Abstract
Human immunodeficiency virus type 1 (HIV-1) contains two copies of genomic RNA that are noncovalently linked via a palindrome sequence within the dimer initiation site (DIS) stem-loop. In contrast to the current paradigm that the DIS stem or stem-loop is critical for HIV-1 infectivity, which arose from studies using T-cell lines, we demonstrate here that HIV-1 mutants with deletions in the DIS stem-loop are replication competent in peripheral blood mononuclear cells (PBMCs). The DIS mutants contained either the wild-type (5′GCGCGC3′) or an arbitrary (5′ACGCGT3′) palindrome sequence in place of the 39-nucleotide DIS stem-loop (NLCGCGCG and NLACGCGT). These DIS mutants were replication defective in SupT1 cells, concurring with the current model in which DIS mutants are replication defective in T-cell lines. All of the HIV-1 DIS mutants were replication competent in PBMCs over a 40-day infection period and had retained their respective DIS mutations at 40 days postinfection. Although the stability of the virion RNA dimer was not affected by our DIS mutations, the RNA dimers exhibited a diffuse migration profile when compared to the wild type. No defect in protein processing of the Gag and GagProPol precursor proteins was found in the DIS mutants. Our data provide direct evidence that the DIS stem-loop is dispensable for viral replication in PBMCs and that the requirement of the DIS stem-loop in HIV-1 replication is cell type dependent.
All retroviruses, including human immunodeficiency virus type 1 (HIV-1), contain two copies of virion genomic RNA (for a review, see references 6, 27, and 38). These virion dimeric RNAs are noncovalently linked at the 5′ end of the RNA genome and undergo rearrangement to form more-stable RNA dimers during the maturation of virion proteins (11, 12, 35, 45).
The packaging of the HIV-1 RNA genome is mediated by the four stem-loop structures found near the 5′ end of the RNA genome (6). These stem-loop structures are also referred to as the dimer initiation site (DIS) stem-loop, splice donor (SD) stem-loop, packaging (Ψ) stem-loop, and Gag initiation stem-loop (1). A highly conserved palindrome sequence (5′GCGCGC3′, also known as the DIS and the kissing-loop) within the DIS stem-loop is important for the formation of viral RNA dimers in vitro (20, 44). Other variants of palindrome DIS-loop sequences (5′GTGCAC3′) and (5′TGCGCA3′) have also been found in a minor proportion of HIV-1 subtypes (46). The fact that the DIS stem-loop forms part of the HIV-1 genomic RNA packaging sequence may, in part, explain the strong correlation between genomic RNA dimerization and virion RNA packaging during HIV-1 assembly (1).
The formation of a dimeric RNA genome in infectious retroviruses can be roughly divided into three different steps. These are (i) the initiation of genomic RNA dimer formation, (ii) the conformational rearrangement of the dimeric RNA (11, 12, 35, 36, 41, 45), and (iii) the stabilization of genomic RNA dimers (11, 12, 35, 36, 45). Steps ii and iii are often collectively referred to as the maturation of RNA dimers, which coincides with the proteolytic cleavage of Gag and GagProPol (11, 12, 35, 36, 45). The proteolytic processing of the primary cleavage site (p2/nucleocapsid [NC]) is particularly important for the stabilization of the dimeric virion RNA genomes (42). Overexpression of a protease (PR)-negative GagProPol in the virion-producing cells generates noninfectious HIV-1 that contains mainly monomeric RNA genomes (40, 43).
While the processing of the primary cleavage site in HIV-1 Gag is important for the stabilization of the virion RNA genome (42), proteolytic processing of virion proteins in HIV-1 Gag only particles is not sufficient to generate dimeric RNA with wild-type (wt) conformation (41). These data suggest that both reverse transcriptase (RT) and integrase (IN) are also important for the formation of wt RNA dimers and that the virion dimeric RNAs assume a number of different conformations throughout the process of virion assembly (41). These variable forms of dimeric RNAs found in HIV-1 are reminiscent of the wt mature, PR-negative immature, and wt rapid-harvest (slow migrating) dimeric RNAs found in the Moloney murine leukemia virus system (12) and the different types of RNA dimers found in in vitro RNA dimerization systems (21, 22).
Although virion RNA packaging is not critical for the production and release of retroviral particles (10, 14, 15, 31-33), it is generally held that virion RNA dimer formation occurs prior to the packaging of genomic RNA (7, 47). While the DIS stem-loop is critical for the dimerization of virion RNA in vitro (20, 44), mutations within this region have variable effects on the dimerization of genomic RNA in the virion (4, 8, 16, 23, 37). It is unclear which RNA sequence drives the initiation of genomic RNA dimerization in the virion. The role of the DIS stem-loop in HIV-1 replication in T-cell lines, such as SupT1 (4, 37), C8166 (16), and MT4 (23, 24, 26), as well as non-T-cell reporter cell lines, such as HOS cells, has been extensively evaluated (8). It is now commonly accepted that the DIS stem-loop structure or the DIS loop is vital for HIV-1 replication (48). In general, these DIS mutants are also defective in RNA packaging (4, 8, 23, 37), and it is thought that the defects of viral replication in these mutants are associated with their defects in genomic RNA packaging and/or dimerization. In addition, it has been shown that the DIS is also important in mediating the complete synthesis of viral cDNA in infected cells (37). However, the replication of HIV-1 DIS stem-loop mutants in primary cells, such as peripheral blood mononuclear cells (PBMCs), has never been examined.
In this study, we provide direct evidence that the DIS stem-loop is dispensable for HIV-1 replication in PBMCs but remains critical for viral replication in SupT1 cells. Deletions of the DIS stem altered the mobility but did not affect the stability of virion RNA dimers. No defect in the processing of HIV-1 Gag and GagProPol precursor proteins was found in these DIS mutant virions. The level of the genomic RNA packaged by the DIS mutant particles was approximately 50% that of the wild type. In addition, the DIS mutants were found to have an increased level of 4-kb singly spliced HIV-1 RNA when compared to the wild type. Our data on virion RNA packaging and dimerization are in agreement with previous reports (4, 8, 16, 23, 37), demonstrating that the DIS stem-loop is important for virion RNA packaging but is not essential for virion RNA dimer formation. We further show that the packaging of wt levels of virion RNA and the DIS stem-loop were not critical for HIV-1 replication in PBMCs.
MATERIALS AND METHODS
Construction of plasmid DNAs.
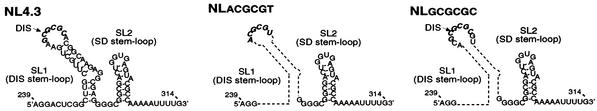
The wt HIV-1 proviral DNA NL4.3 was obtained from the NIH AIDS Reagents Program. The numbering of RNA sequences in Fig. 1 is based on the RNA genome of NL4.3. A 39-nucleotide (nt) sequence encompassing the DIS stem-loop (HIV-1 RNA residues 242 to 280) within the NL4.3 proviral DNA was replaced with the 10-nt palindrome sequence (5′ACGCGCGCGT3′) to generate NLGCGCGC that has the wt DIS sequence (Fig. 1). Similarly, NLACGCGT was constructed by replacing the same 39-nt sequence (HIV-1 RNA residues 242 to 280) within the NL4.3 proviral DNA with an arbitrarily chosen 6-nt palindrome sequence (5′ACGCGT3′) to generate NLACGCGT (Fig. 1). Both mutants were constructed via site-directed PCR mutagenesis using specific PCR primers as previously described (17, 18, 40-42). DNA sequencing was performed to confirm the presence of the desired mutations and the absence of spontaneous mutations via PCR mutagenesis.
FIG. 1.
Schematic representation of the DIS stem-loop sequences in the HIV-1 RNA genome. The numerical values are based on the RNA nucleotide position of NL4.3. The deletion sequences are indicated as dotted lines. The italic font highlights the DIS sequences or the palindrome sequences within the stem-loop. This figure is modified from a figure in the study by Berkhout (1).
Virus production.
Mutant and wt HIV-1 were produced by transfection of proviral DNA into 293T cells as previously described (30). Viral particles were isolated 36 h posttransfection. Briefly, supernatants were centrifuged for 30 min at 2,000 × g, 4°C (Beckman) to remove cellular debris. The clarified supernatants were either frozen at −70°C or used immediately for further analysis. Cells were washed twice with 1× Tris-buffered saline (TBS) (50 mM Tris [pH 7.4], 150 mM NaCl) followed by protein extraction using lysis buffer containing 1× TBS, 10 μl of Nonidet P-40 per ml, 20 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, and 1 μM leupeptin.
Infectivity assay.
PBMCs were isolated from buffy packs (supplied by the Red Cross Blood Bank, Melbourne) as described previously (9). PBMCs were then stimulated with phytohemagglutinin (PHA) (10 μg/ml; Murex Diagnostics) for 3 days and cultured in RPMI 1640 medium containing 10% fetal calf serum (RF10) and 10 units of interleukin-2 (Boehringer-Mannheim) per ml. SupT1 cells (kindly provided by Dale McPhee, The Burnet Institute, Australia) were cultured in RF10. Infectivity was tested on virus stocks obtained from either transfection or long-term infection of PBMCs (see below). Sample virus stocks with equivalent levels of RT activity, as determined by a micro-RT assay (13), were mixed with 105 PBMCs or SupT1 cells in a 96-well tissue culture plate. Eight 10-fold serial dilutions of each virus were tested in triplicate. Supernatants were collected on days 3, 7, 10, and 14 postinfection and subsequently stored at −70°C. Viral infectivity was measured by monitoring the production of viral RT activity by using a micro-RT assay (13). The infectivities of wt and mutant viral particles were quantified by using a 50% tissue culture infective dose (TCID50) method as previously described (9).
Measurement of virion RNA packaging and virion RNA dimer formation.
Pelleted wt and mutant virions derived from transfected 293T cells were normalized according to p24 levels (HIV-1 p24 antigen immunoassay; Abbott Laboratories). Virion RNAs were extracted as previously described (11, 12). Virion RNAs were electrophoretically separated on a denatured agarose gel to examine the impact of DIS mutations on virion RNA packaging (40). A dilution series of wt virion RNA was used to construct a standard curve to quantify the amount of genomic RNA being packaged into the mutant HIV-1 for the equivalent levels of virion-associated p24.
The stability and the conformation of virion RNA dimers isolated from wt and DIS mutant HIV-1 derived from both transfected 293T cells and infected PBMCs were assessed. PHA-stimulated PBMCs (2 × 107) were infected with wt and DIS mutant HIV-1 for 10 days as described below. Virion RNA was isolated, and a native RNA dimerization gel assay was utilized as previously described (11, 12). Briefly, dimeric RNAs were heated at 4, 25, 37, 42, 48, and 52°C for 10 min in an RNA dimerization buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 25 mM NaCl), immediately followed by a quick chill on ice. The dimeric and monomeric RNAs were then separated by electrophoresis (18, 40-43). The electrophoretically separated RNAs were then transferred onto nitrocellulose (Amersham) as previously described (18, 40-43). The virion RNA on the nitrocellulose was detected by use of a 150-nt in vitro-synthesized radioactive riboprobe (pGEM7zHIV-1), which is complementary to the R-U5 regions of the HIV-1 genomic RNA (NL4.3 RNA sequences nt 77 to 227) (18, 40-43).
Intracellular viral protein analysis.
Cell lysates were rapidly freeze-thawed three times to weaken the cellular membrane. Cell debris was subsequently removed by centrifugation for 30 min at 4°C, 20,000 × g. The transfection efficiency of the samples was determined by measuring the level of enhanced green fluorescent protein from the reporter plasmid by use of a Bio Imaging Analyser (Fuji Photo Film Co.). Cellular protein from each sample, normalized for equivalent levels of enhanced green fluorescent protein, was mixed with 3 μl of sample buffer (100 mM Tris [pH 6.8], 3% sodium dodecyl sulfate [SDS], 33% glycerol, and 0.03% bromophenol blue), denatured for 10 min at 95°C, and resolved by SDS-10% polyacrylamide gel electrophoresis (PAGE). Resolved proteins were transferred to a nitrocellulose membrane (Amersham). The membrane was blocked for 2 h in 3% casein dissolved in 1× TBS containing 0.3% Tween 20 (TBST) and probed overnight at 4°C with pooled HIV-1 seropositive patient sera. After three washes with 1× TBST buffer, the membrane was incubated with anti-human horseradish peroxidase-conjugated secondary antibody (DAKO) for 2 h at room temperature. An enhanced chemiluminescence technique (Amersham) was used for visualization of HIV-1 proteins present in the cellular lysates.
Virion purification and protein analysis.
Supernatants from transfected cells were purified and concentrated by ultracentrifugation through a 20% sucrose cushion by using a Beckman ultracentrifuge L-90 model (SW 41 rotor) at 100,000 × g for 1 h at 4°C. Pellets were resuspended in 50 μl of TBS lysis buffer. Equal amounts of virion protein (as determined by virion-associated p24) from each sample were mixed with 3 μl of sample buffer containing 5 mM β-mercaptoethanol, heated for 10 min at 95°C. Virion proteins were then resolved by SDS-10% PAGE as described above. The resolved virion protein samples were transferred onto nitrocellulose membranes by electrophoresis using a Bio-Rad transfer apparatus. Virion HIV-1 protein profiles of the samples were determined by Western analysis as described above.
Long-term culture and sequencing analysis of the viral genome in the infected PBMCs.
PHA-stimulated PBMCs were infected with wt and DIS mutant HIV-1 derived from transfected 293T cells. Virus stocks were treated with DNase to prevent contamination of plasmid DNA prior to the initial infection. PBMCs infected with wt and mutant HIV-1 were maintained in culture for 10 days. Cell-free culture fluids were then collected, and RT activities were compared. Cell pellets were collected and stored at −70°C for DNA sequencing. Collection of culture fluids and cell pellets at day 10 of infection marked the end of the first passage of wt and mutant HIV-1 in primary cells. A total of four passages in PBMCs were performed in parallel for wt and DIS mutant HIV-1. Equivalent amounts of viruses (as determined by the levels of RT activity) were used as input viruses in each of the four passages of PBMC infection. Similar levels of RT activities were detected in the culture fluids collected from wt and DIS mutant HIV-1-infected PBMCs within each passage. The resultant viral supernatants from each passage were tested for infectivity in SupT1 cells as described above.
DNA was extracted from infected PBMCs by incubating cell pellets with 1× DNA extraction buffer (20 mM Tris-HCl [pH 8.0], 50 mM KCl, 0.45% NP-40, 0.45% Tween 20, and 60 μg of proteinase K/ml) at 37°C for 16 h. Heating at 95°C for 10 min subsequently inactivated the proteinase K in the samples. Viral DNA fragments containing nt 37 to 517, nt 947 to 1197, and nt 1387 to 1627 (numbering corresponds to the RNA sequences) were amplified via specific PCR primers pairs containing ApaI and EcoRI sequences. The three sets of primers were DIS/MA sense (5′CCC GAA TTC CTG AGC CTG GGA GCT CTC TGG C3′) and DIS/MA antisense (5′CCC GGG CCC ACG CGT CTA GCT CCC TGC TTG CCC3′) (set 1); CA sense (5′CCC GAA TTC GAG ACC ATC AAT GAG GAA GCT GCA GAA TGG GAT3′) and CA antisense (5′CCC GGG CCC ACG CGT TTT GGT CCT TGT CTT ATG TCC AGA ATG C3′) (set 2); and p2/NC sense (5′CCC GAA TTC AGG GAG TGG GGG GAC CCG GCC ATA AAG3′) and p2/NC antisense (5′CCC GGG CCC ACG CGT AGC CTG TCT CTC AGT ACA ATC TTT C3′) (set 3). The DIS stem-loop region and the selected matrix (MA), capsid (CA), p2 and NC coding sequences were monitored because compensatory mutations have been found in these regions after long-term culturing of partial DIS stem-loop deleted mutants in MT4 cells (25, 26). PCR products were digested with the restriction enzymes ApaI and EcoRI and cloned into the pGEM7z vector for DNA sequencing. Sequencing was performed with an automating fluorescence DNA sequencer (Applied Biosystems) at the Baker Institute, Melbourne, Australia. For each passage of a given wt or mutant virus, four to five separate clones were sequenced with each primer set.
RESULTS AND DISCUSSION
The DIS stem-loop is not required for HIV-1 replication in PHA-stimulated PBMCs.
Previous work has shown that the DIS stem-loop is critical for HIV-1 replication in T-cell lines (4, 8, 16, 23, 37), but the role of DIS in HIV-1 replication in PBMCs has not been examined. Two DIS mutants (NLGCGCGC and NLACGCGT) were used in this study. NLGCGCGC has the natural palindrome DIS sequence (5′GCGCGC3′) in place of the 39-nt DIS stem-loop in NL4.3. NLACGCGT has an arbitrary palindrome sequence, 5′ACGCGT3′, in place of the 39-nt DIS stem-loop in NL4.3 (Fig. 1). Mutant and wt HIV-1 were generated by transfecting the indicated proviral DNAs into 293T cells. Parallel infections were carried out by using the T-cell line SupT1 and PHA-stimulated PBMCs. Mutant and wt HIV-1 collected from the supernatant of the transfected 293T cells were normalized for RT activity prior to infection. Equivalent amounts of wt and DIS mutant virions were used to infect both PBMCs and SupT1 cells. The replication kinetics and TCID50 of wt and DIS mutants were determined through infections with PBMCs from eight different donors and six independent SupT1 infections.
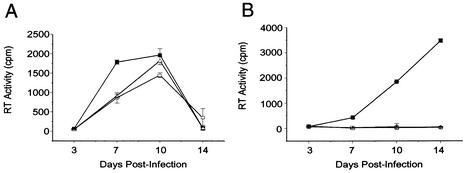
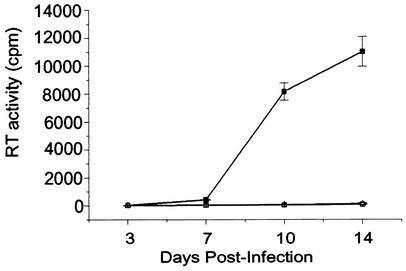
The HIV-1 DIS mutants were replication defective in SupT1 cells (Fig. 2B), which is consistent with reported data from assays using T-cell lines (37). However, NLGCGCGC and NLACGCGT were replication competent in PBMCs (Fig. 2A). The TCID50 of each virus was measured to assess the relative infectivity among wt (NL4.3) and the DIS mutants in both PBMCs and Sup T1 cells (Table 1). Our data show that for equivalent levels of RT activity the HIV-1 DIS mutants were consistently replication competent in PBMCs from eight donors (Table 1). In contrast, the parallel infection study in SupT1 cells showed that the same NLGCGCGC and NLACGCGT virus stocks were approximately 1,000 to 10,000 times less infectious than the wt HIV-1 (Table 1).
FIG. 2.
Replication kinetics of wt HIV-1 and DIS stem-loop mutants in PHA-stimulated PBMCs (A) and T-cell line Sup T1 (B). PHA-stimulated freshly isolated PBMCs or SupT1 cells were infected with either NL4.3 wt (▪) or mutant (NLGCGCGC [Δ] and NLACGCGT [□]) viruses. Supernatants were collected 3, 7, 10, and 14 days after infection, and RT activity in each sample was measured. Results represent means and standard deviations of triplicate samples and are representative of eight and six sets of experiments for PBMCs and Sup T1, respectively.
TABLE 1.
Virion infectivitya of wt and DIS mutant HIV-1
| Cells | TCID50 (10−3 cpm) of RT activity in experiment no.:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| PBMCs | ||||||||
| NL4.3 | 150,000 | 3,200 | 560,000 | 320,000 | 320 | 560 | 320,000 | 56,000 |
| NLACGCGT | 320,000 | 560 | 32,000 | 180,000 | 180 | 56 | 32,000 | 5,600 |
| NLGCGCGC | 5,600 | 56 | 32,000 | 320,000 | 32 | 56 | 460,000 | 32,000 |
| SupT1 | ||||||||
| NL4.3 | 32,000 | 3,200 | 3,200 | 1,800 | 180,000 | 320,000 | ||
| NLACGCGT | 3.2 | 0.56 | 0.56 | 3.2 | 3.2 | 0.32 | ||
| NLGCGCGC | 3.2 | 3.2 | 0.56 | 3.2 | 5.6 | 0.56 | ||
TCID50 was measured as described in Materials and Methods. cpm, counts per minute.
The DIS stem-loop is important for virion RNA packaging and the formation of discrete RNA dimers.
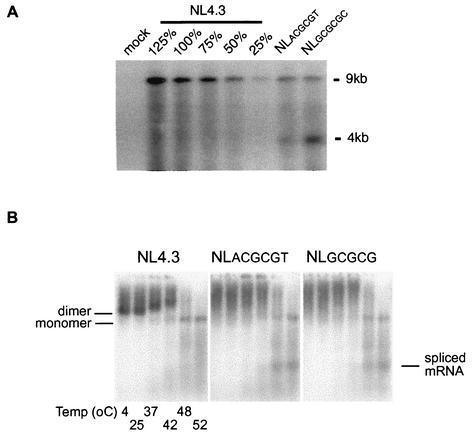
Cell-free in vitro RNA binding studies have shown that the DIS loop sequence (Fig. 1, RNA nt 257 to 262) is critical for viral RNA dimerization (20, 44). Previous reports have shown that deletions similar to those used in this study (NLGCGCGC and NLACGCGT) reduce the packaging of virion RNA and/or impair the formation of discrete RNA dimers (4, 8, 16, 23, 37). Genomic RNA packaging and dimerization analyses were carried out with our DIS mutants to verify the demonstrated impact of DIS mutations on virion RNA genomes. The virion packaging of genomic RNA in these DIS mutants was reduced to approximately 50% of that of the wt normalized for the same amount of p24 proteins (Fig. 3A). While the amount of virion genomic RNA packaged for both of the DIS mutants was reduced, there was an increased level of 4-kb viral RNA compared to that of the wild type (Fig. 3A). One possibility for this phenomenon is that RNA packaging-deficient virus-like particles may nonspecifically incorporate excess singly spliced viral RNA and/or cellular RNA to compensate for the reduction of genomic RNA packaging during retroviral assembly (28, 34). These data also highlighted that reduction of genomic RNA packaging by 50% compared to the wt was sufficient for the replication of HIV-1 DIS mutants in primary cells.
FIG. 3.
(A) Mutations in DIS stem-loop inhibit virion packaging of genomic RNA. Virion particles were produced by transfecting the indicated proviral DNA into 293T cells. Virion RNA samples that were normalized via a quantitative p24 assay were separated by electrophoresis on a denatured agarose gel. A dilution series of wt virion RNA was used to construct a standard curve. The impact of mutations on virion RNA packaging was visualized by Northern analysis and quantified by phosphorimaging. Results are representative of three sets of experiments. (B) The conformation but not the stability of HIV-1 virion RNA dimers from transfected 293T cells was affected by mutations of the DIS stem-loop. Virion RNA dimers were isolated from transfected 293T cells and prepared as described in Materials and Methods. Viral monomeric and dimeric RNA species were separated on a 1% native agarose gel via electrophoresis after the samples were heated for 10 min at various temperatures (4, 25, 37, 42, 48, and 52°C, lanes 1 to 6, respectively). Viral RNAs were visualized by Northern analysis using a radioactive riboprobe that specifically recognizes HIV-1 RNA. Results are representative of five sets of experiments.
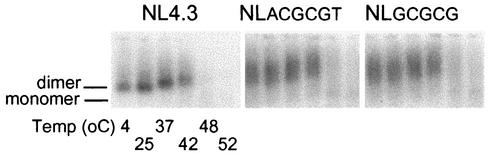
RNA dimerization analysis of wt and DIS mutant RNA derived from transfected 293T cells demonstrated that while the formation of discrete RNA dimers was impaired in each of the DIS mutants, the stability of the virion RNA dimers, as determined by the first appearance of the monomeric RNA band after heating at 42°C, was not affected (Fig. 3B). Increased levels of spliced mRNA are visible in each of the DIS mutants compared to the levels observed with the wt when heated to 48 or 52°C, which is consistent with the increased level of 4-kb viral RNA observed for the DIS mutants in the RNA packaging analysis using a denaturing RNA gel. These results suggest that the diffuse RNA dimer may, at least in part, be a consequence of the spliced RNA that is packaged within the DIS mutant virions. To determine whether the conformation of the DIS mutant RNA dimers was rescued following successful infection of PBMCs, RNA dimerization analysis was carried out with virion RNA isolated after wt and DIS mutants had been used to infect PBMCs for 10 days (Fig. 4). Again, the formation of discrete RNA dimers was impaired in each of the DIS mutants while the stability of the virion RNA dimers was not altered. The finding that the conformation but not the stability of the RNA dimers was affected by DIS mutations is in agreement with the hypothesis that other non-DIS stem-loop RNA sequences are involved in the process of HIV-1 RNA dimer formation (1, 4), and retroviral RNA dimerization may rely on multiple segments of the retroviral RNA genomes (29, 36, 39).
FIG. 4.
The conformation but not the stability of HIV-1 virion RNA dimers isolated from PBMCs was affected by mutations of the DIS stem-loop. Virion RNA dimers were isolated from infected PBMCs and prepared as described in Materials and Methods. Viral monomeric and dimeric RNA species were separated on a 1% native agarose gel by electrophoresis after the samples had been heated for 10 min at various temperatures (4, 25, 37, 42, 48, and 52°C, lanes 1 to 6, respectively). Viral RNAs were visualized by Northern analysis with a radioactive riboprobe that specifically recognizes HIV-1 RNA. Results are representative of three sets of experiments.
Mutations in the HIV-1 DIS stem-loop structure do not alter the processing of viral proteins or yield compensatory mutations in p2 and NC sequences in the viral genome of the HIV-1-infected PBMCs.
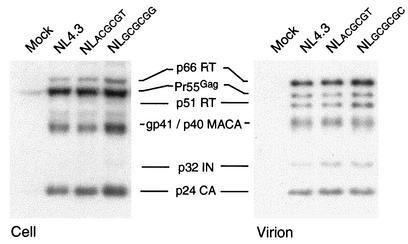
It has been reported that the DIS deletion is associated with defects in precursor protein processing and this processing defect can be rescued by compensatory mutations in p2 and NC sequences (25). Furthermore, compensatory mutations in the matrix and capsid can also rescue the defects of viral infectivity in DIS deletion mutants (26). Western blot analysis of intracellular viral protein (Fig. 5) and pelleted virion proteins from 293T cells transfected with the various DIS mutants showed that the virion protein profiles were similar to those of wt virus (Fig. 5), suggesting that our DIS mutations did not influence the processing of viral proteins.
FIG. 5.
Mutations in the HIV-1 DIS stem-loop do not alter the intracellular viral protein and virion protein profile. Intracellular (293T cells) and virion proteins were resolved by SDS-10% PAGE. The viral proteins were visualized by Western blotting using pooled HIV-1-positive patient sera and anti-human horseradish peroxidase-conjugated secondary antibody. Results are representative of five sets of experiments.
DNA sequencing was performed for wt- and DIS mutant (NLGCGCGC and NLACGCGT)-infected PBMC cultures 40 days postinfection in three discrete experiments. Four to five clones were sequenced for each sample, and in all cases the sequences of the clones were identical. Previous studies have reported that compensatory mutations are required to rescue the infectivity of DIS-deleted HIV-1 mutants in T cells (25, 26); no such mutation was found in the corresponding MA, CA, p2, and NC sequences in the viral genome of NLGCGCGC- and NLACGCGT-infected primary cells. The respective DIS deletions were maintained in the NLGCGCGC- and NLACGCGT-infected PBMC cultures 40 days postinfection (data not shown). DNA sequencing (as described in Materials and Methods) revealed no changes to the DNA sequence when compared to the input viruses. To rule out the possibility of contamination with a replication-competent virus in the NLGCGCGC- and NLACGCGT-infected PBMC cultures, replication kinetics were determined for SupT1 cells by using supernatant collected from each passage of two sets of the long-term infections (eight in total). Although replication competent in PBMCs, viruses present in the supernatants of NLGCGCGC- and NLACGCGT-infected PBMC cultures were replication defective in SupT1 cells (Fig. 6), which indicates that there was no contamination. These results correspond with the DNA sequencing data showing that the sequences of the NLGCGCGC and NLACGCGT input viruses were unaltered after 40 days in PBMCs. Consequently, our data show that there is no strong selective pressure for a wt DIS stem-loop sequence to support HIV-1 replication in PBMCs.
FIG. 6.
Replication kinetics of wt HIV-1 and DIS stem-loop mutants passaged through PBMCs in Sup T1 cells. SupT1 cells were infected with either NL4.3 wt (▪) or mutant viruses (NLGCGCGC [Δ] and NLACGCGT [□]) that had been passaged through PBMCs. Supernatants were collected 3, 7, 10, and 14 days after infection, and RT activity in each sample was measured. Results represent means and standard deviations of triplicate samples and are representative of eight sets of experiments.
In this study, we have demonstrated that the requirement of the HIV-1 DIS stem-loop in virus replication is cell type dependent. The finding that the DIS deletion mutants cannot replicate in SupT1 cells while the DIS is largely dispensable for replication in PBMCs suggests the involvement of a DIS-dependent cellular factor. Our data also demonstrate that wt levels of genomic RNA packaging are not critical for HIV-1 replication in primary cells.
Paillart et al. (37) have shown that the DIS stem-loop is important for the synthesis of cDNA during reverse transcription in the T-cell line SupT1. The cell type-dependent effects of DIS deletions on HIV-1 replication suggest that this DIS-dependent cellular factor may directly or indirectly bind to the DIS stem-loop to enhance HIV-1 replication, perhaps by assisting in the synthesis of cDNA at the early stage of the HIV-1 replication cycle. However, since the DIS stem-loop is part of the complex RNA structure in the 5′ untranslated region, which is important at multiple stages of HIV-1 replication, the impact of DIS mutations on other aspects of HIV-1 replication, such as RNA splicing and the regulation of protein translation, should also be considered.
Using an in vitro system, Berkhout et al. (3, 5) and Huthoff and Berkhout (19) have shown that the HIV-1 5′ leader RNA sequences assume different conformations at various stages of viral replication. It has been suggested that the conformation of the 5′ leader RNA may play a part in regulating viral replication (2). Fu et al. have previously shown that the HIV-1 dimeric virion RNA genome assumes two distinct conformations before and after virion particle maturation (11). Distinct conformations of RNA dimers can also be found in other retroviruses (12, 35, 36, 45). We have recently shown that in addition to these two conformations of RNA dimers, HIV-1 virion RNA can assume a number of different dimeric conformations depending on the presence or the absence of HIV-1 PR, RT, and IN (41). These data support the notion that the reverse-transcription and dimerization reactions may be coupled through conformational changes within the leader RNA (2). Our data also support the suggestion that in addition to the ascribed roles of genomic RNA packaging and the formation of discrete RNA dimers, the DIS stem-loop is also involved in other aspects of HIV-1 replication.
Acknowledgments
We thank John Mills for helpful criticism and review of the manuscript.
J. Mak is a recipient of an NHMRC research grant and a Monash Logan fellowship. M. K. Hill is a recipient of a Burnet Centenary postdoctoral fellowship and an amfAR postdoctoral fellowship. M. Shehu-Xhilaga was a recipient of the NHMRC Ph.D. training scholarship and is a recipient of the NHMRC postdoctoral fellowship. S. M. Campbell is a recipient of the NHMRC Ph.D. training scholarship. P. Poumbourios is supported by NHMRC. S. M. Crowe is supported by a grant from the Australian Council on HIV, AIDS and Related Diseases through the (Australian) National Centre in HIV Virology Research and the BI Research Fund. This work is also supported in part by grants to J. Mak from the Clive and Vera Ramaciotti Foundation, the Honda Foundation, and the Cecilia Kilkeary Foundation.
REFERENCES
- 1.Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54:1-34. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout, B., M. Ooms, N. Beerens, H. Huthoff, E. Southern, and K. Verhoef. 2002. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem. 277:19967-19975. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout, B., and J. L. B. Van Wamel. 2000. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA 6:282-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout, B., and J. L. B. van Wamel. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkhout, B., N. L. Vastenhouw, B. I. Klasens, and H. Huthoff. 2001. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA 7:1097-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging, p. 177-218. In H.-G. Kräusslich (ed.), Current topics in microbiology and immunology: morphogenesis and maturation of retroviruses, vol. 214. Springer, Heidelberg, Germany. [DOI] [PubMed]
- 7.Cheung, K. S., R. E. Smith, M. P. Stone, and W. K. Joklik. 1972. Comparison of immature (rapid harvest) and mature Rous sarcoma virus particles. Virology 50:851-864. [DOI] [PubMed] [Google Scholar]
- 8.Clever, J. L., and T. G. Parslow. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowe, S. M., N. J. Vardaxis, S. J. Kent, A. L. Maerz, M. J. Hewish, M. S. McGrath, and J. Mills. 1994. HIV infection of monocyte-derived macrophages in vitro reduces phagocytosis of Candida albicans. J. Leukoc. Biol. 56:318-327. [DOI] [PubMed] [Google Scholar]
- 10.Dupraz, P., S. Oertle, C. Méric, P. Damay, and P.-F. Spahr. 1990. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J. Virol. 64:4978-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goff, S., P. Traktman, and D. Baltimore. 1981. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J. Virol. 1:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorelick, R. J., J. S. M. Nigida, J. J. W. Bess, L. O. Arthur, L. E. Henderson, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haddrick, M., A. L. Lear, A. J. Cann, and S. Heaphy. 1996. Evidence that a kissing loop structure facilitates genomic RNA dimerisation in HIV-1. J. Mol. Biol. 259:58-68. [DOI] [PubMed] [Google Scholar]
- 17.Hill, M. K., C. W. Hooker, D. Harrich, S. M. Crowe, and J. Mak. 2001. Gag-Pol supplied in trans is efficiently packaged and supports viral function in human immunodeficiency virus type 1. J. Virol. 75:6835-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill, M. K., M. Shehu-Xhilaga, S. M. Crowe, and J. Mak. 2002. Proline residues within the spacer peptide p1 are important for HIV-1 infectivity, protein processing, and genomic RNA dimer stability. J. Virol. 76:11245-11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huthoff, H., and B. Berkhout. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laughrea, M., and L. Jette. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464-13474. [DOI] [PubMed] [Google Scholar]
- 21.Laughrea, M., and L. Jette. 1997. HIV-1 genome dimerization: kissing-loop hairpin dictates whether nucleotides downstream of the 5′ splice junction contribute to loose and tight dimerization of human immunodeficiency virus RNA. Biochemistry 36:9501-9508. [DOI] [PubMed] [Google Scholar]
- 22.Laughrea, M., and L. Jetté. 1996. Kissing-loop model of HIV-1 genomic dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimeric formation. Biochemistry 35:1589-1598. [DOI] [PubMed] [Google Scholar]
- 23.Laughrea, M., L. Jette, J. Mak, L. Kleiman, C. Liang, and M. A. Wainberg. 1997. Mutations in the kissing loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J. Virol. 71:3397-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, C., L. Rong, E. Cherry, L. Kleiman, M. Laughrea, and M. A. Wainberg. 1999. Deletion mutagenesis within the dimerization initiation site of human immunodeficiency virus type 1 results in delayed processing of the p2 peptide from precursor proteins. J. Virol. 73:6147-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang, C., L. Rong, M. Laughrea, L. Kleiman, and M. A. Wainberg. 1998. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initiation site can restore viral replication. J. Virol. 72:6629-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, C., L. Rong, Y. Quan, M. Laughrea, L. Kleiman, and M. A. Wainberg. 1999. Mutations within four distinct Gag proteins are required to restore replication of human immunodeficiency virus type 1 after deletion mutagenesis within the dimerization initiation site. J. Virol. 73:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linial, M. L., and A. D. Miller. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. Immunol. 157:125-152. [DOI] [PubMed] [Google Scholar]
- 28.Luban, J., and S. P. Goff. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 68:3784-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ly, H., and T. G. Parslow. 2002. Bipartite signal for genomic RNA dimerization in Moloney murine leukemia virus. J. Virol. 76:3135-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak, J., M. Jiang, M. A. Wainberg, M.-L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Méric, C., and S. P. Goff. 1989. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J. Virol. 63:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Méric, C., E. Gouilloud, and P. F. Spahr. 1988. Mutations in Rous sarcoma virus nucleocapsid protein p12(NC): deletions of Cys-His boxes. J. Virol. 62:3328-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Méric, C., and P. F. Spahr. 1986. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J. Virol. 60:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retroviral particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oertle, S., and P. F. Spahr. 1990. Role of the gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J. Virol. 64:5757-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortiz-Conde, B. A., and S. H. Hughes. 1999. Studies of the genomic RNA of leukosis viruses: implications for RNA dimerization. J. Virol. 73:7165-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paillart, J. C., L. Berthoux, M. Ottmann, J. L. Darlix, R. Marquet, B. Ehresmann, and C. Ehresmann. 1996. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 70:8348-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rein, A. 1994. Retroviral RNA packaging: a review. Arch. Virol. Suppl. 9:513-522. [DOI] [PubMed] [Google Scholar]
- 39.Sakuragi, J.-I., and A. T. Panganiban. 1997. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vitro. J. Virol. 71:3250-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shehu-Xhilaga, M., S. M. Crowe, and J. Mak. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shehu-Xhilaga, M., M. K. Hill, J. Marshall, J. Kappes, S. M. Crowe, and J. Mak. 2002. The conformation of the mature dimeric human immunodeficiency virus type 1 RNA genome requires packaging of Pol protein. J. Virol. 76:4331-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shehu-Xhilaga, M., H. G. Kraeusslich, S. Pettit, R. Swanstrom, J. Y. Lee, J. A. Marshall, S. M. Crowe, and J. Mak. 2001. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type1 RNA dimer maturation. J. Virol. 75:9156-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shehu-Xhilaga, M., J.-Y. Lee, S. M. Campbell, J. A. Marshall, S. M. Crowe, and J. Mak. 2002. Overexpression and incorporation of GagPol precursor does not impede packaging of HIV-1 tRNA(Lys3) but promotes intracellular budding of virus-like particles. J. Biomed. Sci. 9:697-705. [DOI] [PubMed] [Google Scholar]
- 44.Skripkin, E., J.-C. Paillart, R. Marquet, B. Ehresmann, and C. Ehresmann. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization. Proc. Natl. Acad. Sci. USA 91:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart, L., G. Schatz, and V. M. Vogt. 1990. Properties of avian retrovirus particles defective in viral protease. J. Virol. 64:5076-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.St. Louis, D. C., D. Gotte, E. Sanders-Buell, D. W. Ritchey, M. O. Salminen, J. K. Carr, and F. E. McCutchan. 1998. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J. Virol. 72:3991-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoltzfus, C. M., and P. N. Snyder. 1975. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J. Virol. 64:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanstrom, R., and J. W. Wills. 1997. Retroviral gene expression. II. Synthesis, processing, and assembly of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.