Abstract
Bacterial delivery systems are gaining increasing interest as potential vaccination vectors to deliver either proteins or nucleic acids for gene expression in the recipient. Bacterial delivery systems for gene expression in vivo usually contain small multicopy plasmids. We have shown before that bacteria containing a herpesvirus bacterial artificial chromosome (BAC) can reconstitute the virus replication cycle after cocultivation with fibroblasts in vitro. In this study we addressed the question of whether bacteria containing a single plasmid with a complete viral genome can also reconstitute the viral replication process in vivo. We used a natural mouse pathogen, the murine cytomegalovirus (MCMV), whose genome has previously been cloned as a BAC in Escherichia coli. In this study, we tested a new application for BAC-cloned herpesvirus genomes. We show that the MCMV BAC can be stably maintained in certain strains of Salmonella enterica serovar Typhimurium as well and that both serovar Typhimurium and E. coli harboring the single-copy MCMV BAC can reconstitute a virus infection upon injection into mice. By this procedure, a productive virus infection is regenerated only in immunocompromised mice. Virus reconstitution in vivo causes elevated titers of specific anti-MCMV antibodies, protection against lethal MCMV challenge, and strong expression of additional genes introduced into the viral genome. Thus, the reconstitution of infectious virus from live attenuated bacteria presents a novel concept for multivalent virus vaccines launched from bacterial vectors.
Live, attenuated bacteria have a potential to serve as vaccine vectors for the oral delivery of foreign proteins (29) or DNA (11, 13, 23). Oral vaccination procedures do not require educated personnel, and lyophilized bacterial preparations can be kept and distributed at room temperature, without the costly and difficult necessity of keeping the preparations cold. Moreover, oral vaccination can be performed simultaneously on large numbers of subjects, which should make it an efficient strategy for livestock vaccination.
Live antiviral vaccines are generally considered more efficient than subunit or inactivated vaccines because they are more likely to induce a broad range of immune responses to the expressed gene products and provide a better protection. Furthermore, since these vaccines replicate in the recipient, the immunity they confer should be long lasting (39). Therefore, a bacterial vaccination vector delivering an attenuated, yet infectious virus may present the basis for efficient vaccines that are easy to store, distribute, and administer.
Herpesviruses are important pathogens for humans and livestock. We have previously shown that the large genomes of herpesviruses (up to 230 kb) can be cloned as bacterial artificial chromosomes (BACs) into Escherichia coli (1, 2, 24). Others have demonstrated that BAC DNA encoding the genome of a herpesvirus can induce specific protective immunity (35, 36). However, in these experiments, the BAC DNA was isolated from bacteria by column chromatography and diluted in phosphate-buffered saline (PBS) prior to injection into chickens (36) or adsorbed to gold particles and injected into mice with a gene gun (35).
For this study, we decided to test whether the infection of animals with bacteria could lead to reconstitution of a replication competent virus from a bacterial vector in vivo and whether this could lead to protective immunity.
The cytomegaloviruses (CMVs), ubiquitous members of the betaherpesvirus subgroup, are marked by strict species specificity, tropism for hematopoietic tissue and secretory glands, and slow replication. The infection of mice with murine CMV (MCMV) shares many aspects with human CMV infection and thus serves as a biological model for experimental vaccine developments (16, 22, 26, 42). With double-stranded DNA genomes of 120 to 230 kbp and up to 220 genes, herpesviruses have some of the largest genomes of viruses that infect mammals (25). Many herpesviral genes are not essential for viral replication and could be exchanged for genes from unrelated species to generate recombinant viruses. Therefore, the deletion of viral genes that are not essential for the vaccination success and the insertion of genes from other infectious agents are attractive concepts for the development of live recombinant vaccines. For this purpose, we introduced a gene from another virus into the MCMV genome and determined the presence of the gene product in murine sera. Here we show that infectious MCMV can be reconstituted in vivo directly from bacteria that carry their genomes. We also show that viral reconstitution leads to protective immunity and to strong expression of additional transgenes inserted into the viral genome.
MATERIALS AND METHODS
Plasmids and bacterial strains.
Plasmid pRep4-HBs was generated by inserting the gene of the hepatitis B virus surface antigen (HBsAg) into pRep4 (Invitrogen) as described (3). To obtain MCMV-HBs, the HBs gene bracketed by a Rous sarcoma virus promoter and a simian virus 40 polyadenylation signal was excised from pRep4-HBs and inserted into the ie2 gene locus of the cloned full-length MCMV BAC, pSM3fr (37), as previously described (3). Plasmid pGB2Ωinv-hly (17) was kindly provided by C. Grillot-Courvalin (Unité des Agents Antibactériens, Institut Pasteur, Paris, France).
Plasmid and BAC DNA was introduced by electroporation into the standard E. coli BAC host, DH10B [genotype: F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara leu)7697 galU galK rpsL endA1 nupG]. MCMV-HBs was transferred by conjugation from DH10B to serovar Typhimurium LT2 strains SA1970 (metA22 trpC2 hisF1009 rpsL120 xylR1 recA1 srl-202::Tn10), TT521 (recA1 rpsL srl-202::Tn10), and TT9081 [his-644(del:OGDCBHAF) srl-202::Tn10 recA1]. The serovar Typhimurium strains were obtained from K. E. Sanderson (Salmonella Genetic Stock Centre, University of Calgary, Calgary, Canada).
Mice and viruses.
BALB/c, C57BL/6, and 129SvEv gamma interferon receptor-null (IFN-γR0/0) mice were bred under conventional barrier housing conditions at the central breeding facility of the Rijeka Medical Faculty Animal house. All animal experiments described here received approval by the Ethical Committee at the University of Rijeka. Sex- and strain-matched, 6- to 9-week-old mice were used. Stocks of tissue culture and salivary gland-derived (SGD) MCMV, derived from MW97.01 (37), were prepared as previously described (4).
Virus reconstitution.
Fresh bacterial cultures were grown overnight in Luria-Bertani medium supplied with antibiotics for selective growth. DNA transfer from bacteria to mammalian cells was performed as previously described (5). Two microliters of bacterial culture was used to inoculate NIH 3T3 fibroblasts grown in 96-well dishes without antibiotics. This corresponded to approximately 300 bacteria per fibroblast. The cell culture dishes were spun for 5 min at 1,000 × g and incubated for 2 h at 37°C. The cell culture medium was subsequently replaced with fresh medium supplemented with ampicillin and gentamicin (100 μg/ml each). For infection of mice, bacterial cultures were centrifuged at 2,000 × g for 8 min at 4°C and serially diluted in cold sterile PBS, at concentrations ranging from 2 × 109 to 2 × 106 CFU/ml (bacterial stocks were titrated in parallel on blood agar plates). Five hundred microliters of diluted bacteria (amounts between 106 to 109 CFU) were injected intraperitoneally (i.p.) into each mouse. Lungs were obtained from mice infected with serovar Typhimurium on 28th day postinfection (dpi) and tested for MCMV reconstitution by plaque assay on murine embryonic fibroblasts (MEFs) as previously described (32). The mice infected with E. coli were immunosuppressed with anti-CD4 and anti-CD8 antibodies (9) (1 mg each) on the 6th and 13th dpi. Mice were additionally immunosuppressed by intramuscular (i.m.) injection of 6.25 μg of hydrocortisone on alternate days from day 13 postinfection onwards. Blood samples were collected from tail veins of infected mice on days 12 and 20 postinfection. HBsAg was detected in murine serum by use of a commercial microtiter enzyme-linked immunosorbent assay (ELISA) kit, containing wells coated with anti-HBs antibodies (catalog no. 931801; Ortho Diagnostic Systems) according to manufacturer's instructions. Salivary glands, lungs, and spleens from individual mice were collected on dpi 21 and tested for MCMV reconstitution as previously described (32). Each experiment was performed at least twice.
Immunization and challenge procedures.
Overnight bacterial cultures were centrifuged and resuspended in cold sterile PBS at final concentrations of 5 × 109 CFU/ml. One hundred μl of bacterial suspension (5 × 108 CFU) was injected either i.m. into gluteal muscles or subcutaneously (s.c.) into the soft tissue of the dorsum. For i.p. injection bacteria were diluted 1:10 in sterile PBS and injected in a volume of 500 μl (∼108 CFU). Positive-control mice were infected with 105 PFU of tissue culture-grown BAC-derived MCMV. No immunosuppressive procedure was applied. Blood samples were collected from the tail vein, and sera from individual mice were serially diluted in PBS. The immunization against MCMV and antibody production efficiency was tested by indirect ELISA using MCMV-infected MEFs as the antigen source, as previously described (20). In brief, microtiter plates were coated with lysates of either uninfected or MCMV-infected MEFs. Serially diluted sera were applied in parallel to both kinds of plates, which was followed by wash steps and an incubation with peroxidase-conjugated anti-mouse antibodies. Signals obtained from the plates coated with uninfected MEF lysates were treated as unspecific background signal and subtracted from the signals obtained from the plates coated with lysates of MCMV-infected MEFs. The protective effect of the anti-MCMV immunization was tested by challenge with lethal doses of SGD MCMV. Mice received i.p. injections with 2 50% lethal doses (LD50) of SGD MCMV (i.e., 5 × 104 PFU for 129 IFN-γR0/0 mice) on dpi 28. Animals were checked daily for survival for 2 weeks following the challenge. Survivors were monitored for an additional 3 months. Each experiment was performed at least twice.
RESULTS
Virus reconstitution in vivo by serovar Typhimurium carrying the MCMV BAC.
In our previous studies, the DH10B strain of E. coli served as the host for the MCMV BAC. As DH10B is not commonly used as vector for DNA immunization, we introduced an MCMV BAC into recombinase A-deficient (recA mutant) laboratory strains of Salmonella enterica serovar Typhimurium shown in Table 1. recA mutant bacterial strains were selected to ensure the stability of repetitive sequences present in herpesvirus genomes (31). The rationale for the usage of serovar Typhimurium was their natural ability to invade mammalian cells. Therefore, they should represent a better vehicle for DNA transfer in vivo than E. coli. The clones that received the MCMV BAC were selected by resistance to chloramphenicol and tetracycline. To confirm that the clones in question contained the complete MCMV BAC, BAC DNA was extracted and introduced into fibroblasts to give rise to infectious virus as previously described (6). To check whether bacteria could transfer viral DNA directly into mammalian cells in cell culture, we inoculated NIH 3T3 cells with WB241 bacteria. Five days after inoculation we observed the formation of characteristic viral plaques, indicating direct reconstitution of MCMV from bacteria.
TABLE 1.
List of modified bacterial strains used in this study
| Bacterial vector | Inserted plasmid
|
|||
|---|---|---|---|---|
| Name | Species and strain | MCMV-HBs BAC | pRep4-HBs | pGB2Ωinv-hly |
| Eco/M (WB231) | E. coli K-12, DH10B | + | − | − |
| Eco/M/inv (WB232) | E. coli K-12, DH10B | + | − | + |
| Eco/P/inv (WB233) | E. coli K-12, DH10B | − | + | + |
| WB240 | S. enterica serovar Typhimurium LT2, SA1970 | + | − | − |
| WB241 | S. enterica serovar Typhimurium LT2, TT521 | + | − | − |
| WB242 | S. enterica serovar Typhimurium LT2, TT9081 | + | − | − |
recA mutant bacteria are attenuated in vivo because they cannot repair the DNA damage caused by reactive oxygen species (ROS) in endolysosomal compartments of professional phagocytes (7, 8). ROS are strongly induced in macrophages by IFN-γ (15, 27). As serovar Typhimurium has a preference to infect macrophages in vivo (14, 33), this could lead to damage of the viral DNA before the virus would initiate its replication. In IFN-γR0/0 mice (18), ROS production in macrophages is not induced (21). Thus, we assumed that the use of IFN-γR0/0 mice would increase the chance to test the possibility of herpesvirus reconstitution from bacteria in vivo.
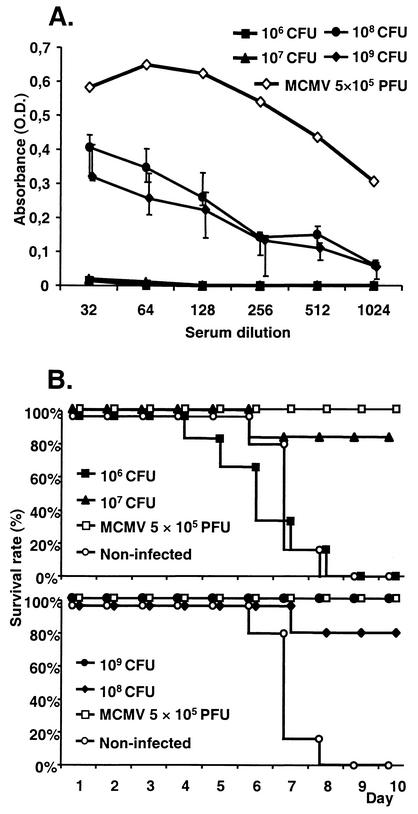
Serovar Typhimurium strain WB241 (Table 1) was grown overnight as described in Materials and Methods and injected i.p. into IFN-γR0/0 mice at doses indicated in the Fig. 1. Control mice were infected with 5 × 105 PFU of BAC-derived MCMV (37). Mock-infected mice were used as negative controls. The ability to mount a specific humoral response to MCMV was taken as an indication of virus protein expression. Mice infected s.c. with 108 or 109 CFU of bacteria displayed elevated titers of anti-MCMV antibodies in their sera on the 28th dpi, whereas mice infected with 107 CFU or less showed no anti-MCMV antibodies (Fig. 1A). Therefore, the infection with 108 CFU of bacteria or more induced humoral immunity against MCMV, whereas infection with 107 CFU or less did not. The very same mice were challenged on dpi 28 with 105 PFU of SGD MCMV (4 LD50) and monitored for survival (Fig. 1B). All mice infected with 109 CFU and 83% of mice infected with 108 CFU of bacteria, as well as all of the virus-infected controls, survived the challenge. Surprisingly, 83% of the mice infected with 107 CFU survived the challenge as well, despite the fact that only one mouse in this group seroconverted. (Fig. 1A). All mice infected with 106 CFU as well as the uninfected control mice died by day 9 following challenge. Therefore, the infection of mice with serovar Typhimurium carrying MCMV BACs, induced specific antibody response and protective immunity against MCMV.
FIG. 1.
129SvEv IFN-γR0/0 mice were infected with the indicated CFU of WB240 (six mice per group). Positive-control mice were infected with 5 × 105 PFU of wild-type MCMV, and negative controls were mock infected. (A) Sera were collected at dpi 28, serially diluted up to 1:1,024, and assayed for anti-MCMV antibodies by ELISA. Median and quartile absorbance values at 492 nm are displayed. Pooled sera from a group of mice that received 5 × 105 PFU of MCMV were used as positive controls. (B) On the same day, mice were challenged with 2 LD50 of salivary gland-passaged MCMV and monitored for survival. Survival curves for the first 10 days following challenge are shown. Exclusively for presentation purposes, the survival curves are presented in two separate graphs. Error bars, quartiles.
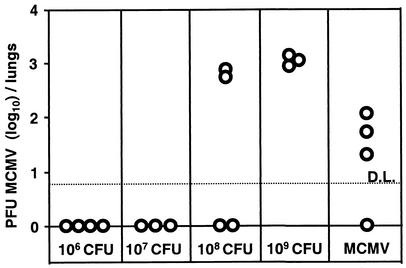
In order to assess whether the immunization was caused by the reconstitution of the infectious viral process or merely due to expression of isolated viral genes from transferred DNA, mice were infected according to the protocol described above, and infectious viral titers in lungs were determined at dpi 28. The animals in the groups infected with 108 or 109 CFU of WB241 contained infectious virus in their organs as seen by plaque assay on MEFs (Fig. 2). The specificity of these plaques for MCMV was confirmed by staining with a monoclonal antibody against the pp89 protein, the product of the IE1 MCMV gene (not shown). In mice infected with 107 CFU or less we could not detect virus. Therefore, the infection with 108 CFU of serovar Typhimurium carrying the MCMV BAC leads to reconstitution of live virus in vivo. Notably, the same number of bacteria that were needed for specific antibody formation was needed to initiate an active viral infection process. Therefore, the ability to mount a specific antibody response could serve as an indirect indicator for virus reconstitution in vivo.
FIG. 2.
129 SvEv IFN-γR0/0 mice were i.p. infected with the indicated doses of WB240 or with 5 × 105 PFU of MCMV. Mice were sacrificed on dpi 28, and lung homogenates were assayed on MEFs for infectious MCMV titers. Circles represent virus titers in the organs of individual mice. D.L., limit of detection.
Next, we tested whether bacteria carrying MCMV BACs could induce the immunization of immunocompetent mice. We infected groups of BALB/c mice with 108 CFU of WB241 bacteria and looked for infectious virus in lungs and spleen and for specific antibody titers to MCMV in sera at 28 dpi. As expected, mice did not show elevated antibody titers (data not shown), nor could infectious virus be isolated from their organs. Therefore, we concluded that the serovar Typhimurium vectors used here could not effectively induce the reconstitution of the viral infectious process in IFN-γR+/+ mice.
Virus reconstitution in vivo by E. coli carrying MCMV BAC.
In previous experiments, we have shown that the cloned MCMV genome can be transferred directly from its E. coli host to cultured fibroblasts to initiate the production of infectious virus in these cells (5). This requires the introduction of the plasmid pGB2Ωinv-hly (17), which directs the expression of the invasin gene of Yersinia pseudotuberculosis and the hemolysin O gene from Listeria monocytogenes in E. coli, and the presence of β1-integrin molecules on the surface of the recipient fibroblasts. We wanted to test to which extent these bacteria could also be used as vectors to deliver the MCMV genome to somatic cells of a mouse and to initiate the infectious cycle in vivo. The rationale for this was that the in vivo infection of nonphagocytic cells like fibroblasts, which do not express ROS, would give an opportunity to recA mutant bacterial vectors to reconstitute viral infection in IFN-γR+/+ animals.
Moreover, we wanted to analyze the efficiency of the expression of a transgene from a recombinant herpesvirus. Thus, we used a recombinant MCMV BAC, MCMV-HBs, which contains the HBsAg gene driven by a Rous sarcoma virus promoter (3). In a previous study we have shown that levels in serum of a secreted virus-encoded protein such as the HBsAg reflect MCMV productivity in vivo and thus can be used to monitor the course of infection over time. Bacteria containing a high-copy-number plasmid with the identical eukaryotic HBs expression cassette were used to control for possible HBsAg expression in the absence of viral replication and to compare virus reconstitution by bacteria with plasmid DNA delivery by bacteria.
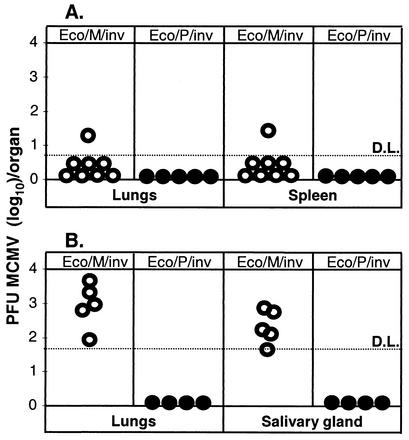
In order to assay for viral reconstitution, BALB/c mice were infected i.p. with 108 CFU of either Eco/M/inv or Eco/P/inv bacteria (Table 1). T lymphocytes were depleted on dpi 6 and 13, and hydrocortisone was applied on alternate days from the 13th dpi onwards in order to suppress the immune response and to increase the chances of establishing a productive viral infection. Lungs, salivary glands, and spleens were collected on the 21st dpi, and organ homogenates were tested by plaque assay on fibroblasts for the presence of infectious MCMV. Only one out of eight mice infected with Eco/M/inv showed reconstitution of infectious virus in the lungs and spleen (Fig. 3A). In repeated experiments in which we used BALB/c or C57BL/6 mice, we did not detect viral reconstitution (data not shown).
FIG. 3.
(A) BALB/c or (B) 129 SvEv IFN-γR−/− mice were i.p. infected with 108 CFU of WB232 (○) or WB233 (•). T lymphocytes were depleted on dpi 6 and 13, and hydrocortisone was applied on alternate days from dpi 13 onwards. Mice were sacrificed on dpi 21, and organ homogenates were assayed on MEFs for infectious MCMV titers. Circles represent virus titer in organs of individual mice. D.L., detection limit.
In a separate experiment, we infected groups of IFN-γR0/0 mice with graded amounts of either Eco/M/inv or Eco/P/inv using the same infection and immune suppression protocol. All mice that received 108 CFU of Eco/M/inv were positive for MCMV plaques in their lungs and salivary glands (Fig. 3B). The mice infected with 107 CFU of Eco/M/inv were negative for MCMV as confirmed by plaque assay. As expected, Eco/P/inv-infected mice showed no evidence for MCMV infection. Therefore, the infection with E. coli carrying the MCMV BAC leads to in vivo reconstitution of infectious virus in IFN-γR0/0 mice but not in IFN-γR+/+ mice. There was no significant difference in the number of bacteria required to launch the infectious process either by serovar Typhimurium or by E. coli.
IFN-γR0/0 mice are more sensitive to virus infection than normal mice. To clarify whether the antiviral activity of IFN-γ alone could explain the lack of reconstitution in IFN-γR+/+ mice, we tried to reconstitute infectious virus from bacteria using B-cell-deficient μMT/μMT mice, which were T cell depleted before infection with bacteria. Mice lacking both B and T cells cannot control MCMV infection (38), whereas IFN-γR0/0 mice can (30). Thus, this system should allow the detection of very low initial amounts of reconstituted virus. However, at dpi 21 we could not detect any virus out of organs of μMT/μMT mice infected with 108 CFU of Eco/M/inv E. coli (data not shown). Therefore, mice very sensitive to MCMV, yet capable of IFN-γ signaling, could not reconstitute infectious virus. This finding argues for an important role of the IFN-γ signaling pathway during the early phase of virus reconstitution in vivo.
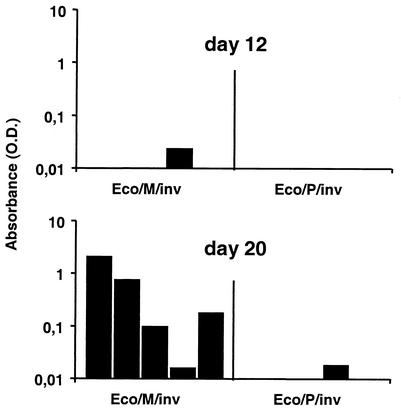
Sera were collected from the very same IFN-γR0/0 mice whose organs were assayed for virus titer and shown in Fig. 3, on dpi 12 and 20 and assayed for the product of the introduced viral transgene. The presence of the viral protein HBsAg was determined by direct ELISA. At day 12, no HBsAg could be detected in any of the groups (Fig. 4). At day 20, four out of five mice infected with Eco/M/inv but none of the mice infected with Eco/P/inv showed detectable amounts of HBsAg in their sera (Fig. 4). Notably, the amount of HBsAg correlated with the viral yield in the salivary gland in individual mice, which was determined in parallel (r = 0.91). Therefore, the MCMV-HBs reconstitution is associated with a considerable HBsAg expression in the sera of animals. Animals that received the HBs gene alone as a multicopy plasmid did not produce measurable HBsAg concentrations. The absence of HBsAg at dpi 12 indicates that HBsAg was expressed to detectable levels only upon virus reconstitution. In fact, no infectious virus was detected when mice infected under identical conditions with Eco/M/inv were sacrificed at dpi 14 (data not shown).
FIG. 4.
129 SvEv IFN-γR0/0 mice were i.p. infected with 108 CFU of either Eco/M/inv or Eco/P/inv. T lymphocytes were depleted on dpi 6 and 13, and hydrocortisone was applied on alternate days from day 13 onwards. Sera were collected at days 12 and 20 and assayed for the presence of HBsAg by direct ELISA. At day 12, no HBsAg could be detected in any of the groups. At day 20, four out of five mice infected with Eco/M/inv and none of the mice infected with Eco/P/inv showed detectable amounts of HBs in their sera.
Virus reconstitution in vivo by bacteria carrying the MCMV BAC is independent of bacterial invasion genes.
Virus reconstitution from E. coli was restricted to IFN-γR0/0 mice, as it was the case with serovar Typhimurium vectors. Therefore, we assumed that this process was occurring in professional phagocyte cells. The additional genes supplied for active bacterial invasion and virus reconstitution in vitro might therefore be dispensable for virus reconstitution in vivo. In order to test this hypothesis, bacteria carrying the MCMV BAC, but not the pGB2Ωinv-hly plasmid, were tested for DNA transfer and viral reconstitution.
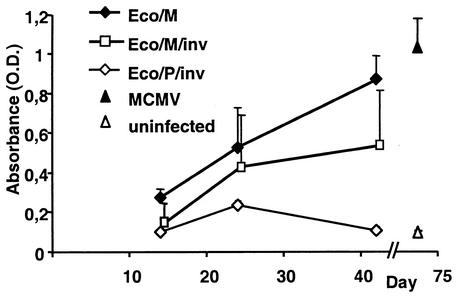
Groups of IFN-γR0/0 mice infected with 108 CFU of either Eco/M/inv, Eco/M, or Eco/P/inv bacteria were monitored for MCMV specific antibody production by IFN-γR0/0 mice over time. We observed that five of five of the mice infected with Eco/M and four of five mice infected with Eco/M/inv bacteria mounted an antibody response (Fig. 5). The MCMV-specific antibody titer was only slightly elevated over the values from control mice at dpi 14 but rose by dpi 28 and at dpi 42 was comparable to titers obtained from MCMV infected mice at dpi 70. Control mice infected with Eco/P/inv did not mount a specific immune response. Eco/M bacteria, which lack the pGB2Ωinv-hly invasion plasmid, induced a strong MCMV-specific antibody response. The presence of pGB2Ωinv-hly did not enhance the immunization rate. This was in accordance with the observation that infectious MCMV could be isolated from organs of Eco/M-infected mice (not shown). Taken together, these data indicate that successful virus reconstitution in vivo was independent of the genes encoded by pGB2Ωinv-hly, whereas the pGB2Ωinv-hly plasmid was required for BAC transfer and virus reconstitution in cell culture.
FIG. 5.
129 SvEv IFN-γR0/0 mice were infected i.p. with 108 CFU of indicated bacteria (five mice per group). Sera were obtained at 14, 24, and 42 dpi, serially diluted in PBS, and assayed for anti-MCMV antibodies by ELISA. Arithmetic means and standard deviations of absorbance values at 492 nm for the 1:64 dilution step are shown. Sera from mice infected with 105 PFU of MCMV were obtained at dpi 70 and used as positive controls (▴). Sera from uninfected mice (▵) were used as negative controls.
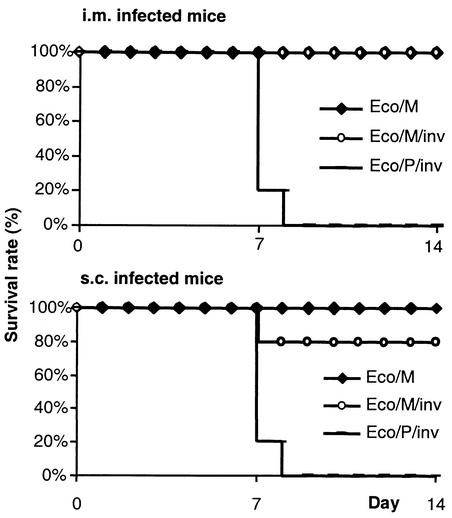
Next, we tested whether the immunization procedure by E. coli was also protective against a challenge with a lethal dose of MCMV. Groups of 10 IFN-γR0/0 mice were immunized by inoculation with 109 CFU of either Eco/M or Eco/M/inv. Control mice were inoculated with Eco/P/inv. Half of each group received the bacteria i.m. and the other half received them s.c. On the 28th dpi, seroconversion occurred in most of the Eco/M or Eco/M/inv infected mice (8 of 10 in each group), but in none of the controls, as detected by ELISA (data not shown). On the same day mice were challenged with 2 LD50 of SGD MCMV and monitored for survival. All five mice infected i.m. with Eco/M/inv and four of five of mice infected s.c. with Eco/M/inv, as well as all animals infected with Eco/M (10 of 10) survived the challenge (Fig. 6). All mice from the control groups died by dpi 8 or 9. The results indicate that mice immunized by bacteria carrying the MCMV BAC were protected against a challenge with lethal doses of virus.
FIG. 6.
129 SvEv IFN-γR0/0 mice were i.m. or s.c. infected with 109 CFU of Eco/M or Eco/M/inv bacteria. Control mice were infected with Eco/P/inv. At 28 dpi, mice were challenged with 2 LD50 of SGD MCMV and monitored for survival. Survival curves for the first 14 days following challenge are shown.
DISCUSSION
The cloning of herpesvirus genomes as bacterial artificial chromosomes allows fast and accurate mutagenesis procedures of basically any viral gene in the prokaryotic E. coli host (6, 24). The biological properties of the viral mutants are analyzed upon the reconstitution of the virus in eukaryotic cells. The application of random transposon mutagenesis procedures led to the establishment of libraries of mutant genomes (6). In order to test the biological properties of random mutant libraries, virus reconstitution was achieved by direct inoculation of host cells in vitro by invasive bacteria carrying BACs (5).
Whether these bacteria would also allow virus reconstitution in vivo was the next logical question. Direct in vivo reconstitution of a virus infection from viral DNA is not a new concept. However, successful approaches had been restricted so far to viruses carrying much smaller genomes (around 10 kb), and to the direct injection of DNA (12, 40, 41). Here we show with the example of the 230-kb genome of MCMV that there should be, in principle, no viral genome size limit for virus reconstitution from DNA in vivo.
Suter et al. have shown that injection of herpesvirus BAC DNA can lead to protective immunity in the absence of viral replication (35). Another recent report has shown that the inoculation of Marek's disease virus BAC DNA diluted in PBS into chicken could lead to protective immunity (36). Our data show for the first time that a herpesvirus infection can be launched directly from bacteria carrying the infectious virus genome, omitting the need for technical preparatory work to isolate BAC DNA. We have also demonstrated that additional genes introduced into the virus are effectively expressed (3). Thus, the use of BAC-derived viruses has a potential as vaccine and vaccine vector, and this is also the case when the viral genome is launched from bacteria.
Interestingly, no infectious virus could be isolated from organs before day 21 after injection of bacteria. Since the infection with 105 PFU MCMV leads to detectable viral titers in organs within few days, this implied that the peak of productive viral replication was delayed for at least 2 weeks. This finding was not unexpected, as we have already reported a time lag in the production of infectious virus already upon transfection of cultured cells with MCMV BAC DNA (37). Therefore, at dpi 28, lung virus titers after virus reconstitution from serovar Typhimurium are still high, whereas in mice infected with MCMV, the virus infection was already contained by the immune system. The other evidence for this delay, are the kinetics of anti-MCMV antibody titers. Titers were still low at 14 days postinfection, rose by dpi 28 and even more by dpi 42. Therefore, Direct comparison of antibody titers at dpi 28 shows a marked difference between mice that were infected by bacteria or by the virus (Fig. 1A). However, at dpi 70, antibody titers from vaccinated mice were undistinguishable from mice infected with MCMV (data not shown).
It was surprising that infection with 107 CFU of serovar Typhimurium did not induce a detectable antibody response or production of infectious virus yet protected mice against a lethal challenge with SGD MCMV. Low-frequency DNA transfer events leading to expression of viral genes together with an abortive infection could explain this observation. In that case, the expression of viral genes might induce a protective cellular immunity, but not a strong humoral immune response. Indeed, DNA immunization against the MCMV by a plasmid encoding for an immunodominant viral gene has already been shown to induce protective T-cell immunity in the absence of an antibody response (16). Moreover, it was demonstrated that an additional injection of formalin inactivated viruses could induce a specific humoral immune response (26).
Another unexpected observation was that virus reconstitution in vivo was not linked to the ability of E. coli to invade cells, as opposed to the situation in vitro. This suggested that the bacteria were not actively entering host cells. Therefore, the probable target cells, into which the viral genomes entered and started the viral replication, could be phagocytic cells that actively took up bacteria. The half-life of granulocytes is only a few hours (19), which is too short to allow MCMV reconstitution and replication. Thus, the properties of professional phagocytic cells, i.e., macrophages and dendritic cells, probably dictate the success of the transfer, rather than the invasive properties of the bacteria. Therefore, it was not surprising that viral reconstitution could be achieved only in IFN-γR0/0 mice that are deficient for ROS production. Professional phagocytes in these mice lack activating signals from IFN-γ pathways (21) and therefore can eliminate intracellular pathogens only at a lower rate (10, 18). This indicates that strategies for the improvement of the virus reconstitution efficiency in vivo should focus on the survival of bacteria in phagocytic cells.
To maintain the stability of the herpesvirus genome, the BACs are usually propagated in recA mutant bacterial strains. Originally, we developed the herpesvirus BAC system for the mutagenesis of viral genomes. Recombination-deficient (recA mutant) bacteria are preferred as hosts for the MCMV BAC because the 21 chi sites (34) present in the MCMV genome, as well as repeated viral sequences within the MCMV genome (31), could potentially serve as hot spots for spontaneous recombination (34).
The requirement for recA mutant bacterial strains for propagation of herpesvirus BACs at present appears to be the major obstacle for further development. Since parenteral administration of 108 CFU of recA mutant bacteria was required for successful reconstitution in IFN-γR0/0 mice, the amounts of bacteria required for efficient transfer in fully immunocompetent mice may need to be even higher. Additionally, these strains have not been designed for oral administration and DNA delivery via mucosal routes. On the other hand, recA+ strains of serovar Typhimurium have already been used as vectors for oral DNA vaccination (11, 28). Can recA+ strains be used to increase transfer efficiency? We have seen that viral BAC DNA can be maintained in the recA+ E. coli strain CBTS or GS500, at least for short periods of time, without losing the potential to start a productive infection (5, 24). However, our attempts to introduce an intact MCMV BAC into recA+ strains of serovar Typhimurium have so far not met with success. Perhaps the genome repeats and the chi sequences need to be deleted or modified, in order to increase the stability of the viral genomes in a recA+ host. Careful design of viral mutations could allow the propagation of stable viral mutant genomes in recA+ bacterial strains. If recA+ bacterial strains could be used for oral delivery of viral DNA, this would have a huge potential for livestock vaccination and perhaps even for human vaccines. The observation that a herpesvirus infection can be reconstituted from bacteria in vivo presents the first step in this direction.
Acknowledgments
This work was supported by DFG SPP “New vaccination strategies,” Bayerische Forschungsstiftung “Forimmun,” and Br1730/2.
We thank Torsten Sacher, Markus Wagner, and Zsolt Ruzsics for useful discussions; Tanja Saulig, Mijo Golemac, and Miljana Kricka for technical assistance; and Astrid Krmpotic and Milena Hasan for technical advice.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brune, W., M. Hasan, M. Krych, I. Bubic, S. Jonjic, and U. H. Koszinowski. 2001. Secreted virus-encoded proteins reflect murine cytomegalovirus productivity in organs. J. Infect. Dis. 184:1320-1324. [DOI] [PubMed] [Google Scholar]
- 4.Brune, W., H. Hengel, and U. H. Koszinowski. 1999. A mouse model for cytomegalovirus infection, p. 19.7.1-19.7.13. In J. Coligan, A. Kruisbeen, D. Marguiles, E. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley, New York, N.Y. [DOI] [PubMed]
- 5.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 6.Brune, W., C. Menard, U. Hobom, S. Odenbreit, M. Messerle, and U. H. Koszinowski. 1999. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat. Biotechnol. 17:360-364. [DOI] [PubMed] [Google Scholar]
- 7.Buchmeier, N. A., C. J. Lipps, M. Y. So, and F. Heffron. 1993. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol. Microbiol. 7:933-936. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson, J., and V. S. Carpenter. 1980. The recA+ gene product is more important than catalase and superoxide dismutase in protecting Escherichia coli against hydrogen peroxide toxicity. J. Bacteriol. 142:319-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobbold, S. P., A. Jayasuriya, A. Nash, T. D. Prospero, and H. Waldmann. 1984. Therapy with monoclonal antibodies by elimination of T-cell subsets in vivo. Nature 312:548-551. [DOI] [PubMed] [Google Scholar]
- 10.Dalton, D. K., S. Pitts-Meek, S. Keshav, I. S. Figari, A. Bradley, and T. A. Stewart. 1993. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science 259:1739-1742. [DOI] [PubMed] [Google Scholar]
- 11.Darji, A., C. A. Guzman, B. Gerstel, P. Wachholz, K. N. Timmis, J. Wehland, T. Chakraborty, and S. Weiss. 1997. Oral somatic transgene vaccination using attenuated S. enterica serovar Typhimurium. Cell 91:765-775. [DOI] [PubMed] [Google Scholar]
- 12.Dubensky, T. W., B. A. Campbell, and L. P. Villarreal. 1984. Direct transfection of viral and plasmid DNA into the liver or spleen of mice. Proc. Natl. Acad. Sci. USA 81:7529-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fennelly, G. J., S. A. Khan, M. A. Abadi, T. F. Wild, and B. R. Bloom. 1999. Mucosal DNA vaccine immunization against measles with a highly attenuated Shigella flexneri vector. J. Immunol. 162:1603-1610. [PubMed] [Google Scholar]
- 14.Garcia-del Portillo, F. 2001. Salmonella intracellular proliferation: where, when and how? Microbes Infect. 3:1305-1311. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg, M., L. S. Belkowski, and B. R. Bloom. 1990. Regulation of macrophage function by interferon-gamma. Somatic cell genetic approaches in murine macrophage cell lines to mechanisms of growth inhibition, the oxidative burst, and expression of the chronic granulomatous disease gene. J. Clin. Investig. 85:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Armas, J. C., C. S. Morello, L. D. Cranmer, and D. H. Spector. 1996. DNA immunization confers protection against murine cytomegalovirus infection. J. Virol. 70:7921-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grillot-Courvalin, C., S. Goussard, F. Huetz, D. M. Ojcius, and P. Courvalin. 1998. Functional gene transfer from intracellular bacteria to mammalian cells. Nat. Biotechnol. 16:862-866. [DOI] [PubMed] [Google Scholar]
- 18.Huang, S., W. Hendriks, A. Althage, S. Hemmi, H. Bluethmann, R. Kamijo, J. Vilcek, R. M. Zinkernagel, and M. Aguet. 1993. Immune response in mice that lack the interferon-gamma receptor. Science 259:1742-1745. [DOI] [PubMed] [Google Scholar]
- 19.Janeway, C. A., and P. Travers. 1997. Immunobiology: the immune system in health and disease, 3rd ed. Current Biology, Ltd./Garland Publisihng, Inc., New York, N.Y.
- 20.Jonjic, S., M. del Val, G. M. Keil, M. J. Reddehase, and U. H. Koszinowski. 1988. A nonstructural viral protein expressed by a recombinant vaccinia virus protects against lethal cytomegalovirus infection. J. Virol. 62:1653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamijo, R., D. Shapiro, J. Le, S. Huang, M. Aguet, and J. Vilcek. 1993. Generation of nitric oxide and induction of major histocompatibility complex class II antigen in macrophages from mice lacking the interferon gamma receptor. Proc. Natl. Acad. Sci. USA 90:6626-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacDonald, M. R., X. Y. Li, R. M. Stenberg, A. E. Campbell, and H. W. t. Virgin. 1998. Mucosal and parenteral vaccination against acute and latent murine cytomegalovirus (MCMV) infection by using an attenuated MCMV mutant. J. Virol. 72:442-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina, E., and C. A. Guzman. 2001. Use of live bacterial vaccine vectors for antigen delivery: potential and limitations. Vaccine 19:1573-1580. [DOI] [PubMed] [Google Scholar]
- 24.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 26.Morello, C. S., M. Ye, and D. H. Spector. 2002. Development of a vaccine against murine cytomegalovirus (MCMV), consisting of plasmid DNA and formalin-inactivated MCMV, that provides long-term, complete protection against viral replication. J. Virol. 76:4822-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nathan, C. F., H. W. Murray, M. E. Wiebe, and B. Y. Rubin. 1983. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 158:670-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paglia, P., E. Medina, I. Arioli, C. A. Guzman, and M. P. Colombo. 1998. Gene transfer in dendritic cells, induced by oral DNA vaccination with Salmonella typhimurium, results in protective immunity against a murine fibrosarcoma. Blood 92:3172-3176. [PubMed] [Google Scholar]
- 29.Poirier, T. P., M. A. Kehoe, and E. H. Beachey. 1988. Protective immunity evoked by oral administration of attenuated aroA Salmonella typhimurium expressing cloned streptococcal M protein. J. Exp. Med. 168:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Presti, R. M., J. L. Pollock, A. J. Dal-Canto, A. K. O'Guin, and H. W. Virgin. 1998. Interferon gamma regulates acute and latent murine cytomegalovirus infection and chronic disease of the great vessels. J. Exp. Med. 188:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddehase, M. J., F. Weiland, K. Munch, S. Jonjic, A. Luske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richter-Dahlfors, A., A. M. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, G. R., S. M. Kunes, D. W. Schultz, A. Taylor, and K. L. Triman. 1981. Structure of chi hotspots of generalized recombination. Cell 24:429-436. [DOI] [PubMed] [Google Scholar]
- 35.Suter, M., A. M. Lew, P. Grob, G. J. Adema, M. Ackermann, K. Shortman, and C. Fraefel. 1999. BAC-VAC, a novel generation of (DNA) vaccines: a bacterial artificial chromosome (BAC) containing a replication-competent, packaging-defective virus genome induces protective immunity against herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 96:12697-12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tischer, B. K., D. Schumacher, M. Beer, J. Beyer, J. P. Teifke, K. Osterrieder, K. Wink, V. Zelnik, F. Fehler, and N. Osterrieder. 2002. A DNA vaccine containing an infectious Marek's disease virus genome can confer protection against tumorigenic Marek's disease in chickens. J. Gen. Virol. 83:2367-2376. [DOI] [PubMed] [Google Scholar]
- 37.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Syst. excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welsh, R. M., J. O. Brubaker, M. Vargas-Cortes, and C. L. O'Donnell. 1991. Natural killer (NK) cell response to virus infections in mice with severe combined immunodeficiency. The stimulation of NK cells and the NK cell-dependent control of virus infections occur independently of T and B cell function. J. Exp. Med. 173:1053-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitley, R. J., and B. Roizman. 2002. Herpes simplex viruses: is a vaccine tenable? J. Clin. Investig. 110:145-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Will, H., R. Cattaneo, G. Darai, F. Deinhardt, H. Schellekens, and H. Schaller. 1985. Infectious hepatitis B virus from cloned DNA of known nucleotide sequence. Proc. Natl. Acad. Sci. USA 82:891-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Will, H., R. Cattaneo, H. G. Koch, G. Darai, H. Schaller, H. Schellekens, P. M. van Eerd, and F. Deinhardt. 1982. Cloned HBV DNA causes hepatitis in chimpanzees. Nature 299:740-742. [DOI] [PubMed] [Google Scholar]
- 42.Ye, M., C. S. Morello, and D. H. Spector. 2002. Strong CD8 T-cell responses following coimmunization with plasmids expressing the dominant pp89 and subdominant M84 antigens of murine cytomegalovirus correlate with long-term protection against subsequent viral challenge. J. Virol. 76:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]