Abstract
In common with other herpesviruses, the human cytomegalovirus (HCMV) DNA polymerase contains a catalytic subunit (Pol or UL54) and an accessory protein (UL44) that is thought to increase the processivity of the enzyme. The observation that antisense inhibition of UL44 synthesis in HCMV-infected cells strongly inhibits viral DNA replication, together with the structural similarity predicted for the herpesvirus processivity subunits, highlights the importance of the accessory protein for virus growth and raises the possibility that the UL54/UL44 interaction might be a valid target for antiviral drugs. To investigate this possibility, overlapping peptides spanning residues 1161 to 1242 of UL54 were synthesized and tested for inhibition of the interaction between purified UL54 and UL44 proteins. A peptide, LPRRLHLEPAFLPYSVKAHECC, corresponding to residues 1221 to 1242 at the very C terminus of UL54, disrupted both the physical interaction between the two proteins and specifically inhibited the stimulation of UL54 by UL44. A mutant peptide lacking the two carboxy-terminal cysteines was markedly less inhibitory, suggesting a role for these residues in the UL54/UL44 interaction. Circular dichroism spectroscopy indicated that the UL54 C-terminal peptide can adopt a partially α-helical structure. Taken together, these results indicate that the two subunits of HCMV DNA polymerase most likely interact in a way which is analogous to that of the two subunits of herpes simplex virus DNA polymerase, even though there is no sequence homology in the binding site, and suggest that the UL54 peptide, or derivatives thereof, could form the basis for developing a new class of anti-HCMV inhibitors that act by disrupting the UL54/UL44 interaction.
Human cytomegalovirus (HCMV) is a human pathogen responsible for a variety of severe diseases in immunocompromised patients, including pneumonia, gastrointestinal disease, and retinitis in transplant recipients and in patients with AIDS (7). HCMV is also a major cause of congenital defects in newborn children (51). Antiviral agents currently licensed for the treatment of HCMV infections include ganciclovir, foscarnet, and cidofovir, which all inhibit the HCMV DNA polymerase (21). Ganciclovir and cidofovir are nucleoside analogs which function as DNA chain terminators, whereas foscarnet inhibits viral DNA polymerase through binding to its pyrophosphate binding site (12). There is renewed interest in the search for new HCMV inhibitors because of the emergence of drug-resistant viral strains, particularly in immunocompromised patients (42), and because some of these antiviral agents, e.g., ganciclovir and foscarnet, have toxic side effects. Since the HCMV DNA polymerase represents a major target for antiviral chemotherapy, further studies on this enzyme could be important in developing novel drugs.
HCMV DNA replication has not been as fully characterized as that of herpes simplex virus (HSV); however, HCMV homologs to six of the seven proteins essential for HSV type 1 (HSV-1) replication (53) have been identified (41). These homologs include the two subunits of the DNA polymerase, which is composed of a catalytic subunit (Pol or UL54) (23, 28) and an accessory protein (UL44 or infected-cell protein 36) (17). The HCMV DNA polymerase has a number of characteristics in common with the HSV-1 counterpart. The UL54 protein, which is the homolog of HSV-1 UL30, possesses DNA-dependent DNA polymerase activity (33) as well as 3′-5′ exonuclease activity (38). Both these activities are dependent on salt concentration (11), in a manner very similar to that observed for the HSV-1 DNA polymerase (22). The UL44 accessory subunit is analogous to the HSV-1 UL42 protein, which is the processivity subunit of HSV DNA polymerase. Like HSV-1 UL42, HCMV UL44 has been shown to bind double-stranded DNA with high affinity and to stimulate the DNA polymerase activity, possibly by increasing the processivity of the enzyme (17, 52). Although the HCMV accessory protein has little sequence homology to UL42 and to the accessory proteins of other herpesviruses, it has been predicted to possess a similar “processivity fold” structure (54).
For the HSV-1 UL30/UL42 interaction, there is excellent evidence that the interaction is essential for viral replication and thus a valid target for antiviral drugs (13, 14, 48). Moreover, we recently showed that an oligopeptide corresponding to the C-terminal 27 amino acids (aa) of UL30, when delivered into HSV-1-infected cells by a protein carrier, is able to inhibit viral replication by disruption of the UL30/UL42 interaction (30). This study established proof of principle for blocking the HSV-1 DNA polymerase subunit interaction as an antiviral strategy. The observation that antisense inhibition of UL44 synthesis in HCMV-infected cells strongly inhibits viral DNA replication (43) highlights the importance of the accessory protein for virus growth.
UL54 and UL44 have been expressed both in an in vitro-coupled transcription-translation reticulocyte lysate system (11) and in insect cells infected with recombinant baculoviruses (16, 17). However, these approaches, due to low expression level (11) and relatively low yields and activity of the products (16), did not provide a reliable source of enzyme suitable for detailed studies. A functional analysis of the accessory subunit demonstrated that both the UL54- and the DNA-binding activity reside within the N-terminal 309 residues of the UL44 sequence (52). However, the catalytic subunit has not been subject to such a detailed analysis, nor has the UL44-binding site been identified.
In this work, we report the recloning of UL54 into a recombinant baculovirus and a new purification scheme for baculovirus-expressed UL44. Both purified proteins were recovered in yields sufficient for biochemical analysis. We also report the characterization of the physical and functional interaction between the catalytic and accessory subunit of HCMV DNA polymerase and the identification of peptides from the C terminus of UL54 which can efficiently disrupt the physical interaction between UL44 and UL54 and specifically inhibit the stimulation of UL54 by UL44. These findings could aid the development of a new strategy for anti-HCMV chemotherapy based on disruption of the UL54/UL44 interaction.
MATERIALS AND METHODS
Cells and viruses.
Spodoptera frugiperda (Sf9) cells were maintained in TC100 medium (Invitrogen Life Technologies) supplemented with 5% (vol/vol) fetal calf serum (FCS), penicillin (100 U/ml), streptomycin (100 μg/ml) and, neomycin (50 μg/ml). The Autographa californica nuclear polyhedrosis virus recombinant P9, expressing UL44 epitope-tagged at the C terminus with the 3-amino-acid motif EEF, was a gift from P. F. Ertl (GlaxoSmithKline, Stevenage, United Kingdom).
Recombinant baculovirus expressing HCMV gene UL54 under the control of the polyhedrin promoter was generated as follows. A 3.7-kb fragment spanning nucleotides 76863 to 80631 of HCMV DNA (9) and containing the HCMV UL54 open reading frame was amplified by PCR from a cloned copy of the HindIII F fragment of HCMV strain AD169. The primers used were 5′-ATTATCTAGACCGCTATGTTTTTCAACCCG-3′ and 5′-TATATCTAGACATCATCACCGTCCCCAGTCA-3′, which contained XbaI sites (underlined). The PCR-generated fragment was cleaved with XbaI and initially cloned into the XbaI site of pUC19. The XbaI fragment was then recloned into the XbaI site of the baculovirus transfer vector pAcYMX1 (47) downstream of the polyhedrin promoter to generate plasmid PY54. The entire XbaI fragment was sequenced to confirm the presence of the authentic UL54 gene. The transfer plasmid PY54 was cotransfected with Bsu36I-cleaved DNA (Clontech) of the parental baculovirus AcPAK6 (2) into Sf9 cells and resulting recombinant baculovirus AcUL54 was isolated as described by Kitts et al. (27). The presence of the desired UL54 gene was confirmed by Southern blot analysis. Preparation and titration of virus stocks have already been described (8, 35).
Purification of proteins. (i) UL54.
UL54 was purified first by phosphocellulose and then by DNA cellulose chromatography as described previously for the HSV-1 homolog UL30 (19, 34).
(ii) UL44.
For UL44, a new purification scheme was developed that did not depend on the epitope tag affinity chromatography previously used (17). Sf9 cells (2.5 × 109) were infected with recombinant baculovirus P9 at a multiplicity of infection of 10 PFU per cell and harvested 48 h postinfection. Infected cells were washed three times with cold phosphate-buffered saline (PBS), resuspended in 50 mM HEPES (pH 7.0)-0.25 M NaCl-1% Nonidet P-40 containing protease inhibitors (Complete protease inhibitor cocktail tablets, Roche Applied Science), and incubated on ice for 30 min. Insoluble material was removed by centrifugation for 1 h at 80,000 × g at 4°C. The cell extract was dialyzed extensively against 20 mM HEPES (pH 7.5)-10% glycerol-50 mM NaCl containing protease inhibitors (Complete protease inhibitor cocktail tablets; Roche Applied Science). Protein purification was conducted at room temperature with the assistance of an AKTApurifier fast protein liquid chromatography system (Amersham Pharmacia Biotech), and all fractions were collected at 0°C. For the first purification step, the protein extract was applied to a 10-ml double-stranded DNA cellulose column (Sigma) equilibrated with 20 mM Tris-HCl (pH 8.0)-50 mM NaCl-0.5 mM dithiothreitol-0.5 mM EDTA-10% glycerol. Proteins were eluted with a linear salt gradient from 50 mM to 0.7 M NaCl. The presence of UL44 in column fractions was determined by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) analysis. After dialysis against 20 mM Tris-HCl (pH 8.0) containing 0.5 mM dithiothreitol, 0.5 mM EDTA, 10% glycerol, and 50 mM NaCl plus protease inhibitors (Complete protease inhibitor cocktail tablets; Roche Applied Science), the sample was loaded onto a Mono Q (MQ) column (HR 5/5; Amersham Pharmacia Biotech) equilibrated with 20 mM Tris-HCl (pH 8.0)-50 mM NaCl and eluted with a gradient of 50 mM to 0.5 M NaCl in 20 mM Tris-HCl (pH 8.0). Fractions containing UL44 were pooled and diluted 1 in 10 with 50 mM Tris-HCl (pH 7.0)-1 M ammonium sulfate. The last step of purification utilized a phenyl-Sepharose hydrophobic interaction column (RESOURCE PHE; Amersham Pharmacia Biotech). Proteins were eluted with a 1 M to 50 mM ammonium sulfate gradient in 50 mM Tris-HCl (pH 7.0). Fractions were analyzed by SDS-10% PAGE, and those containing purified UL44 were pooled, aliquoted, and stored at −80°C until use.
Oligopeptides.
Peptides were synthesized by continuous-flow 9-fluorenylmethoxy carbonyl chemistry using standard protocols as previously described (36). Preparative reverse-phase high-pressure liquid chromatography was used to purify monomeric peptides (39). The molecular masses of the peptides were determined by matrix-assisted laser desorption-time of flight mass spectrometry and corresponded to the expected values. Peptides 6 and 19 of UL42 have been described previously (39). The 27-mer peptide corresponding to residues 1209 to 1235 of HSV-1 UL30 has also been previously described (34). Peptide concentrations were confirmed by quantitative amino acid analysis.
UL54-UL44 interaction enzyme-linked immunosorbent assay (ELISA).
Microtiter wells (Immulon 1; Dynatech) were coated with 0.2 μg of purified UL54 and blocked with 2% bovine serum albumin (Sigma) in PBS for 1 h. After washing with PBS containing 0.3% Tween 20, either different amounts of purified UL44 alone (dissolved in PBS) or 0.5 μg of UL44 mixed with peptides (dissolved in 100 mM Tris-HCl [pH 8.0] containing 0.1% Tween 20) was added and incubated for 1 h at 37°C. Following further washes, the wells were incubated with monoclonal antibody (MAb) YL1/2 (diluted 1:50 in PBS containing 2% FCS) for 1 h at 37°C. MAb YL1/2, which recognizes the EEF epitope inserted at the C terminus of UL44 (46), was purchased from Serotec Ltd. Plates were then washed and incubated with horseradish peroxidase (HRP)-conjugated anti-rat antibody (Sigma) (diluted 1:500 in PBS containing 2% FCS). After the washes, the chromogenic substrate 2,2′-azino-bis(3-ethylbenzthiazinzoline-6-sulfonic acid (ABTS; Dynatech) in citrate phosphate buffer (pH 4.0) containing 0.01% hydrogen peroxide was added, and absorbance was read at 405 nm on a Multiskan plate reader (Titertek, ICN Biomedicals).
DNA polymerase assays.
Stimulation of UL54 activity by UL44 was measured by incorporation of [3H]dTTP into either a poly(dA)-oligo(dT) or an activated calf thymus DNA template. One hundred femtomoles of purified UL54 and 200 to 400 fmol of purified UL44 protein were mixed and preincubated at 37°C for 30 min. The reaction was initiated by addition of a reaction mixture containing 75 mM Tris-HCl (pH 8.0), 6.5 mM MgCl2, bovine serum albumin (400 μg/ml), 1.67 mM β-mercaptoethanol, 150 mM KCl, 1.6 μM [3H]dTTP, and either poly(dA)-oligo(dT) (10 μg/ml; Amersham Pharmacia Biotech) or activated calf thymus DNA (125 μg/ml; Amersham Pharmacia Biotech) plus a 40 μM concentration (each) of dATP, dCTP, and dGTP. Samples were taken after 10, 20, and 30 min of incubation at 37°C and spotted onto DE81 filters (Whatman), previously soaked in 0.1 M EDTA and air dried. The filters were washed three times in 5% Na2HPO4, two times in water, and two times in methanol and then were dried. Radioactivity was measured with an LKB 1025 scintillation counter.
DNA precipitation assay.
Activated calf thymus DNA (Sigma) was labeled with 32P, incubated with peptide for 15 min, and then centrifuged to pellet any DNA-peptide complexes as described (39).
Circular dichroism (CD) spectroscopy.
Lyophilized peptides were resuspended in water and then diluted in 50 mM phosphate buffer (pH 8.0) or in 20 or 95% (vol/vol) 2,2,2-trifluoroethanol (TFE) (Fluka)-H2O solution at a peptide concentration of 50 μM. Wavelength scans were recorded with a Jasco J-715 spectropolarimeter at 20°C in a 0.1-cm-path-length cuvette. All spectra were recorded in the wavelength range 195 to 255 nm using 2-nm bandwidth and 1 s of time constant at a scan speed of 50 nm/min. The signal-to-noise ratio was improved by accumulation of at least 10 scans. All spectra are reported in terms of mean residue molar ellipticity [θ]R (degrees · centimeter2 · decimole−1). Data processing was carried out using a J-700 software package.
To analyze secondary structure content in peptides, three different methods (3, 10, 20) have been used. The results provided by all the methods were consistent with each other in the limit of experimental errors.
RESULTS
UL54-UL44 physical interaction.
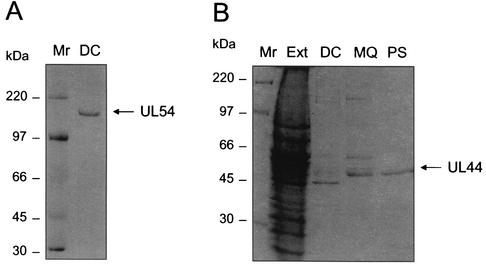
In order to detect the physical interaction between UL54 and UL44, an interaction ELISA was developed using proteins purified from insect cells infected with recombinant baculoviruses. UL54 was purified according to the procedure previously used for the HSV-1 homolog UL30 (19, 34), which yielded a stable and active protein. SDS-PAGE analysis of the purified protein demonstrated the presence of a protein band of the expected molecular mass (Fig. 1A). To provide additional evidence for the authenticity of the expressed UL54 protein, an antiserum (designated 144) was raised in rabbits against the C-proximal 15 aa (residues 1226 to 1240) of the predicted UL54 sequence (9). The peptide was made as a multiply antigenic peptide (49) with the structure (HLEPAFLPYSVKAHE)4K3A, as such peptides have been shown to generate sera with higher antiprotein titers (36). The antiserum, but not the preimmune serum, reacted with the expressed protein (data not shown).
FIG. 1.
Purified UL44 and UL54 preparations. Proteins were expressed in insect cells infected with recombinant baculoviruses and purified by column chromatography as described in Materials and Methods. Samples after the final step of UL54 purification (A) and after each step of UL44 purification (B) were analyzed on a 5% or 10% polyacrylamide SDS-PAGE gel, respectively. Abbreviations: Mr, marker; Ext, protein extract from Sf9 cells infected with recombinant baculovirus P9; DC, after double-stranded DNA cellulose column; MQ, after MQ column; PS, after phenyl-Sepharose column. The position of the proteins (UL54 and UL44) and the molecular masses (in kilodaltons) of the markers are indicated on the right and left, respectively.
For UL44, a new purification scheme was developed that did not depend on the epitope-tag affinity chromatography previously used (17). The new procedure involved (i) double-stranded DNA cellulose, (ii) MQ ion-exchange, and (iii) phenyl-Sepharose hydrophobic interaction chromatography. This approach for purification of UL44, which is less expensive than that previously reported (17) but still allows rapid and efficient isolation of baculovirus-expressed UL44 protein, yielded a highly pure and stable protein. SDS-PAGE analysis of the product after each purification step is shown in Fig. 1B.
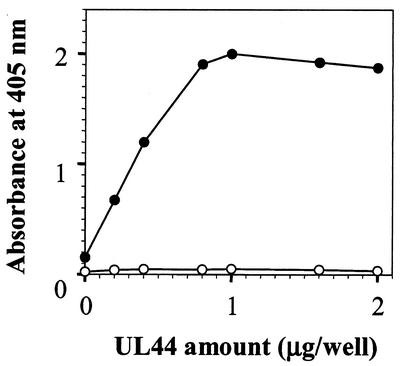
In the interaction ELISA we developed, UL44 binding to UL54-coated microtiter wells was detected using MAb YL1/2, which recognizes the EEF epitope inserted at the C terminus of UL44 (46). In preliminary experiments, the amount of UL54, UL44, YL1/2, and anti-rat HRP-conjugated antibody were optimized. Figure 2 shows that the amount of YL1/2 bound was dependent on both the presence of UL54 and UL44, since no signal was detected in UL54-uncoated wells after addition of UL44. The figure also shows that, for amounts of UL44 up to approximately 1 μg/well, an increase in the amount of UL44 produced an increase in absorbance and that the relation between the absorbance and the amount of UL44 added was linear up to 0.8 μg. These results confirm that UL54 and UL44 can form a stable complex and demonstrate that no other HCMV protein is required for the UL54/UL44 complex formation.
FIG. 2.
UL54/UL44 interaction assay. The UL44 protein was added to microtiter wells precoated with UL54 (0.2 μg) (•) or uncoated wells (○). Bound UL44 was detected with MAb YL1/2, which was in turn detected with an HRP-conjugated anti-rat antibody.
The physical interaction between UL54 and UL44 can be blocked by peptides from the C terminus of UL54.
To investigate whether the C terminus of UL54 plays a role in the interaction with UL44, as previously reported for the interaction of HSV-1 homolog UL30 with UL42 (5, 6, 13, 15, 34, 48, 50, 54), overlapping peptides spanning residues 1161 to 1242 of UL54 (Table 1) were synthesized and tested for inhibition of the physical interaction between UL54 and UL44 using the interaction ELISA described above.
TABLE 1.
Peptides used in this study
| Peptide no. | Corresponding residues in UL54 | Sequence | IC50a (μM) |
|---|---|---|---|
| 1 | 1221-1242 | LPRRLHLEPAFLPYSVKAHECC | 11 |
| 2 | 1206-1225 | PGGETARKDKFLHMVLPRRL | >500 |
| 3 | 1191-1210 | EQVLKAVTNVLSPVFPGGET | 280 |
| 4 | 1176-1195 | YVREHGVPIHADKYFEQVLK | >500 |
| 5 | 1161-1180 | PPSAVCNYEVAEDPSYVREH | >500 |
| 6 | 1223-1242 | RRLHLEPAFLPYSVKAHECC | 20 |
| 7 | 1221-1241 | LPRRLHLEPAFLPYSVKAHEC | 75 |
| 8 | 1221-1240 | LPRRLHLEPAFLPYSVKAHE | >500 |
Concentration of peptide required to reduce UL44 binding by 50%.
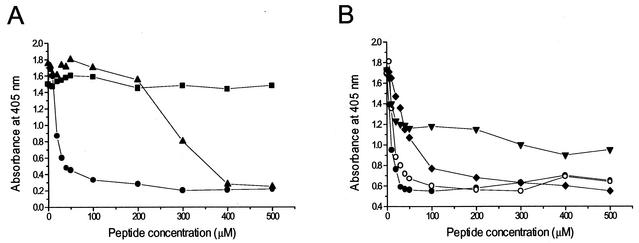
Our results indicated that peptide 1, corresponding to the C-terminal 22 aa of UL54, was the most potent inhibitory peptide, with a 50% inhibitory concentration (IC50) for inhibition of the UL54/UL44 interaction of 11 μM (Fig. 3A). Of the other peptides, only peptide 3 showed some degree of inhibition, even though it was markedly less potent than peptide 1. A peptide slightly shorter than peptide 1 (peptide 6) had an inhibitory effect similar to that of peptide 1 (Fig. 3B). All the other peptides did not significantly inhibit the UL54/UL44 interaction (Fig. 3A and data not shown). These results indicate that the C terminus of UL54 plays a role in its physical interaction with the accessory protein UL44.
FIG. 3.
Inhibition of the physical interaction between UL54 and UL44 by C-proximal and C-terminal UL54 peptides as measured by an interaction ELISA. (A) Inhibitory activity of peptides corresponding to residues 1221 to 1242 (peptide 1 [•]), 1191 to 1210 (peptide 3 [▴]), and 1161 to 1180 (peptide 5 [▪]) of UL54. (B) Inhibitory activity of peptides corresponding to residues 1221 to 1242 (peptide 1 [•]), 1223 to 1242 (peptide 6 [○]), 1221 to 1241 (peptide 7 [⧫]), and 1221 to 1240 (peptide 8 [▾]) of UL54.
The inhibitory UL54 C-terminal peptide possesses two carboxy-terminal cysteines that are unusual among the C termini of herpesvirus DNA polymerases. In order to investigate whether the two carboxy-terminal cysteines are important for the disrupting activity of the C-terminal peptide, peptides 7 and 8 were also synthesized, which lack the last one or both the C-terminal cysteines, respectively. As shown in Fig. 3B, peptide 7 was still inhibitory, but its IC50 was higher than that of peptide 1. Peptide 8 was still less inhibitory and the highest concentration of peptide used still left more than half the activity. Therefore, the C-terminal cysteines are important for the inhibitory activity of the UL54 peptide, suggesting a role for these residues in the UL54/UL44 interaction.
Peptides from the C terminus of UL54 specifically inhibit the stimulation of UL54 activity by UL44.
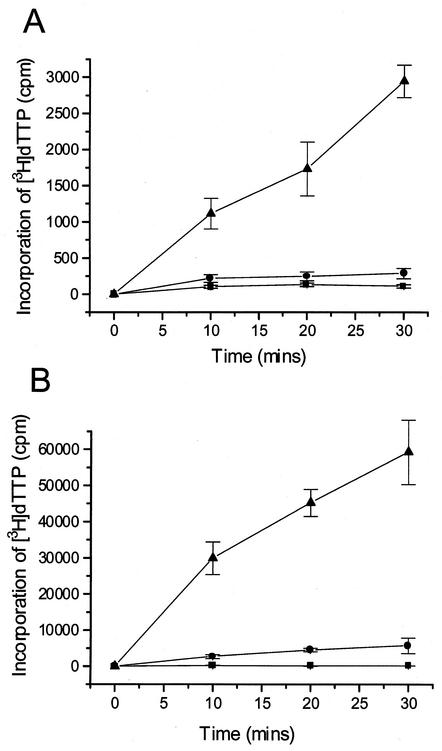
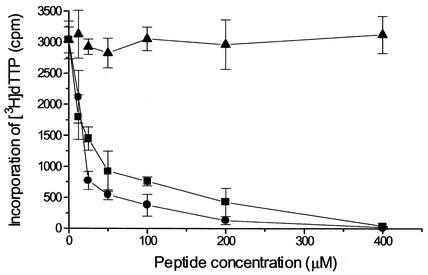
The peptides corresponding to the C-terminal region of UL54 were also tested for their ability to interfere with the functional interaction of UL54/UL44. The functional interaction was measured by the incorporation of [3H]dTTP into a poly(dA)-oligo(dT) or into an activated calf thymus DNA template. Based on several experiments, the UL44 protein stimulated the incorporation activity of UL54 by between 2- and 10-fold with poly(dA)-oligo(dT) as a template and by between 3- and 10-fold with calf thymus DNA as a template. One such experiment is shown in Fig. 4. Peptide 1, corresponding to the C-terminal residues 1221 to 1242 of UL54, inhibited the activity of the UL54/UL44 complex with an IC50 of approximately 20 μM (Fig. 5). At a concentration of 400 μM, this peptide completely inhibited the holoenzyme. Similar inhibition was observed using activated calf thymus DNA as a template (data not shown). The inhibitory effect was peptide-specific, since a control peptide corresponding to the C-terminal residues 1209 to 1235 of HSV-1 UL30 had no effect on the HCMV enzyme (Fig. 5). In addition, HCMV peptide 1 did not inhibit DNA synthesis activity of HSV-1 UL30/UL42 complex (data not shown). Surprisingly, peptide 5, which was inactive in the physical interaction assay (Fig. 3), was strongly inhibitory in the functional assay (IC50 of approximately 15 μM, Fig. 5). All the other peptides showed no significant inhibitory activity (IC50 > 500 μM; data not shown).
FIG. 4.
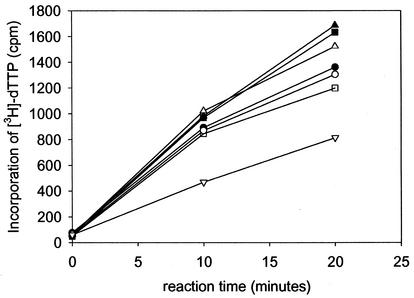
Stimulation of the activity of the catalytic subunit UL54 by the polymerase accessory protein UL44. The effect of UL44 on UL54 activity was examined by measuring incorporation of [3H]dTTP into two different templates: poly(dA)-oligo(dT) (A) and activated calf thymus DNA (B). Symbols: ▪, no protein; •, UL54; ▴, UL54 + UL44. Error bars show standard deviations.
FIG. 5.
Inhibition of the UL54-UL44 functional interaction by C-terminal UL54 peptides. Effect of synthetic peptides on incorporation of [3H]dTTP into a poly(dA)-oligo(dT) by UL54/UL44 complex was tested. The peptides correspond to residues 1221 to 1242 (▪) and residues 1161 to 1180 (•) of HCMV UL54. A peptide corresponding to C-terminal 27 residues of HSV-1 UL30 (aa 1209 to 1235 [▴]) was used as a control. The graph shows the average of data from three independent experiments together with the standard deviation (error bars).
To investigate whether the HCMV peptides might inhibit the HCMV polymerase by inhibiting the activity of the catalytic subunit rather than its interaction with UL44, we tested the effect of the peptides on incorporation of [3H]dTTP into the poly(dA)-oligo(dT) template by UL54 alone. Peptide 1 only weakly inhibited the catalytic subunit with an IC50 of approximately 400 μM (Fig. 6), a value 20-fold higher than that observed for the holoenzyme (Fig. 5). Peptide 5 also showed no significant inhibitory activity on UL54 alone (data not shown). We conclude that inhibition of the UL54/UL44 activity is not due to inhibition of the catalytic subunit.
FIG. 6.
Effect of the UL54 C-terminal peptide on activity of the catalytic subunit of HCMV DNA polymerase. The incorporation of [3H]dTTP into a poly(dA)-oligo(dT) template by UL54 alone was measured in the presence of different concentrations of peptide 1, corresponding to residues 1221 to 1242 of UL54. Symbols: ▴, no peptide; ▪, 12.5 μM; ▵, 25 μM; •, 50 μM; ○, 100 μM; □, 200 μM; ▿, 400 μM.
In order to determine whether the peptides interfered with the functional interaction of UL54 and UL44 by binding to and sequestering DNA template, their ability to precipitate DNA was tested using peptide concentrations of 100 μM. The results are shown in Table 2. Peptides 6 and 19 from UL42 were used as controls and, as previously reported (39), peptide 19 precipitated DNA, whereas peptide 6 did not. Neither peptide 1 nor peptide 5 from C terminus of HCMV UL54 precipitated DNA at the concentration tested. We conclude that these UL54 C-terminal and -proximal peptides do not interact with DNA and thus their inhibitory effect on HCMV DNA polymerase activity is not mediated by sequestering the template.
TABLE 2.
Precipitation of DNA by peptides
| Peptide | Description | % DNA remaining in solution (mean ± SD) |
|---|---|---|
| None | 100a | |
| 19b | Residues 89-102 of HSV-1 UL42 | 4 ± 1 |
| 6b | Residues 23-28 of HSV-1 UL42 | 109 ± 10 |
| 1 | Residues 1221-1242 of HCMV UL54 | 98 ± 9 |
| 5 | Residues 1161-1180 of HCMV UL54 | 102 ± 9 |
The radioactivity remaining in solution in the absence of peptide was 42,098 cpm/10 μl.
Peptides previously described by Owsianka et al. (39).
Analysis of the secondary structure of the UL54 C-terminal peptide.
CD and nuclear magnetic resonance studies on UL30 C-terminal peptides indicated that these peptides are partially helical and suggested that C terminus of HSV-1 Pol could fold into a helix-loop-helix structure (5, 15). We therefore investigated whether the UL54 C-terminal peptide possesses a similar secondary structure.
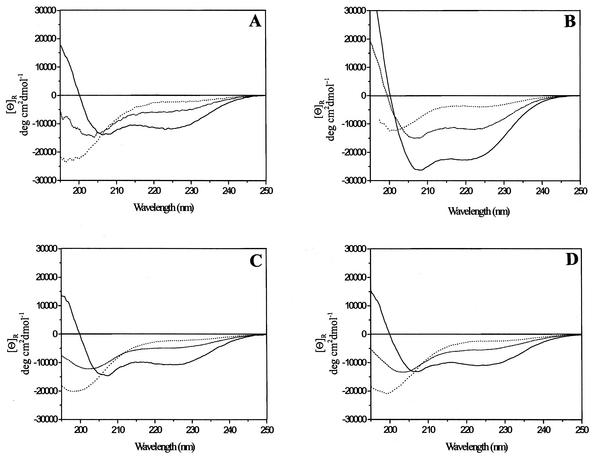
The CD spectrum of peptide 1 in aqueous solution showed a profile characteristic of a mostly random coil conformation (26), with a weak maximum at 218 nm and an intense minimum at 195 nm (Fig. 7A, dotted line). However, on the basis of the Chou-Fasman theoretical methods for secondary structure prediction, a few residues of both the N and C terminus of UL54 peptide were expected to be helical, while the central part of the sequence had higher probability to give turn rather than helix as a consequence of the presence of two proline residues in this region. Therefore, in an attempt to determine whether the UL54 C-terminal peptide could adopt an ordered structure, we investigated the conformational properties of peptide 1 in TFE, a secondary structure-inducing solvent. TFE is the most commonly used agent for stabilizing α-helical structure in peptides (29, 44). This solvent does not induce secondary structure indiscriminately, but regions of the polypeptide chain that are helical in the native state are stabilized preferentially (25, 37, 45). In light of these issues, we studied the effect of increasing amount of TFE on the chiroptical properties of peptide 1.
FIG. 7.
Analysis of the structure of UL54 inhibitory peptide. CD spectra of peptide 1 (A), corresponding to residues 1221 to 1242 of HCMV UL54, of a 27-mer peptide corresponding to residues 1209 to 1235 of HSV-1 UL30 (B) and of peptide 7 (C) and peptide 8 (D), corresponding to residues 1221 to 1241 and 1221 to 1240 of HCMV UL54, respectively. CD spectra of peptides in 50 mM phosphate buffer (pH 8.0) (dotted line), in 20% (vol/vol) TFE-water (dashed line), and in 95% (vol/vol) TFE-water (solid line) are reported. Double minima at 222 and 208 nm and a maximum at 192 nm are characteristic of α-helix spectra, while a weak maximum at 215 nm and an intense minimum at 195 nm are characteristic of random coils (26).
The CD spectrum of peptide 1 in 20% (vol/vol) TFE-water showed a partially ordered structure (Fig. 7A, dashed line), with a helical content of ∼14%. The helicity of the peptide increased upon further addition of TFE, reaching a maximum in 95% (vol/vol) TFE-water (Fig. 7A, solid line). Under this condition, a helical content of ∼38% was estimated, which approximately corresponds to 8 residues being involved in such conformation. The degree of helicity of peptide 1 did not vary substantially over the concentration range of 10 to 100 μM (data not shown), suggesting that the peptide does not aggregate in solution. CD spectra of peptide 7 and peptide 8 (Fig. 7C and D), which lack the last or both the two C-terminal cysteines, respectively, showed levels of α-helix substantially similar to those of peptide 1, suggesting that the deletion of the C-terminal cysteines does not cause a significant structural change in peptide 1 structure.
For a comparison, CD spectra of a peptide corresponding to the C-terminal 27 residues of UL30 were recorded under the same conditions. In aqueous solution, such a peptide is not completely random, showing a helical content of 11% (Fig. 7B, dotted line). As observed for the UL54 C-terminal peptide, the helicity of UL30 peptide increases upon addition of TFE, with 33 and 70% of α-helical content in 20% and 95% TFE-water, respectively (Fig. 7B, dashed and solid line).
DISCUSSION
In this work we show the inhibition of HCMV DNA polymerase by peptides that mimic the binding site of the catalytic subunit, UL54, to the accessory protein, UL44. To our knowledge, this is the first report demonstrating successful inhibition of HCMV DNA polymerase by disruption of the interaction between the two enzyme subunits. Although this new antiviral strategy has been already proposed for inhibition of HSV-1 DNA polymerase by Marsden and coworkers (30, 32, 34) and others (5, 6, 15), no evidence existed that a similar strategy could be feasible also for the HCMV DNA polymerase. Therefore, we decided to investigate the HCMV Pol-UL44 interaction both for mechanistic information and because we believe that it could be an attractive target for antiviral drugs.
In order to localize the UL44-binding site on HCMV Pol, overlapping peptides spanning C-terminal region of UL54 were synthesized and tested for inhibition of the interaction between UL54 and UL44 by two independent methods. First, we investigated the importance of sequences within the extreme C terminus of UL54 in its physical interaction with UL44 by means of an interaction ELISA. We found that a peptide corresponding to the C-terminal 22 aa of UL54 (residues 1221 to 1242) blocked the physical interaction between the two proteins. The concentration of peptide required to inhibit the UL54/UL44 physical interaction by 50% was approximately 11 μM, a value comparable to that observed, 2 to 30 μM, for inhibition of the HSV-1 UL30/UL42 interaction by UL30 C-terminal peptides (15, 34). Interestingly, the UL54 C terminus presents two carboxy-terminal cysteines which are unusual among herpesvirus DNA polymerases and are important for the inhibitory activity of the peptide. Removal of one cysteine from the extreme C terminus of peptide 1 diminished the capability of the peptide to interfere with the physical interaction between HCMV Pol and UL44, while deletion of the two cysteines substantially impaired the inhibitory activity of the peptide, while having no effect on peptide structure as evidenced by CD spectroscopy. These results strongly suggest that these residues of UL54 are directly involved in the interaction with UL44, whether by binding to UL44 directly, or by interaction with some other region of UL54 to stabilize a structure within UL54 itself that can then bind to UL44. Nevertheless, although the extreme C terminus of UL54 seems to be highly important for binding to UL44, upstream residues may still play a role.
Next, we analyzed the ability of the peptides spanning the UL54 C-terminal region to inhibit enzyme activity in vitro. The results demonstrated that the C-terminal 22-mer also efficiently inhibits the UL54/UL44 functional interaction. The concentration of C-terminal peptide required to inhibit HCMV Pol/UL44 activity by 50% was very similar to the IC50 for inhibition of the physical interaction (20 versus 11 μM). Inhibition of the HCMV Pol/UL44 activity by C-terminal peptide appeared to be specific since at concentration as high as 1 mM, the HCMV peptide did not inhibit the HSV-1 Pol/UL42 interaction (data not shown), and a peptide corresponding to the 27 C-terminal residues of HSV-1 Pol (aa 1209 to 1235) did not inhibit the HCMV Pol/UL44 interaction. The observation that the HCMV peptide 1 inhibited the functional UL54/UL44 interaction is most simply interpreted by the direct involvement of the corresponding regions of UL54 in UL44 binding. Our data indicate that inhibition of the functional interaction was not due to the peptide binding to the DNA template, nor was it a consequence of the peptide inhibiting HCMV Pol in the absence of UL44.
A surprising finding was that a peptide comprising residues 1161 to 1180 of UL54 (peptide 5) also inhibited the activity of the HCMV Pol/UL44 complex (IC50 15 μM), having no significant effect on the physical interaction. The mechanism of inhibition of HCMV DNA polymerase by peptide 5 remains to be explained, since we found no evidence that the inhibitory effect was due to dissociation of the UL54/UL44 interaction, binding to DNA, or inhibition of UL54 catalytic activity. We speculate that following interaction with UL44, UL54 undergoes some conformational change that cannot occur in the presence of peptide 5. Peptide 5 corresponds to a C-proximal region of UL54 which not only shares considerable sequence homology with corresponding regions of other herpesvirus DNA polymerases (1), but also contains a highly conservedmotif, (N/-)(Y/-)(E/D/Q)X(A/S)EDPX(Y/H/F)(V/A)XX(H/N/L), corresponding to residues 1167 to 1180 (in which X is any amino acid and a dash indicates the absence of an amino acid), found also in eukaryotic DNA polymerase δ (4, 24). Such conserved features make very unlikely that such a peptide or peptidomimetic derivatives could represent the basis for clinically useful compounds, since they are unlikely to be specific for the viral enzyme.
Our findings on the interaction between HCMV Pol and UL44 highlight several features that are reminiscent of the interaction between HSV-1 Pol and UL42. First, the extreme C terminus of HCMV Pol is crucial for interacting with UL44, as is the C terminus of HSV-1 for its interaction with UL42. Remarkably, despite this functional analogy of the carboxy terminus of HCMV Pol to that of HSV-1 Pol, there is almost no sequence homology, and neither the HSV-1 nor the HCMV catalytic subunits are stimulated by the noncognate accessory protein (data not shown). This provides an interesting example of evolution of protein-protein interactions. Noteworthy, the extreme C-terminal region of HCMV Pol is not highly conserved among any other herpesvirus or cellular DNA polymerase, making it particularly attractive as a starting point for the de novo design of new specific anti-HCMV drugs. Second, the C-terminal region of UL54 can fold into a helical structure, as already reported for the C terminus of HSV-1 UL30 (5, 54). In fact, CD spectroscopy of peptide 1 in TFE-water indicated that it has some propensity to adopt a partially α-helical structure. The helicity of the peptide was apparently independent of peptide concentration suggesting that it is monomeric, as was found for the C-terminal UL30 peptides (15). Although we cannot exclude the possibility that the C-terminal 22 residues of HCMV Pol fold into a different conformation in the intact protein, it would be surprising if the α-helical character of the region was not preserved. In fact, a comparison of the nuclear magnetic resonance results on the UL30 C-terminal peptides (5) with the crystal structure of an UL30 C-terminal peptide bound to UL42 (54), indicates that also this peptide is less well ordered in solution and becomes better ordered upon binding with UL42. Third, we recently demonstrated the presence of a functional nuclear localization signal, RRLHL, corresponding to residues 1223 to 1227 of HCMV UL54, within the C-terminal inhibitory peptide, and a functionally equivalent sequence, corresponding to residues RRMLHR, was found also in the C terminus of the catalytic subunit UL30 of HSV-1 (31). Fourth, like HSV-1 UL42, HCMV UL44 is a DNA-binding protein (18) that is essential for viral replication (40, 43), specifically associates with Pol, and stimulates long-chain DNA synthesis (17, 52). The Pol- and DNA-binding activities of UL44 reside within the N-terminal two-thirds; as well as for UL42, the C terminus is dispensable (52). Fold recognition programs predict that the overall structure of HCMV UL44 resembles that of UL42, again despite a lack of sequence homology (54).
Taken together, these observations indicate that HCMV Pol and UL44 most likely interact in a way which is analogous to that of HSV-1 Pol and UL42, suggesting that the UL54/UL44 interaction could be an excellent target for the development of new drugs as well as the UL30/UL42 interaction. A route to rational drug design stems from our identification of a peptide that specifically inhibits DNA synthesis by HCMV DNA polymerase. This finding heralds the prospect that such a peptide, or shorter derivatives, could form the basis for the synthesis of peptidomimetics that inhibit HCMV replication. Encouragement for this approach has come from recent studies based on mutational and calorimetric analysis of UL30/UL42 interaction (5, 6), suggesting that disruption of one or a few hydrogen bonds, which could be accomplished by a small molecule, is sufficient to disrupt the interaction. This suggestion found recent support with the identification of small inhibitory molecules able to block this functional interaction in vitro as well as virus replication (B. D. Pilger, C. Cui, and D. M. Coen, personal communication). It is our hope that this route will lead to the discovery of clinically useful anti-HCMV drugs and will suggest similar strategies for inhibiting other herpesvirus DNA polymerases or, more in general, other viral targets (32).
Acknowledgments
We thank Peter F. Ertl for kindly providing the recombinant baculovirus P9. We also thank Sun Yi for cloning in preparation of the recombinant baculovirus AcUL54, Alessandro Case for assistance with CD spectroscopy, and Medhat Khattar and William Turnell for analysis of sequences homologous to peptide 5. We gratefully acknowledge Donald M. Coen for communicating results prior to publication and for critical reading of the manuscript.
This work was supported by program funding from the Medical Research Council (United Kingdom) to H.S.M., by grant 40C.68 from the Istituto Superiore di Sanità of Italy, by PRIN-EX40% (2001-2002), by FIRB and 1% project I.R.C.C.S. San Matteo (2001) to G.P., and by Progetto di Ricerca di Ateneo (2000) and MURST EX60% to A.L.
REFERENCES
- 1.Berthomme, H., S. J. Monahan, D. S. Parris, B. Jacquemont, and A. Epstein. 1995. Cloning, sequencing, and functional characterization of the two subunits of the pseudorabies virus DNA polymerase holoenzyme: evidence for specificity of interaction. J. Virol. 69:2811-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, D. H. 1992. Baculovirus expression vectors. Semin. Virol. 3:253-264. [Google Scholar]
- 3.Bohm, G., R. Muhr, and R. Jaenicke. 1992. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 5:191-195. [DOI] [PubMed] [Google Scholar]
- 4.Boulet, A., M. Simon, G. Faye, G. A. Bauer, and P. M. Burgers. 1989. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 8:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridges, K. G., Q. Hua, M. R. Brigham-Burke, J. D. Martin, P. Hensley, C. E. Dahl, P. Digard, M. A. Weiss, and D. M. Coen. 2000. Secondary structure and structure-activity relationships of peptides corresponding to the subunit interface of herpes simplex virus DNA polymerase. J. Biol. Chem. 275:472-478. [DOI] [PubMed] [Google Scholar]
- 6.Bridges, K. G., C. S. Chow, and D. M. Coen. 2001. Identification of crucial hydrogen-bonding residues for the interaction of herpes simplex virus DNA polymerase subunits via peptide display, mutational, and calorimetric approaches. J. Virol. 75:4990-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2492-2523. In B. N. Fields, D. M. Knipe, P. M. Howlet, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 8.Brown, M., and P. Faulkner. 1977. A plaque assay for nuclear polyhedrosis viruses using a solid overlay. J. Gen. Virol. 36:361-364. [Google Scholar]
- 9.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. H., J. T. Yang, and K. H. Chau. 1974. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry 13:3350-3359. [DOI] [PubMed] [Google Scholar]
- 11.Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1997. Expression of the catalytic subunit (UL54) and the accessory protein (UL44) of human cytomegalovirus DNA polymerase in a coupled in vitro transcription/translation system. Protein Expr. Purif. 11:209-218. [DOI] [PubMed] [Google Scholar]
- 12.Coen, D. M. 1992. Molecular aspects of antiherpesvirus drugs. Semin. Virol. 3:3-12. [Google Scholar]
- 13.Digard, P., W. Bebrin, K. Weisshart, and D. M. Coen. 1993. The extreme C-terminus of herpes simplex virus DNA polymerase is crucial for functional interaction with processivity factor UL42 and for viral replication. J. Virol. 67:398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Digard, P., C. S. Chow, L. Pirrit, and D. M. Coen. 1993. Functional analysis of the herpes simplex virus UL42 protein. J. Virol. 67:1159-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Digard, P., K. P. William, P. Hensley, I. S. Brooks, C. E. Dahl, and D. M. Coen. 1995. Specific inhibition of herpes simplex virus DNA polymerase by helical peptides corresponding to the subunit interface. Proc. Natl. Acad. Sci. USA 92:1456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ertl, P. F., M. S. Thomas, and K. L. Powell. 1991. High level expression of DNA polymerases from herpesviruses. J. Gen. Virol. 72:1729-1734. [DOI] [PubMed] [Google Scholar]
- 17.Ertl, P. F., and K. L. Powell. 1992. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J. Virol. 66:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson, W., T. L. Murphy, and C. Roby. 1981. Cytomegalovirus-infected cells contain a DNA-binding protein. Virology 111:251-262. [DOI] [PubMed] [Google Scholar]
- 19.Gottlieb, J., A. I. Marcy, D. M. Coen, and M. D. Challberg. 1990. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J. Virol. 64:5976-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenfield, N., and G. D. Fasman. 1969. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8:4108-4116. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths, P. D. 2001. Cytomegalovirus therapy: current constraints and future opportunities. Curr. Opin. Infect. Dis. 14:765-768. [DOI] [PubMed] [Google Scholar]
- 22.Hart, G. J., and R. E. Boehme. 1992. The effect of the UL42 protein on the DNA polymerase activity of the catalytic subunit of the DNA polymerase encoded by herpes simplex virus type 1. FEBS Lett. 305:97-100. [DOI] [PubMed] [Google Scholar]
- 23.Heilbronn, R., G. Jahn, A. Burkle, U. K. Freese, B. Fleckenstein, and H. zur Hausen. 1987. Genomic localization, sequence analysis, and transcription of the putative human cytomegalovirus DNA polymerase gene. J. Virol. 61:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, J., and D. K. Brathwaite. 1991. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 19:4045-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jasanoff, A., and A. R. Fersht. 1994. Quantitative determination of helical propensities from trifluoroethanol titration curves. Biochemistry 33:2129-2135. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, W. C. 1990. Protein secondary structure and circular dichroism: a practical guide. Proteins 7:205-214. [DOI] [PubMed] [Google Scholar]
- 27.Kitts, P. A., M. D. Ayres, and R. D. Possee. 1990. Linearisation of baculovirus DNA enhances recovery of the recombinant virus expression vectors. Nucleic Acids Res. 18:5667-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouzarides, T., A. T. Bankier, S. C. Satchwell, K. Weston, P. Tomlinson, and B. G. Barrell. 1987. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J. Virol. 61:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehrman, S. R., J. L. Tuls, and M. Lund. 1990. Peptide alpha-helicity in aqueous trifluoroethanol: correlations with predicted alpha-helicity and the secondary structure of the corresponding regions of bovine growth hormone. Biochemistry 29:5590-5596. [DOI] [PubMed] [Google Scholar]
- 30.Loregian, A., E. Papini, B. Satin, H. S. Marsden, T. R. Hirst, and G. Palù. 1999. Intranuclear delivery of an antiviral peptide mediated by the B subunit of Escherichia coli heat-labile enterotoxin. Proc. Natl. Acad. Sci. USA 96:5221-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loregian, A., E. Piaia, E. Cancellotti, E. Papini, H. S. Marsden, and G. Palù. 2000. The catalytic subunit of herpes simplex virus type 1 DNA polymerase contains a nuclear localization signal in the UL42-binding region. Virology 273:139-148. [DOI] [PubMed] [Google Scholar]
- 32.Loregian, A., H. S. Marsden, and G. Palù. 2002. Protein-protein interactions as targets for antiviral chemotherapy. Rev. Med. Virol. 12:239-262. [DOI] [PubMed] [Google Scholar]
- 33.Mar, E. C., P. C. Patel, and E. S. Huang. 1981. Human cytomegalovirus-associated DNA polymerase and protein kinase activities. J. Gen. Virol. 57:149-156. [DOI] [PubMed] [Google Scholar]
- 34.Marsden, H. S., M. Murphy, G. L. McVey, K. A. MacEachran, A. M. Owsianka, and N. D. Stow. 1994. Role of the carboxy terminus of herpes simplex virus type 1 DNA polymerase in its interaction with UL42. J. Gen. Virol. 75:3127-3135. [DOI] [PubMed] [Google Scholar]
- 35.Matsuura, Y., R. D. Possee, H. A. Overton, and D. H. Bishop. 1987. Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J. Gen. Virol. 68:1233-1250. [DOI] [PubMed] [Google Scholar]
- 36.McLean, G. W., A. M. Owsianka, J. H. Subak-Sharpe, and H. S. Marsden. 1991. Generation of anti-peptide and anti-protein sera: effect of peptide presentation on immunogenicity. J. Immunol. Methods 137:149-157. [DOI] [PubMed] [Google Scholar]
- 37.Nelson, J. W., and N. R. Kallenbach. 1989. Persistence of the alpha-helix stop signal in the S-peptide in trifluoroethanol solutions. Biochemistry 28:5256-5261. [DOI] [PubMed] [Google Scholar]
- 38.Nishiyama, Y., K. Maeno, and S. Yoshida. 1983. Characterization of human cytomegalovirus-induced DNA polymerase and the associated 3′-to-5′ exonuclease. Virology 124:221-231. [DOI] [PubMed] [Google Scholar]
- 39.Owsianka, A. M., G. Hart, M. Murphy, J. Gottlieb, R. Boehme, M. Challberg, and H. S. Marsden. 1993. Inhibition of herpes simplex virus type 1 DNA polymerase activity by peptides from the UL42 accessory protein is largely nonspecific. J. Virol. 67:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reusser, P. 1996. Herpesvirus resistance to antiviral drugs: a review of the mechanisms, clinical importance and therapeutic options. J. Hosp. Infect. 33:235-248. [DOI] [PubMed] [Google Scholar]
- 43.Ripalti, A., M. C. Boccuni, F. Campanini, and M. P. Landini. 1995. Cytomegalovirus-mediated induction of antisense mRNA expression to UL44 inhibits virus replication in astrocytoma cell line: identification of an essential gene. J. Virol. 69:2047-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanford, D. G., C. Kanagy, J. L. Sudmeier, B. C. Furie, B. Furie, and W. W. Bachovchin. 1991. Structure of the propeptide of prothrombin containing the gamma-carboxylation recognition site determined by two-dimensional NMR spectroscopy. Biochemistry 30:9835-9841. [DOI] [PubMed] [Google Scholar]
- 45.Sonnichsen, F. D., J. E. Van Eyk, R. S. Hodges, and B. D. Sykes. 1992. Effect of trifluoroethanol on protein secondary structure: an NMR and CD study using a synthetic actin peptide. Biochemistry 31:8790-8798. [DOI] [PubMed] [Google Scholar]
- 46.Stammers, D. K., M. Tisdale, S. Court, V. Parmar, C. Bradley, and C. K. Ross. 1991. Rapid purification and characterisation of HIV-1 reverse transcriptase and RNAse H engineered to incorporate a C-terminal tripeptide-tubulin epitope. FEBS Lett. 293:298-302. [DOI] [PubMed] [Google Scholar]
- 47.Stow, N. D. 1992. Herpes simplex virus type 1 origin-dependent DNA replication in insect cells using recombinant baculoviruses. J. Gen. Virol. 73:313-321. [DOI] [PubMed] [Google Scholar]
- 48.Stow, N. D. 1993. Sequences at the C-terminus of the herpes simplex virus type 1 UL30 protein are dispensable for DNA polymerase activity but not for viral origin-dependent DNA replication. Nucleic Acids Res. 21:87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tam, J. P. 1988. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 85:5409-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tenney, D. J., P. A. Micheletti, J. T. Stevens, R. K. Hamatake, J. T. Matthews, A. R. Sanchez, W. W. Hurlburt, M. Bifano, and M. G. Cordingley. 1993. Mutations in the C terminus of herpes simplex virus type 1 DNA polymerase can affect binding and stimulation by its accessory protein UL42 without affecting basal polymerase activity. J. Virol. 67:543-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trincado, D. E., G. M. Scott, P. A. White, C. Hunt, L. Rasmussen, and W. D. Rawlinson. 2000. Human cytomegalovirus strains associated with congenital and perinatal infections. J. Med. Virol. 61:481-487. [DOI] [PubMed] [Google Scholar]
- 52.Weiland, K. L., N. L. Oien, F. Homa, and M. W. Wathen. 1994. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 34:191-206. [DOI] [PubMed] [Google Scholar]
- 53.Wu, C. A., N. J. Nelson, D. J. McGeoch, and M. D. Challberg. 1988. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J. Virol. 62:435-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuccola, H. J., D. J. Filman, D. M. Coen, and J. M. Hogle. 2000. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell 5:267-278. [DOI] [PubMed] [Google Scholar]