Abstract
Reiterated terminal sequences of Epstein-Barr virus (EBV) DNA are numerically heterogeneous among infectious virions, providing a viral measure of clonality in infected cells. After in vitro infection, carcinoma cells bearing EBV episomes with fewer terminal repeats (TRs) proliferated faster. In single-cell clones, TR number varied inversely to the quantity of latent membrane protein 2A (LMP2A) transcripts whose unspliced precursors cross joined TRs. Thus, EBV clonality may reflect selection for a TR number that optimizes LMP2A-enhanced tumor progression, with infection occurring after epithelial cell transformation.
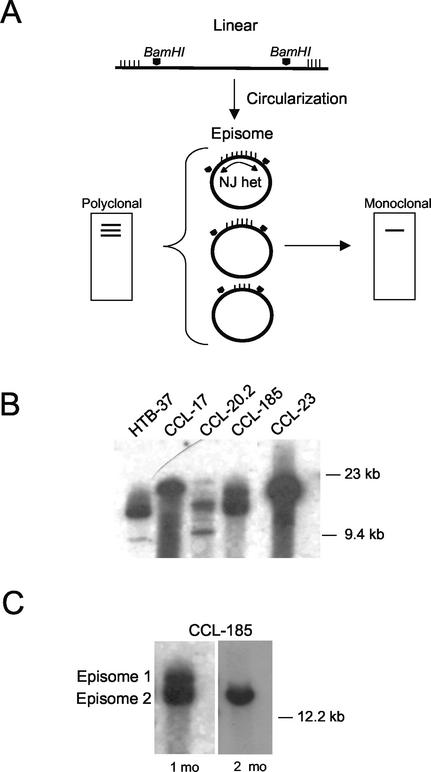
Epstein-Barr virus (EBV) has been linked to numerous human tumors of diverse tissue origin: nasopharyngeal carcinoma, Burkitt's lymphoma, Hodgkin's lymphoma, gastric carcinoma, T-cell lymphomas, leiomyosarcoma, and breast cancer (24). Although the association of EBV with cancers such as gastric carcinoma (∼15%), Hodgkin's lymphoma (40 to 60%), and sporadic Burkitt's lymphoma (∼30%) is less than complete, assignment of a causal role to EBV has been based in part on the perception that these diseases are clonal expansions of a single EBV-infected cell (2, 23). Upon entry, linear EBV DNA circularizes by homologous recombination of its terminal repeats (TRs), producing fused termini of unique length for each circularization event (13). Unlike the length heterogeneity found with a mixed-cell population at primary infection, fused TRs with an identical number of repeat units denote expansions of a single infected progenitor (Fig. 1A). Detection of a single form of EBV in all tumor cells indicates that cellular proliferation occurred after EBV infection and argues for an etiologic role for virus in these tumors (20, 23).
FIG. 1.
Evolution of EBV clonality after in vitro infection of carcinoma cells. (A) Schematic of analysis of EBV termini (23). Circularization of the linear genome by random homologous recombination of TRs yields cells containing episomes of varying TR number (vertical hatch marks). BamHI-restricted total cellular DNA, when hybridized on Southern blot to the NJ het fragment of EBV DNA, yields multiple high-molecular-weight bands representing fused repeats (polyclonal); a single high-molecular-weight band indicates clonal expansion of a single cell (monoclonal). Pentagonal arrowheads indicate BamHI restriction sites. (B) Analysis of EBV termini at passage 2 of human carcinoma cell lines infected with EBV Neor under G418 selection. (C) Episomal loss from CCL-185 cells after 2 months in culture.
What is less evident from clonality data is whether EBV infection facilitates tumor initiation or subsequent malignant progression (22, 32), an issue central to understanding viral pathogenesis. The fact that EBV is present prior to clonal expansion is intuitive evidence for a viral role in an early, initiating event. Indeed, it has been argued that if EBV were to infect preexisting neoplastic lesions, one would expect either a polyclonal pattern of TRs or the lack of EBV altogether in some tumor cells. Here, we present evidence from carcinoma cell lines infected in vitro that suggests that this need not be the case. Instead, EBV clonality postinfection may be a consequence of a selective growth advantage conferred by viral episomes with a minimal number of TRs.
Our experiments stemmed from the unexpected observation, also made by others, that in vitro infection of established epithelial cell lines frequently gives rise to a clonal pattern by EBV TR analysis (3, 15). To determine the basis for this outcome, we infected established human carcinoma cell lines with a recombinant EBV whose expression of the neomycin-resistance gene would force retention of all viral genomes in cells maintained under antibiotic pressure. EBV Neor (a gift of L. Hutt-Fletcher, University of Missouri—Kansas City) was generated by insertion of a neomycin resistance cassette in the BDLF3 open reading frame of the EBV strain Akata (1). Viral stocks were produced by treatment of host cells with anti-human immunoglobulin G (1, 30). Eight carcinoma cell lines (HTB-37 [colon], CCL-17 [oral mucosa], CCL-20.2 [conjunctiva], CCL-185 [lung], CCL-23 [laryngeal], CCL-2 [cervix], HTB-35 [cervix], and HT-29 [gastric]; American Type Culture Collection, Rockville, Md.), which were determined not to express the B-lymphocyte EBV receptor CD21 (data not shown), were incubated for 24 h with 0.5 ml of Akata culture supernatant containing EBV Neor and then resuspended in Dulbecco's modified Eagle medium containing 350 μg of G418/ml. When G418-resistant cells became confluent, they were examined by Southern blot analysis for the configuration of EBV termini by using the BamHI NJ het fragment of EBV DNA as the probe as described previously (23) (Fig. 1A). Five of the eight cell lines were successfully infected and contained EBV episomes, corroborating similar evidence from other laboratories indicating an as-yet-unknown epithelial cell receptor (11). When forced under antibiotic pressure to retain virus, infected cell lines demonstrated a polyclonal pattern at early passage (Fig. 1B). However, infections that were initially polyclonal rapidly evolved to predominance by single episomes over time in culture (Fig. 1C).
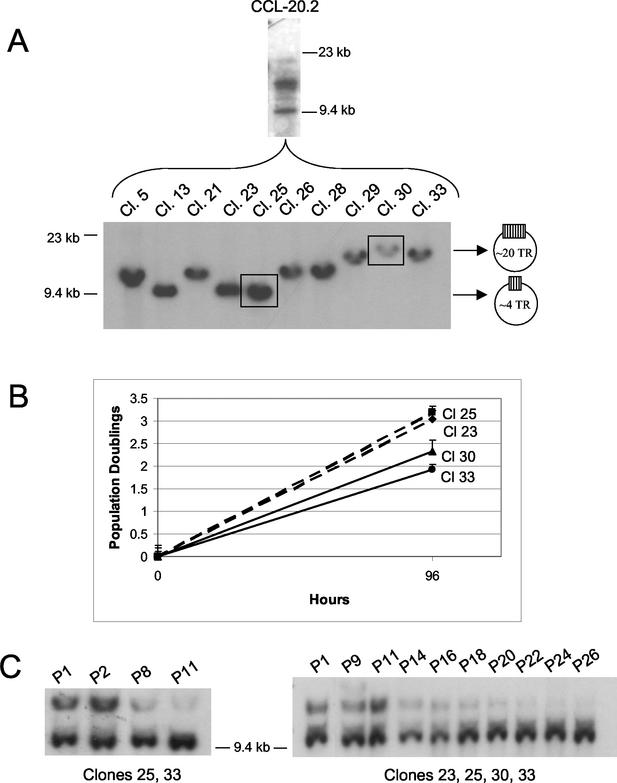
Because episomes with minimal TRs seemed to prevail in bulk culture (Fig. 1C), we examined whether the length of joined termini correlated with the host cell population doubling time. First, single-cell clones of the infected CCL-20.2 epithelial cell line were derived by cell sorting (FACSVantage SE; Becton Dickinson, San Jose, Calif.) and the relative size of joined EBV DNA ends was determined by analysis of EBV termini. Unlike the infected parental cell line that was polyclonal at early passage by virtue of the distinct TR size attributable to each cell, resultant cell clones contained uniform episomal populations with TRs that ranged in size from approximately 4 to 20 repeat units (Fig. 2A). When seeded at 8 × 103 cells/ml in the presence of G418, two separate clones with the fewest repeats (clones 23 and 25) had an average population doubling time of 31 h, while clones with the most repeats (clones 30 and 33) doubled in 46 h (Fig. 2B). By comparison, individual CCL-20.2 clones not infected with virus did not differ in doubling times, ruling out clonal variation as an explanation for differences in growth rates (data not shown). Moreover, when equivalent cell numbers from infected clones 23, 25, 30, and 33 were combined in mixed culture, clones containing episomes with the fewest TRs emerged as the predominant cell population within as few as eight passages (Fig. 2C).
FIG. 2.
TR number in relation to epithelial cell proliferation and EBV clonal emergence. (A) Polyclonality of CCL-20.2 EBV Neor as shown by EBV termini analysis of bulk culture (CCL-20.2) and constituent single-cell clones (Cl). (B) Population doublings of CCL-20.2 EBV Neor single-cell clones bearing episomes with the most (clones 30 and 33) versus those with the least (clones 23 and 25) TRs. Population doublings were determined in the presence of G418 prior to cell confluence (at 96 h) in three separate experiments by using the equation (Nt96/Nt0)/2, where N is the number of cells. (C) Episomal dominance in recombined clonal populations with time in culture. Single-cell clones of CCL-20.2 EBV Neor, containing episomes of divergent TR unit length, were mixed in equal numbers and passaged at a 1:5 split every 4 days under G418 selection. Shown at left is a Southern blot of BamHI-restricted cellular DNA from a mixed culture of clones 25 and 33 sampled over a 6-week period and probed with the NJ het fragment of EBV DNA; shown at right is a mixed culture of clones 23, 25, 30, and 33 over 13 weeks of growth. P, passage.
Malignancies associated with EBV are often grouped according to which of nine EBV latency proteins are expressed in tumor tissue (24). The restricted expression patterns in nasopharyngeal carcinoma, gastric carcinoma, and Hodgkin's lymphoma typically include one or both latent membrane proteins, LMP1 and LMP2A, each of which affects epithelial cell growth and differentiation pathways (4, 7, 29, 31). With either protein, TR number has at least the potential to affect levels of expression. For example, LMP2A is encoded by a spliced gene with exons located at either end of the viral genome (Fig. 3A). Consequently, its open reading frame is created only after circularization of the linear DNA molecule following viral entry into cells (14, 28). Transcription proceeds across fused TRs, whose 500-bp units may vary by as many as 15 to 20 reiterations in infected lymphocyte clones from a single individual (2). In the case of LMP1, there is a transcriptional enhancer located in the viral origin of plasmid replication, oriP, that acts across the TRs to increase expression of LMP1 (9). Moreover, the activity of an LMP1 TATA-less promoter located in the first TR and functional in epithelial cells might be influenced by TR number (27).
FIG. 3.
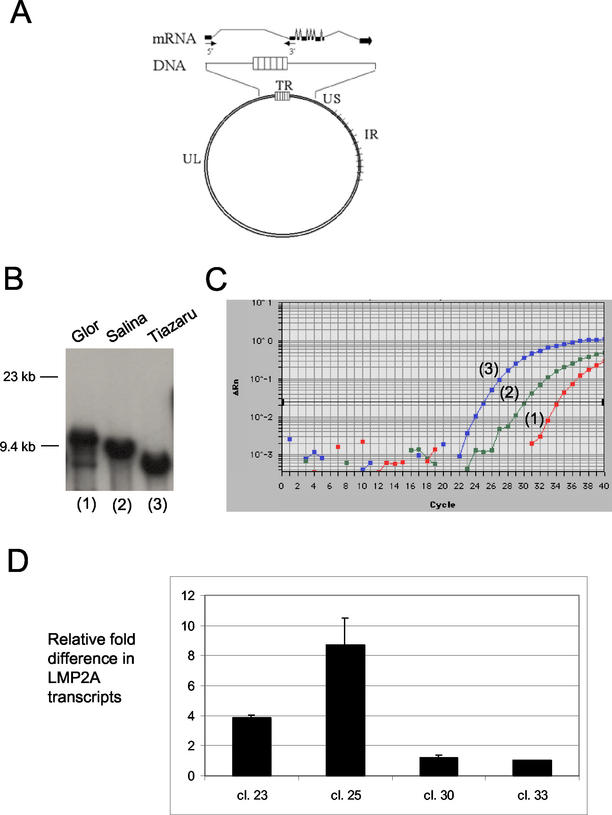
Length of fused TRs and LMP2A expression levels. (A) Schematic of LMP2A exons (black boxes) in relation to intervening TRs (14, 28). Arrows labeled 5′ and 3′ represent the forward and reverse PCR primers, respectively, used to quantify LMP2A gene expression. UL, unique long DNA sequences; US, unique short; IR, internal repeats. (B) Fused TR in three Burkitt's lymphoma cell lines shown by Southern blot analysis, ranging from ∼14 reiterations in Glor to ∼2 in Tiazaru (estimates take into account 7.5 kb of unique DNA flanking the 500-bp repeats in theBamHI restriction fragment). (C) Real-time RT-PCR amplification profile of LMP2A mRNA in the cell lines in panel B (labeled 1 to 3), with RT-negative and EBV-negative control reactions falling below threshold levels. Data analysis was performed using ABI Prism 7700 sequence detection system software (PE Applied Biosystems), with cycle threshold (CT) values being determined by automated threshold analysis. The fluorescence intensity (ΔRn) collected in real time for each sample was plotted against the number of PCR cycles. The black horizontal line represents the threshold setting, set at 10 standard deviations above the baseline. The CT value is defined as the fractional cycle number in which the fluorescence generated within a reaction has crossed the threshold. This value indicates that a sufficient number of amplicons have accumulated to be at a statistically significant point above the baseline. CT values inversely correlate with target gene expression. The comparative CT method (ΔΔCT), with the GAPDH gene expression as the cellular RNA control, was used to assess relative differences in LMP2A transcript levels between single-cell clones according to the manufacturer's protocol. (D) Relative differences (n-fold) in LMP2A mRNA levels in CCL-20.2 epithelial cell clones. Shown are average values from two separate experiments in which samples were run in duplicate as in panel C and transcript levels were normalized to EBV copy number as determined by real-time PCR for the BamHI K region of EBV DNA (10). Clones 23 and 25 contain EBV episomes with the least TRs; clones 30 and 33 have the most TRs.
Because levels of LMP2A have been shown to differ significantly between B-cell lines (21), we first quantified by reverse transcriptase (RT) PCR the LMP2A mRNA levels in three endemic Burkitt's lymphoma lines (Glor, Salina, and Tiazaru; gift of A. B. Rickinson, University of Birmingham, Birmingham, United Kingdom) containing episomes with diminishing TR number as shown by analysis of EBV termini (Fig. 3B). In brief, for cDNA synthesis, 5 μg of total RNA was reverse transcribed with Moloney murine leukemia virus RT (Gibco BRL, Rockville, Md.) by using random hexamer primers. LMP2A-specific primers were selected (Primer Express 1.5; PE Applied Biosystems, Foster City, Calif.) such that the forward primer spanned the boundary between the first and second exons (5′-AGCGGGCAGAGGAAG-3′, base coordinate 166444; National Center for Biotechnology Information GenBank accession no. AJ507799) and the reverse primer was located in exon 2 (5′-AAGAGGTAGGGCGCAACAATT-3′, base coordinate 102). The TaqMan fluorogenic system (PE Applied Biosystems), in which real-time amplification was measured by cleavage of a fluorescent dye-labeled LMP2A-specific probe (5′ FAM-TCCAGTATGCCTGCCTG-TAMRA 3′, base coordinate 64) by the 5′-to-3′ exonuclease activity of Taq DNA polymerase, was used as described elsewhere (10). Reactions were performed in a 50-μl volume with 50 to 500 ng of cDNA by using TaqMan Universal master mix (PE Applied Biosystems) and universal thermocycler conditions according to the manufacturer's instructions (ABI Prism 7700 sequence detection system; PE Applied Biosystems). Template-negative and RT-negative reactions served as controls. All samples were run in duplicate, together with reactions to quantify expression of an internal control gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (PE Applied Biosystems), to normalize for any differences in the amount of total RNA added.
By quantitative RT-PCR, LMP2A mRNA levels were found to vary inversely to TR number in Burkitt's lymphoma lines derived from three individuals (Fig. 3C). The differences in LMP2A mRNA (n-fold), normalized to GAPDH, were 1, 39, and 247 for the Glor, Salina, and Tiazaru cell lines, respectively. No product was obtained in the RT-negative or template-negative controls. To ensure that levels of transcripts reflected intervening TR length, not disparate EBV genome equivalents within cell lines, viral copy number was determined by real-time PCR of the BamHI K region of EBV DNA as previously described (10). Transcript levels normalized to EBV DNA copy number did not substantially alter these results (data not shown).
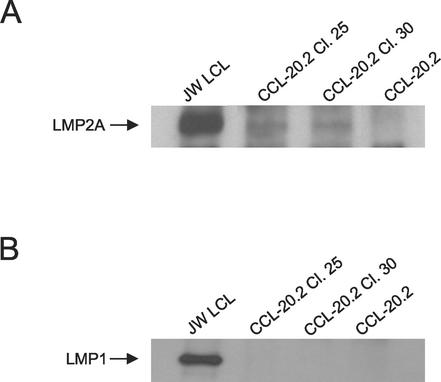
Unlike the monoclonal Burkitt's lymphoma lines originating from different hosts, single-cell clones derived from the experimentally infected epithelial cell line CCL20.2 (Fig. 2A) afforded a common cell background in which to measure the relation of length of joined termini to LMP2A transcript level. In epithelial cell clones, a similar inverse relationship was observed wherein increased size of joined termini corresponded with lower levels of LMP2A mRNA (Fig. 3D). Higher levels of LMP2A mRNA correlated precisely with accelerated rates of epithelial cell proliferation (Fig. 2B). In addition, immunoprecipitation of LMP2A protein from the two epithelial cell clones that exhibited an eightfold difference in LMP2A mRNA levels revealed an approximately twofold difference in LMP2A protein by densitometric analysis (Fig. 4A). Repeat immunoprecipitation of the spent substrates was negative, indicating that all LMP2A protein was blotted in the original analysis.
FIG. 4.
Latent membrane protein expression in epithelial clones containing EBV episomes of divergent TR number. (A) Immunoblot of LMP2A protein expression in infected CCL-20.2 single-cell clones with the most (clone 30) and the least (clone 25) TRs. Cells were lysed with Triton X-100 radioimmunoprecipitation assay buffer, and immunoprecipitation was performed on 1 mg of protein from each epithelial cell sample (100 μg of protein from the lymphoblastoid cell control) with LMP2A-specific monoclonal antibody 14B7 (gift of E. Kremmer, Institute for Immunology, Munich, Germany) and protein A-protein G agarose beads (Calbiochem, La Jolla, Calif.) (8). Protein was transferred to Immobilon-P membranes (Millipore, Bedford, Mass.) after electrophoresis, incubated with 14B7, and visualized by enhanced chemiluminescence (Amersham, Piscataway, N.J.). JW LCL, lymphoblastoid cell line positive control; CCL-20.2, uninfected negative control. (B) Immunoblot of LMP1 protein in CCL-20.2 single-cell clones. Whole-cell lysates were immunoprecipitated as described for panel A with the LMP1-specific monoclonal antibody S-12 (17), and immunoblotting was performed with the CS1-4 LMP1 monoclonal antibody (gift of L. S. Young, University of Birmingham) (26).
In contrast to LMP2A mRNA, LMP1 mRNA was not detected in any of the epithelial cell clones by RT-PCR with primers that would amplify transcripts initiated at both LMP1 promoters (5′-AGACAAGTAAGCACCCGAAGAT-3′, base coordinate 168505; 5′-GCCCTTTGTATACTCCTACTGATG-3′, base coordinate 168773; and probe 5′ FAM-CCCTCCTGCTCATCGCTCCTGGA-TAMRA 3′) (data not shown). Consistent with the absence of transcripts, LMP1 protein was also not detected by immunoblotting (Fig. 4B).
Although the inverse correlation between LMP2A transcription and TR unit length held for both lymphoid and epithelial cell types, the actual biologic impact in epithelial cells may be quite different. The ability of LMP2A to induce epithelial cell transformation contrasts with its effect in B cells, where it is thought to contribute to maintenance of viral latency but not to be required for cell transformation (16, 18, 29). The mechanism by which TR unit length influences LMP2A transcript level is yet to be defined. One possibility is that CpG methylation, evident within the GC-rich TRs, inhibits RNA transcript elongation, as has been shown previously for 3′ structural regions of genes including the herpes simplex thymidine kinase gene (5, 12, 25).
Our results, demonstrating clonal emergence of epithelial cells bearing episomes with minimal TRs, do not infer that all EBV-associated carcinomas contain episomes of a uniformly sparse TR number. Indeed, the sizes of fused EBV termini vary considerably between individual tumors (20, 23). Instead, these data support the notion that nascent tumors infected early after clonal expansion, though initially polyclonal with respect to EBV episomal TR number, rapidly select for malignant subclones bearing episomes with comparatively few TRs relative to the range within that tumor.
Infection of preexisting neoplasia by endogenous EBV runs contrary to the current view of EBV tumorigenesis, yet it is consistent with the long-standing observation that onset of EBV malignancies in healthy virus carriers may be preceded for months to years by elevated antibody titers to EBV lytic antigens, indicating viral reactivation (6, 19, 33). Implicit in such a reversed order of events is that EBV participates in tumor progression, not initiation, a notion corroborated by the contribution of LMP2A to epithelial cell growth (29). Indeed, our results linking optimal LMP2A expression to minimal intervening TR sequences and ensuing epithelial cell clonal emergence are consistent with experimental data from a mouse tumorigenicity model showing selection for cells with high levels of LMP2A in the newly derived tumors (29). Cell transformation with the extant possibility of subsequent EBV infection provides a logical explanation for the less-than-complete association of virus with malignancies of similar histogenetic phenotype and is a sequence of events entirely compatible with the detection of clonal EBV in diverse human cancers.
Acknowledgments
We thank the Research Core Facility (Louisiana State University Health Sciences Center) for assistance with cell sorting and real-time PCR analysis; A. B. Rickinson, L. S. Young, E. Kremmer, R. Longnecker, and L. Hutt-Fletcher for cells and reagents; P. Schuetze for technical support; and Y. J. Gan for thoughtful discussions.
This study was supported by NIH grants CA67372 and DE12187.
REFERENCES
- 1.Borza, C. M., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus recombinant lacking expression of glycoprotein gp150 infects B cells normally but is enhanced for infection of epithelial cells. J. Virol. 72:7577-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, N. A. C.-R. Liu, Y.-F. Wang, and C. R. Garcia. 1988. B-cell lymphoproliferations and lymphomagenesis are associated with clonotypic intracellular terminal regions of the Epstein-Barr virus. J. Virol. 62:962-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chodosh, J., Y. J. Gan, V. P. Holder, and J. W. Sixbey. 2000. Patterned entry and egress by Epstein-Barr virus in polarized CR2-positive epithelial cells. Virology 266:387-396. [DOI] [PubMed] [Google Scholar]
- 4.Dawson, C. W., A. B. Rickinson, and L. S. Young. 1990. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature 344:777-780. [DOI] [PubMed] [Google Scholar]
- 5.Deobagkar, D. D., M. Liebler, M. Graessmann, and A. Graessman. 1990. Hemimethylation of DNA prevents chromatin expression. Proc. Natl. Acad. Sci. USA 87:1691-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De-Thé, G., A. Geser, N. E. Day, P. M. Tukei, E. H. Williams, D. P. Beri, P. G. Smith, A. G. Dean, G. W. Bornkamm, P. Feorino, and W. Henle. 1978. Epidemiological evidence for causal relationship between Burkitt's lymphoma from Ugandan prospective study. Nature 274:756-761. [DOI] [PubMed] [Google Scholar]
- 7.Fahraeus, R., L. Rymo, J. S. Rhim, and G. Klein. 1990. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein-Barr virus. Nature 345:447-449. [DOI] [PubMed] [Google Scholar]
- 8.Fruehling, S., S. K. Lee, R. Herrold, B. Frech, G. Laux, E. Kremmer, F. A. Grasser, and R. Longnecker. 1996. Identification of latent membrane protein 2A (LMP2A) domains essential for the LMP2A dominant-negative effect on B-lymphocyte surface immunoglobulin signal transduction. J. Virol. 70:6216-6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gahn, T. A., and B. Sugden. 1995. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J. Virol. 69:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan, Y. J., B. I. Razzouk, T. Su, and J. W. Sixbey. 2002. A defective, rearranged Epstein-Barr virus genome in EBER-negative and EBER-positive Hodgkin's disease. Am. J. Pathol. 160:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai, S., J. Nishikawa, and K. Takada. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 72:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keshet, I., J. Yisraeli, and H. Cedar. 1985. Effect of regional DNA methylation on gene expression. Proc. Natl. Acad. Sci. USA 82:2560-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kintner, C. R., and B. Sugden. 1979. The structure of the termini of the DNA of Epstein-Barr virus. Cell 17:661-671. [DOI] [PubMed] [Google Scholar]
- 14.Laux, G., M. Perricaudet, and P. J. Farrell. 1988. A spliced Epstein-Barr virus gene expressed in immortalized lymphocytes is created by circularization of the linear viral genome. EMBO J. 7:769-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, C. T., C. Wannshin, M. M. Hsu, and A. N. Dee. 1997. Clonal versus polyclonal Epstein-Barr virus infection in nasopharyngeal carcinoma cell lines. Lab. Investig. 76:793-798. [PubMed] [Google Scholar]
- 16.Longnecker, R., C. L. Miller, B. Tomkinson, X.-Q. Miao, and E. Kieff. 1993. Deletion of DNA encoding the first five transmembrane domains of Epstein-Barr virus latent membrane proteins 2A and 2B. J. Virol. 67:5068-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann, K. P., D. Staunton, and D. A. Thorley-Lawson. 1985. Epstein-Barr virus encoded protein found in plasma membranes of transformed cells. J. Virol. 55:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, C. L., J. H. Lee, E. Kieff, and R. Longnecker. 1994. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc. Natl. Acad. Sci. USA 91:772-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller, N., A. Evans, N. L. Harris, G. W. Comstock, E. Jellum, K. Magnus, N. Orentreich, B. F. Polk, and J. Vogelman. 1989. Hodgkin's disease and Epstein-Barr virus. Altered antibody pattern before diagnosis. N. Engl. J. Med. 320:689-695. [DOI] [PubMed] [Google Scholar]
- 20.Neri, A., F. Barriga, G. Inghirami, D. M. Knowles, J. Neequaye, I. T. Magrath, and R. Dalla-Favera. 1991. Epstein-Barr virus infection precedes clonal expansion in Burkitt's and acquired immunodeficiency syndrome-associated lymphoma. Blood 77:1092-1096. [PubMed] [Google Scholar]
- 21.Panousis, C. G., and D. T. Rowe. 1997. Epstein-Barr virus latent membrane protein 2 associates with and is a substrate for mitogen-activated protein kinase. J. Virol. 71:4752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathmanathan, R., U. Prasad, R. Sadler, K. Flynn, and N. Raab-Traub. 1995. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N. Engl. J. Med. 333:693-698. [DOI] [PubMed] [Google Scholar]
- 23.Raab-Traub, N., and K. Flynn. 1986. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell 47:883-889. [DOI] [PubMed] [Google Scholar]
- 24.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Field's virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 25.Rountree, M. R., and E. U. Selker. 1997. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 11:2383-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe, M., H. S. Evans, L. S. Young, K. Hennessy, E. Kieff, and A. B. Rickinson. 1987. Monoclonal antibodies to the latent membrane protein of Epstein-Barr virus reveal heterogeneity of the protein and inducible expression in virus-transformed cells. J. Gen. Virol. 68:1575-1586. [DOI] [PubMed] [Google Scholar]
- 27.Sadler, R. H., and N. Raab-Traub. 1995. The Epstein-Barr virus 3.5-kilobase latent membrane protein 1 mRNA initiates from a TATA-less promoter within the first terminal repeat. J. Virol. 69:4577-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sample, J., D. Liebowitz, and E. Kieff. 1989. Two related Epstein-Barr virus membrane proteins are encoded by separate genes. J. Virol. 63:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholle, F., K. M. Bendt, and N. Raab-Traub. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74:10681-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takada, K., and Y. Ono. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 63:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson, J. B., W. Weinberg, R. Johnson, S. Yuspa, and A. J. Levine. 1990. Expression of the BNLF-1 oncogene of Epstein-Barr virus in the skin of transgenic mice induces hyperplasia and aberrant expression of keratin 6. Cell 61:1315-1327. [DOI] [PubMed] [Google Scholar]
- 32.Yeung, W. M., Y. S. Zong, C. T. Chiu, K. H. Chan, J. S. T. Sham, D. T. K. Choy, and M. H. Ng. 1993. Epstein-Barr virus carriage by nasopharyngeal carcinoma in situ. Int. J. Cancer 53:746-750. [DOI] [PubMed] [Google Scholar]
- 33.Zeng, Y., L. G. Zhang, Y. C. Wu, Y. S. Huang, N. Q. Huang, J. Y. Li, Y. B. Wang, M. K. Jiang, Z. Fang, and N. N. Meng. 1985. Prospective studies on nasopharyngeal carcinoma in Epstein-Barr virus IgA/VCA antibody-positive persons in Wuzhou City, China. Int. J. Cancer 36:545-547. [DOI] [PubMed] [Google Scholar]