis of the neurovirulence of type 1 polioviruses isolated from vaccine-associated paralytic patients. Arch. Virol. 141:1047-1054. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H.-M., D.-P. Zheng, L.-B. Zhang, M. S. Oberste, M. A. Pallansch, and O. M. Kew. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 74:11153-11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macadam, A. J., C. Arnold, J. Howlett, A. John, S. Marsden, F. Taffs, P. Reeve, N. Hamada, K. Wareham, J. Almond, N. Cammack, and P. D. Minor. 1989. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 strain of poliovirus in vaccinees. Virology 172:408-414. [DOI] [PubMed] [Google Scholar]
- 34.Macadam, A. J., S. R. Pollard, G. Ferguson, R. Skuce, D. Wood, J. W. Almond, and P. D. Minor. 1993. Genetic basis of attenuation of the Sabin type 2 vaccine strain of poliovirus in primates. Virology 192:18-26. [DOI] [PubMed] [Google Scholar]
- 35.Martín, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minor, P. D., and G. Dunn. 1988. The effect of sequences in the 5′ non-coding region on the replication of polioviruses in the human gut. J. Gen. Virol. 69:1091-1096. [DOI] [PubMed] [Google Scholar]
- 37.Nakano, J. H., M. H. Hatch, M. L. Thieme, and B. Nottay. 1978. Parameters for differentiating vaccine-derived and wild poliovirus strains. Prog. Med. Virol. 24:78-206. [PubMed] [Google Scholar]
- 38.Nathanson, N., and P. Fine. 2002. Poliomyelitis eradication—a dangerous endgame. Science 296:269-270. [DOI] [PubMed] [Google Scholar]
- 39.Nathanson, N., and J. R. Martin. 1979. The epidemiology of poliomyelitis: enigmas surrounding its appearance, epidemicity, and disappearance. Am. J. Epidemiol. 110:672-692. [DOI] [PubMed] [Google Scholar]
- 40.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 41.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 42.Patriarca, P. A., R. W. Sutter, and P. M. Oostvogel. 1997. Outbreaks of paralytic poliomyelitis, 1976-1995. J. Infect. Dis. 175(Suppl. 1):S165-S172. [DOI] [PubMed] [Google Scholar]
- 43.Patriarca, P. A., P. F. Wright, and T. J. John. 1991. Factors affecting the immunogenicity of oral poliovirus vaccine in developing countries: review. Rev. Infect. Dis. 13:926-939. [DOI] [PubMed] [Google Scholar]
- 44.Paul, J. R. 1971. A history of poliomyelitis. Yale University Press, New Haven, Conn.
- 45.Pipkin, P. A., D. J. Wood, V. R. Racaniello, and P. D. Minor. 1993. Characterisation of L cells expressing the human poliovirus receptor for the specific detection of polioviruses in vitro. J. Virol. Methods 41:333-340. [DOI] [PubMed] [Google Scholar]
- 46.Plotkin, S. A., A. Murdin, and E. Vidor. 1999. Inactivated polio vaccine, p. 345-363. In S. A. Plotkin and W. A. Orenstein (ed.), Vaccines, 3rd ed. W.B. Saunders, Philadelphia, Pa.
- 47.Pollard, S. R., G. Dunn, N. Cammack, P. D. Minor, and J. W. Almond. 1989. Nucleotide sequence of a neurovirulent variant of the type 2 oral poliovirus vaccine. J. Virol. 63:4949-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren, R., E. G. Moss, and V. R. Racaniello. 1991. Identification of two determinants that attenuate vaccine-related type 2 poliovirus. J. Virol. 65:1377-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rezapkin, G. V., L. Fan, D. M. Asher, M. R. Fibi, E. M. Dragunsky, and K. M. Chumakov. 1999. Mutations in Sabin 2 strain of poliovirus and stability of attenuated phenotype. Virology 258:152-160. [DOI] [PubMed] [Google Scholar]
- 50.Rico-Hesse, R., M. A. Pallansch, B. K. Nottay, and O. M. Kew. 1987. Geographic distribution of wild poliovirus type 1 genotypes. Virology 160:311-322. [DOI] [PubMed] [Google Scholar]
- 51.Rueckert, R. R., and M. A. Pallansch. 1981. Preparation and characterization of encephalomyocarditis (EMC) virus. Methods Enzymol. 78:315-325. [PubMed] [Google Scholar]
- 52.Sabin, A. B., and L. R. Boulger. 1973. History of Sabin attenuated poliovirus oral live vaccine strains. J. Biol. Stand. 1:115-118. [Google Scholar]
- 53.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 54.Shulman, L., J. Manor, R. Handsher, F. Delpeyroux, M. McDonough, T. Halmut, I. Silberstein, J. Alfandari, J. Quay, T. Fisher, J. Robinov, O. Kew, R. Crainic, and E. Mendelson. 2000. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J. Clin. Microbiol. 38:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shulman, L. M., R. Handsher, S.-J. Yang, C.-F. Yang, J. Manor, A. Vonsover, Z. Grossman, M. Pallansch, E. Mendelson, and O. M. Kew. 2000. Resolution of the pathways of poliovirus type 1 transmission during an outbreak. J. Clin. Microbiol. 38:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sokal, R. R., and F. J. Rohlf. 1995. Biometry, 3rd ed. W. H. Freeman and Co., New York, N.Y.
- 57.Strebel, P. M., R. W. Sutter, S. L. Cochi, R. J. Biellik, E. W. Brink, O. M. Kew, M. A. Pallansch, W. A. Orenstein, and A. R. Hinman. 1992. Epidemiology of poliomyelitis in the United States one decade after the last reported case of indigenous wild virus-associated disease. Clin. Infect. Dis. 14:568-579. [DOI] [PubMed] [Google Scholar]
- 58.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 59.Sutter, R. W., S. L. Cochi, and J. L. Melnick. 1999. Live attenuated poliovirus vaccine, p. 364-408. In S. A. Plotkin and W. A. Orenstein (ed.), Vaccines, 3rd ed. W.B. Saunders Company, Philadelphia, Pa.
- 60.Sutter, R. W., and R. Prevots. 1994. Vaccine-associated paralytic poliomyelitis among immunodeficient persons. Infect. Med. 11:426-438. [DOI] [PubMed] [Google Scholar]
- 61.Swofford, D. L., G. J. Olsen, D. J. Wadell, and D. M. Hillis. 1996. Phylogenetic inference, p. 407-514. In D. M. Hillis, C. Moritz, and B. K. Mable (ed.), Molecular systematics, 2nd ed. Sinauer Associates, Sunderland, Mass.
- 62.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 63.Technical Consulting. Group to the World Health Organization on the Global Eradication of Poliomyelitis. 2002. “Endgame” issues for the Global Polio Eradication Initiative. Clin. Infect. Dis. 34:72-77. [DOI] [PubMed] [Google Scholar]
- 64.Thorley, B., F. Paladin, and H. Shimizu. 2002. Poliomyelitis due to vaccine-derived polioviruses in the Philippines. Presented at the XIIth International Congress of Virology, Paris 27 July to 1 August 2002.
- 65.van der Avoort, H. G. A. M., B. P. Hull, T. Hovi, M. A. Pallansch, O. M. Kew, R. Crainic, D. J. Wood, M. N. Mulders, and A. M. van Loon. 1995. Comparative study of five methods of intratypic differentiation of polioviruses. J. Clin. Microbiol. 33:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Westrop, G. D., K. A. Wareham, D. M. Evans, G. Dunn, P. D. Minor, D. I. Magrath, F. Taffs, S. Marsden, M. A. Skinner, G. C. Schild, and J. W. Almond. 1989. Genetic basis of attenuation of the Sabin type 3 oral poliovirus vaccine. J. Virol. 63:1338-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.W. H. O. Collaborative Study Group on Oral and Inactivated Poliovirus Vaccines. 1997. Combined immunization of infants with oral and inactivated poliovirus vaccines: results of a randomized trial in The Gambia, Oman, and Thailand. J. Infect. Dis. 175(Suppl. 1):S215-S227. [DOI] [PubMed] [Google Scholar]
- 68.Wood, D. J., R. W. Sutter, and W. R. Dowdle. 2000. Stopping poliovirus vaccination after eradication: issues and challenges. Bull. W. H. O. 78:347-357. [PMC free article] [PubMed] [Google Scholar]
- 69.World Health Organization. 2001. Network strategy for detecting cVDPV is defined. Polio Lab Network Quarterly Update 7:1-2. [Google Scholar]
- 70.World Health Organization. 2002. Progress towards poliomyelitis eradication in Egypt, 2001. Wkly. Epidemiol. Rec. 77:70-74. [PubMed] [Google Scholar]
- 71.World Health Organization. 2003. Progress towards poliomyelitis eradication in Egypt, 2002. Wkly. Epidemiol. Rec. 78:90-94. [PubMed] [Google Scholar]
- 72.World Health Organization. 2003. Progress towards the global eradication of poliomyelitis, 2002. Wkly. Epidemiol. Rec. 78:138-144. [PubMed] [Google Scholar]
- 73.World Health Organization. 2001. Transmission of wild poliovirus type 2—apparent global interruption. Wkly. Epidemiol. Rec. 76:95-97. [PubMed] [Google Scholar]
- 74.World Health Organization. 2003. Wild poliovirus type 2—reference strains isolated in India. Wkly. Epidemiol. Rec. 78:88. [Google Scholar]
- 75.Yang, C.-F., L. De, B. P. Holloway, M. A. Pallansch, and O. M. Kew. 1991. Detection and identification of vaccine-related polioviruses by the polymerase chain reaction. Virus Res. 20:159-179. [DOI] [PubMed] [Google Scholar]
- 76.Yang, C.-F., L. De, S.-J. Yang, J. R. Gómez, J. R. Cruz, B. P. Holloway, M. A. Pallansch, and O. M. Kew. 1992. Genotype-specific in vitro amplification of sequences of the wild type 3 polioviruses from Mexico and Guatemala. Virus Res. 24:277-296. [DOI] [PubMed] [Google Scholar]
- 77.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 78.Yang, Z., and R. Nielsen. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17:32-43. [DOI] [PubMed] [Google Scholar]
- 79.Yoneyama, T., A. Hagiwara, M. Hara, and H. Shimojo. 1982. Alteration in oligonucleotide fingerprint patterns of the viral genome in poliovirus type 2 isolated from paralytic patients. Infect. Immun. 37:46-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoshida, H., H. Horie, K. Matsuura, T. Kitamura, S. Hashizume, and T. Miyamura. 2002. Prevalence of vaccine-derived polioviruses in the environment. J. Gen. Virol. 83:1107-1111. [DOI] [PubMed] [Google Scholar]