Abstract
Women infected with clade A human immunodeficiency virus type 1 harbor a virus population that is genetically diverse in the envelope gene, a fact that contrasts with the homogeneous virus population identified in newly infected men. It is not known whether viral genetic diversity at this early stage of infection is manifested as phenotypic diversity. This is a significant question because phenotypic diversity in the viral population that establishes infection in women may have important implications for pathogenesis and therapeutic intervention. Thus, in this study we compared the biological properties of three pairs of chimeric viruses that contained envelope genes representative of variant groups in each of three infected women—Q23, Q45, and Q47. Envelope chimeras were evaluated for replication in stimulated and resting peripheral blood mononuclear cells alone and in competition, for coreceptor use, and for neutralization sensitivity. All viruses utilized CCR5 exclusively and had a non-syncytium-inducing phenotype on MT-2 cells and in primary culture. There were no significant differences in replication parameters between paired variants in individual cultures. However, in competition experiments, one chimera of each variant pair always dominated. The dominant virus from Q23 and Q47, but not from Q45, infected a significantly higher number of CCR5- and CD4-expressing GHOST cells than the weaker chimeras. Significantly, chimeric viruses from Q47 and Q45 showed markedly different neutralization sensitivity to antibodies to CCR5 and gp120, respectively. These data indicate that distinct envelope genotypes identified in clade A-infected women near seroconversion confer unique phenotypes that affect viral fitness and that may be due, in part, to different requirements for relative configuration of CD4 and CCR5 on infected cells.
Virus transmission from an infected donor to a new host imposes a bottleneck that limits the diversity of the virus population. This phenomenon has important implications for human immunodeficiency virus type 1 (HIV-1) pathogenesis, because a donor may harbor a virus population of up to 10% diversity, but the transmission bottleneck may decrease the diversity in a virus population to near-homogeneity (51, 63, 65). In addition to changes in the genotypic diversity of the virus population, transmission also affects virus phenotype. HIV-1 variants transmitted to a new host are usually macrophage tropic, replicate slowly, are non-syncytium inducing, and utilize CCR5 as a coreceptor (64). As the virus population diversifies in the host, variants acquire different properties that include the capacity to replicate rapidly and induce syncytia in cell lines and to utilize CXCR4 as a coreceptor (53). This phenotypic change occurs in the majority of infections with clade B HIV-1 and is correlated with disease onset, although clinical symptoms do occur without a switch of viral coreceptor utilization (17). Primary isolates that have the capacity to use several coreceptors—dualtropic viruses—have been identified (11, 25, 54, 55). It is significant that virus variants detected over time have both genotypic and phenotypic features that are distinct from characteristics of viruses identified at the time of infection, because this suggests that properties that favor transmission of virus between hosts may be different from those that favor replication within a host.
Although women represent approximately 50% of HIV-1-infected individuals worldwide, the paradigm for transmission dynamics and viral pathogenesis during the early, asymptomatic years of infection is based primarily on studies in male cohorts. In contrast to the homogeneous virus population found in men, multiple variants were detected in the virus population in a cohort of clade A HIV-1-infected women near the time of seroconversion (45). Diversity of the infecting virus swarm was related to gender and not to the clade of HIV-1, because men from the same region harbored a homogeneous virus population at seroconversion (31). More recently, it has been determined that the gender difference in virus diversity between men and women may not relate to differences in diversity in the virus inoculum, because close to the time of infection, viral heterogeneity can be detected in both men and women (29, 31). In men, viral variation is rapidly contained and a clonal virus population emerges, whereas virus diversity is maintained in infected women. The effect of a diverse virus population on prognosis has been debated previously (15, 30, 32, 34, 36, 37, 52, 61). However, the persistence of genetically diverse variants in recently infected women presents a unique opportunity to correlate genetic and biological features and the fate of different viral genotypes transmitted to a naive host, which may lead to a better understanding of virus characteristics responsible for the successful establishment of new infections.
Viral fitness is a parameter that describes the relative ability of a virus to produce infectious progeny in a given environment (19). Viruses that replicate more slowly typically produce fewer progeny and consequently have lower fitness than rapidly replicating viruses. Fitness is often assessed by comparing the replication potential of viral variants in mixed infections. If a variant is highly fit under the growth conditions used, it may emerge as the dominant virus and exclude a less-fit variant (18). However, population sweeps by highly fit variants do not always occur either in vitro or in vivo. Variants with high replicative ability may not increase in the population if they are initially present at low frequency (13). Furthermore, viruses of different fitness can coexist if they occupy different niches. Serial passage of viruses between different cell types may allow the concurrence of variants with different levels of fitness (42) and may be a mechanism by which to constrain evolution of arthropod-borne viruses in vivo (60). It is of interest that in natural infections with HIV-1, variants compartmentalize to different tissues (14, 37, 45, 47) because this segregation may reflect different levels of fitness of variants in these compartments. In addition to dominance or coexistence, interaction among variants in vivo may be mutualistic (26, 59), thus endowing the viral population with features not attributable to individual genotypes. Competition among variants, therefore, is a useful means to assess relative virus fitness levels and can provide insight into population interactions that may not be revealed by studying virus replication in isolation.
Our previous analyses of genetic diversity (45) and evolution (47) of clade A HIV-1 from newly infected women were based on portions of the envelope gene that included the first three variable regions. To investigate the biological implications of genetic diversity in the seroconversion virus population, we first cloned a full-length infectious virus from subject Q23 and then created a library of viruses chimeric in the gp120 portion of env, which represented envelope sequence diversity identified at different time points. The natural history of virus infection in subject Q23 indicated that one of the infecting variants (ScA) eventually rose to dominance and replaced variant ScB (46, 47). gp120 diversity among these seroconversion variants was 1.1%, and the primary differences in amino acid sequence were in the V1 loop. In vitro, two important properties distinguished ScA from ScB variants: (i) ScA, but not ScB, could infect resting peripheral blood mononuclear cells (PBMC) following a low multiplicity of infection pulse exposure of autologous dendritic cells and (ii) resting cells infected with ScA proliferated more robustly than either ScB or uninfected cells following stimulation with immobilized α-CD3 (46). These results demonstrated that genetically distinct variants detected near the time of infection in one individual also had different biological properties.
The chimeras used in previously reported experiments contained the transmembrane protein, gp41, of infectious clone Q23-17, which was recovered 1 year after seroconversion and was used as the backbone for all chimera construction. Interaction between gp120 and gp41 dictates important biological properties of the virus, including the stability of the gp120-gp41 interaction (38), infectivity, and fusion (44). Evolution may act in concert on the two functional cleavage products of the gp160 polyprotein. Thus, to further explore biological properties of the diverse clade A variants identified in recently infected women, we created additional chimeric viruses that contained the entire extracellular coding region of the envelope gene.
In this study, we tested the hypothesis that envelope variants derived from the genetically divergent virus population identified near the time of infection in women conferred distinct phenotypic properties on a virus. Two envelope sequences that represented diverse variant groups in the viral population identified in subjects Q23, Q45, and Q47 were used to create three pairs of envelope chimeras. Virus replication kinetics in both individual and mixed culture, receptor utilization, and neutralization profiles were determined for each clade A envelope chimera pair. Our results indicate that one member of each of the three pairs of chimeras had greater fitness in competition assays. Although all chimeras used CCR5 as a coreceptor, enhanced fitness in competition assays correlated with the ability to infect more CCR5 GHOST cells in two of three cases. Q45 and Q47 chimeras were notably different in their susceptibility to neutralization with anti-CCR5 and anti-gp120 antibodies, respectively. These data support the hypothesis that divergent envelope sequences that coexist in clade A-infected women near the time of infection confer significantly different biological properties on variant viruses.
MATERIALS AND METHODS
Reagents.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: human recombinant interleukin-2 (rIL-2) from Maurice Gately, Hoffmann-La Roche, Inc. (28); CCR5-purified mouse anti-human monoclonal antibody (2D7) from BD PharMingen, San Diego, Calif. (62); GHOST cells, both parental and transfectants, from Vineet N. KewalRamani and Dan R. Littman (39); monoclonal antibody to HIV-1 gp120 (IgG1b12) from Dennis Burton and Carlos Barbas (4, 7, 8, 48); and HIV-1 gp120 monoclonal antibody (2G12) from Hermann Katinger (6, 12, 21, 35, 57). Antibodies to the following cell surface markers were purchased from BD PharMingen: CD3, CD4, CD8, CCR5, CXCR4, CD25, CD44, CD45RA, CD45RO, and CD62L. Isotype control monoclonal antibodies were mouse immunoglobulin G1 (IgG1) K (BD PharMingen), mouse IgG, (Southern Biotechnology Associates, Inc., Birmingham, Ala.), and mouse IgG2a (R & D Systems, Inc., Minneapolis, Minn.). Human IL-2 from human lymphocytes was obtained from Roche Diagnostic Corp. (Indianapolis, Ind.). HIV-1 p24 antigen assay was purchased from Beckman Coulter (Miami, Fla.). Tissue culture reagents including penicillin, streptomycin, phosphate-buffered saline (PBS), RPMI 1640 medium, and Dulbecco's modified Eagle’s medium were from BioWhittaker (Walkersville, Md.). Fetal calf serum (FCS) was obtained from HyClone (Logan, Utah).
Chimera constructions.
The envelope genes used to construct chimeras described in this report were amplified in a nested PCR using first-round primer pairs 5676F (GCAGAAGACAGTGGCAATGAGAGT) and 8629R (GTCCCTGGCCCTGGTGTG) and second-round primer pairs 5693F (TGAGAGTGATGGGGATACAGAGGA) and 8267R (TCCCACTATGCTACTTTTTGACCA). Envelope genes used to create chimeras Q23A4, Q23B6, and Q47S6 were derived from cDNA prepared from plasma RNA of each individual as previously described (46). The envelope genes used to create chimeras Q47 M1, Q45D5, and Q45D6 were derived from genomic DNA. Conditions for the first-round amplification were 94°C for 3 min, then 30 cycles of 94°C for 30 s, 56°C for 30 s, and extension at 71°C for 3 min. Second-round PCR conditions were 94°C for 3 min, then 30 cycles of 94°C for 30 s, 60°C for 30 s, and extension at 71°C for 2.75 min. PCR products were cloned into the PCR4 vector (Invitrogen Corp., Carlsbad, Calif.) and screened for appropriate-sized inserts. All plasmid preparation was done with Qiagen miniprep filters. The insert was removed by digestion with NdeI and ClaI, and the fragment was gel purified and ligated into a vector containing a 3′ subclone of the infectious clone Q23-17 (accession number AF004885). After transformation and DNA purification, the 3′ fragment containing the new envelope gene was excised by EcoRI and XhoI digestion and ligated into the full-length viral genome as previously described (46). Viral stocks were generated by transfection of 10 to 15 μg of plasmid DNA into 293T cells. Supernatant was recovered and used to infect 3-day phytohemagglutinin (PHA) blasts as described previously (46). All virus stocks were quantitated by assessing levels of p24gag. In addition, virus stocks containing equivalent amounts of p24gag were mixed in equal proportions and evaluated by quantitative heteroduplex tracking assay (HTA) to confirm that each contained an equivalent amount of viral RNA.
Sequence analysis.
Envelope genes were sequenced in the PCR4 vector and, following chimera construction, cloning sites were sequenced. Sequences were assembled and evaluated with the suite of programs in Lasergene99 (version 4.06) from DNASTAR, Inc. (Madison, Wis.). All sequences were aligned by using CLUSTAL W, and manual adjustments were made to improve the alignment. The isoelectric point for all predicted proteins was determined by using Protean in this sequence package. Pairwise distance analysis was conducted in the Paup* software application (version 4.0b10).
Preparation of PBMC.
Leukocyte packs were obtained from the American Red Cross (Portland, Ore., and Great Falls, Mont.). Lymphocytes were isolated on lymphocyte separation medium (ICN, Costa Mesa, Calif.), washed two or three times in PBS, resuspended in RPMI 1640 medium, and plated in a T75 flask for 1 h to remove adherent cells. Cells were recovered, enumerated, and seeded at 1 × 106 to 2 × 106 cells/ml in medium containing RPMI 1640, 15% FCS, 10 U of human IL-2 per ml, and (1%) each penicillin, streptomycin, and glutamine. Cells were stimulated with 1 to 2 μg of PHA-P (Sigma, St. Louis, Mo.) per ml.
To isolate resting cells, a portion of cells recovered after adherence was resuspended at a concentration of 5 × 106 cells per ml in cold RPMI 1640 containing 10% FCS. These cells were layered over a discontinuous gradient of 30, 40, and 60% Percoll (Amersham-Pharmacia, Piscataway, N.J.). Tubes were centrifuged for 25 min at 2,800 rpm in a Beckman GP centrifuge, GH 3.9 rotor, at 4°C. Resting cells were recovered from the 40 to 60% interface. The phenotype of resting and activated cells was determined by using antibodies to CD62L, CD44, and CD25. Evaluation of stained cells was conducted on a FACSCalibur flow cytometer, and all analyses used Cell Quest software (Becton Dickinson, San Jose, Calif.). In addition to activation markers, the levels of CD3, CD4, CD8, CCR5, CXCR4, CD45RA, and CD45RO expression were evaluated. Some experiments did not include resting cells because an insufficient number of cells was recovered to conduct experiments.
Kinetic assays and growth rate determination.
Virus stocks were incubated with day 3 PHA blasts for 4 to 5 h at 37°C at the concentrations of p24gag indicated in the figure legends. Cells were recovered by centrifugation and washed once in PBS, and 2.5 × 105 cells were plated in 48-well plates in quadruplicate. Every 3 days, 200 μl of medium was removed for determination of p24gag by antigen enzyme-linked immunosorbent assay (ELISA), and the mean production was obtained from the replicate wells. Kinetic assays in resting cells were conducted in an identical manner, but these were done only at high concentrations of virus (i.e., 5 and 1 ng/ml). A wider range of virus concentrations was used to initiate infections with the Q23 chimeras because these viruses replicated to lower titers and were the most variable in their ability to infect cells from different donors. Hence, experiments were conducted in cells from seven donors to obtain sufficient data for analysis.
The slope of the log-transformed growth curve was obtained from plots of log p24gag versus days of infection and used to determine daily growth rate. The y intercept of this line was used to determine the amount of virus produced at day 0, which was used as an estimate of the number of cells initially infected with each virus. As an objective criterion of exponential growth, an R2 value of >0.90 for the trend line was selected. If viruses were not growing exponentially because of saturation, the day 12 value was deleted from the analysis. This situation occurred only in Q45D6-infected cultures infected at viral titers of 5 ng/ml.
Competition assays.
Competition assays were conducted at 5 and 1 ng of total virus inoculum (3 ng/ml for Q23 chimeras) per ml. Stocks were then mixed in proportions of 20:80 or 50:50 of each virus, and infections were carried out as described for kinetic assays. RNA was extracted from each of the mixtures used to initiate the infection (day 0 sample) and from infected culture supernatant on days 6, 9, and 12 with the QiaAmp viral RNA minikit (Qiagen) following the manufacturer's protocol. The cDNA was made from 16 μl of RNA by using 20 ng of primer 8915R (TCCGCCCAAACCACACCT) to initiate the reaction. The mixture was heated for 10 min at 70°C, cooled on ice for 2 min, and then added to the cDNA reaction mix (1× Superscript II buffer [Invitrogen], 0.01 M dithiothreitol, 0.5 mM deoxynucleoside triphosphate, 40 U of RNase inhibitor [Roche Molecular Biochemicals], and 300 U of SuperScript II RNase H reverse transcriptase [Invitrogen]). The reaction mixture was incubated for 1 h at 42°C and inactivated by heating to 70°C for 10 min. A 725-bp fragment of env was amplified from cDNA from each of the competition reactions with primers 6030F (AAGCCTAAAGCCATGTGTAAAGTT) and 6755R (TCTCTAGATCCCCTCCTG). PCR conditions were 94°C for 3 min followed by 35 cycles of denaturing at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 50 s, with a final extension step of 9 min at 72°C.
Quantitative HTA.
A fluorescent single-stranded antisense probe specific for each virus pair was generated by amplifying 100 ng of plasmid with 1 μM fluorescent Hex-labeled 6755R primer and 20 pM 6030F unlabeled primer. PCR conditions were 94°C for 3 min, 94°C for 30 s, 54°C for 30 s, and 72°C for 45 s (35 cycles) and a final extension step of 9 min at 72°C. The probe was purified by using a Qiagen PCR purification kit. Heteroduplex formation reactions were done as described by Delwart et al. (16) with the following modifications. The HTA reactions consisted of 1× annealing buffer (0.05 M NaCl, 5 mM Tris-HCl [pH 8.0], 1 mM EDTA), 6 μl of target PCR product, and 60 ng of probe. Reaction mixtures were denatured at 95°C for 3 min, cooled for 4 min at 4°C, heated to 55°C for 4 min, and chilled again for 4 min at 4°C. Products were resolved on a 6% polyacrylamide gel in a Bio-Rad Protean II gel apparatus at 275 V for 3 h. Gels were visualized with a 520-nm excitation laser on a Fujifilm FLA 3000G fluorescent image analyzer by using Fujifilm Image Reader version 1.8E. Homoduplex and heteroduplex band intensities were quantitated by using Fujifilm Image Gauge version 3.3 software. Standards were generated by mixing quantitated PCR products from amplification of plasmids containing the envelope gene for each of the viral chimeras in known proportions and were run on each gel. Percent heteroduplex for standards was calculated as follows: [IntensityHeteroduplex/(IntensityHeteroduplex + IntensityHomoduplex)] × 100. Proportional representation of each variant was determined from the standard curve.
Coreceptor utilization assays.
This assay was carried out in human osteosarcoma (HOS) cells engineered to express CD4 and one of eight chemokine receptors (GHOST cells) (39). GHOST cells were maintained in basal DMEM containing 10% FCS, 1% each of penicillin, streptomycin, and glutamine, with 500 μg of Geneticin (Invitrogen) and 50 μg of hygromycin (Invitrogen) per ml. Cells transfected with chemokine receptors also contained 1 μg of puromycin (Sigma) per ml. Cells were seeded in 12-well dishes and infected overnight with 10 ng of p24gag of each virus stock per ml. Medium was replaced the following morning. After 60 h, cells were washed, trypsinized, and fixed in 4% paraformaldehyde for 1 h. Cells were then washed and resuspended in 200 μl of PBS containing 2% serum and 1% paraformaldehyde, and 20,000 cells were evaluated by flow cytometry.
Neutralization assays.
Antibody neutralization of clade A chimeras was determined by using GHOST cells, which were maintained as described above. Antibody to CCR5, 2D7, was incubated with cells at a range of concentrations at 37°C for 30 min prior to infection with 10 ng of p24gag of virus per ml. Infections and analyses were conducted as described above. Control cells were preincubated with an isotype control antibody at a concentration of 10 μg/ml. The number of infected cells per nanogram of virus was determined for each virus at each antibody concentration by flow cytometry. The number of cells infected in the presence of control antibodies was set to 100%, and the data are displayed as the number of cells infected in the presence of 2D7 divided by the number of cells infected in the presence of the control antibody. Neutralization assays with antibodies 2G12 and IgG1b12 were conducted in a similar manner, but antibodies at a concentration of 10 μg/ml were preincubated with virus stocks for 30 min at room temperature prior to infection.
Statistical analysis.
Because virus growth in cells from different donors was variable, a nonparametric Wilcoxon signed ranks test was used to determine if growth rate or initial amount of virus produced differed between the paired viruses in each experiment.
Each test for difference in virus kinetics was performed by using the set of paired values obtained from growth in the same donor under the same experimental conditions of cell activation and virus concentration. An analysis was first conducted to determine if the growth rates and number of infected cells differed between the paired variants grown in individual culture. There were only sufficient data available from stimulated-cell infection experiments to conduct this analysis because variants did not always grow in resting cells. A second analysis was conducted to determine if growth rates and initial virus production of a chimera differed in individual and mixed cultures. Because some competition experiments resulted in no virus growth for some variants, pairing individual experiments was not feasible. Therefore, the difference between average values for virus alone and virus in competition obtained from each donor were used for this analysis.
Nucleotide sequence accession numbers.
Sequences reported in this manuscript are deposited in GenBank with accession number AY288084 through AY288087. .
RESULTS
Properties of full-length envelope glycoproteins used to construct chimeras.
Envelope genes representative of the sequence diversity detected in three women recently infected with clade A HIV-1 were chosen from a data set of full-length sequences derived at or near seroconversion from proviral DNA and from plasma viral RNA (Table 1). Sequences were aligned, and pairwise distances were used to group genetically similar variants (46). Two variant groups were identified by using this method, and a group consensus sequence was generated for each. The sequence from each group that was most similar to the group consensus sequence was chosen for chimera construction. The two sequences from Q23 and Q45 were derived from the same time point and the same tissue source. Although the Q47 M1 envelope was derived at a later time point than Q47S6, it only differed by six amino acid substitutions (0.8%) from the sequence targeted for chimera construction, which was detected in plasma at the same time point at which Q47S6 env was derived. Efforts were made to create several chimeras by using envelope genes from each variant group, but most were not infectious. Thus, three pairs of virus chimeras were created that each contained an envelope gene that closely approximated the consensus of each of the genetically similar groups of envelope variants identified from the clade A-infected women.
TABLE 1.
Properties of envelope genes, predicted proteins, and chimeric virusesa
| Virus | Clade | Time from infectionb | pI | N-linked glycosylation sites | Phenotype on MT-2c |
|---|---|---|---|---|---|
| Q23A4 | A | 4 mo | 8.16 | 27 | NSI |
| Q23B6 | A | 4 mo | 8.23 | 27 | NSI |
| Q45D5 | A2 | 8 mo | 8.20 | 26 | NSI |
| Q45D6 | A2 | 8 mo | 8.20 | 27 | NSI |
| Q47M1 | A | 12 mo | 8.25 | 30 | NSI |
| Q47S6 | A | 0 mo | 8.18 | 24 | NSI |
Percent identity between viral pairs is as follows. Q23A4 and Q23B6: DNA, 99.2, and protein, 98.8; Q45D5 and Q45D6: DNA, 98.8, and protein, 98.1; Q47M1 and Q47S6: DNA, 96.8, and protein, 94.1.
Numbers indicate the approximate time from infection that the samples from which the envelope genes used in this study were obtained. Women were evaluated by antibody ELISA for evidence of HIV-1 infection at approximately monthly intervals. The time of infection is estimated to be the date prior to seroconversion that virus was detected by PCR but the individual was seronegative.
NSI, non-syncytium inducing.
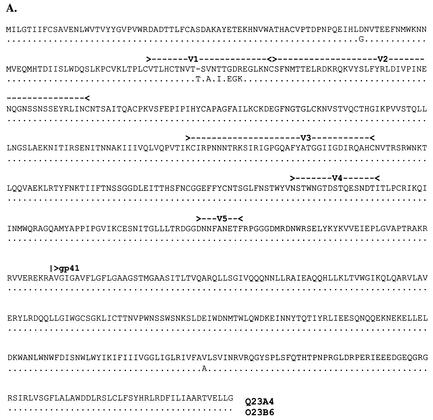
Variation in the pool of Q23 virus envelopes was manifested in the V1 loop. The sequences used to construct Q23 chimeras (i.e., Q23A4 and Q23B6) were chosen to represent the two V1 sequences, which were equally represented in the virus population (46) and differed at only two positions outside this region (Fig. 1A). We attributed the biological differences in viruses bearing envelopes representative of the two coexisting variant groups previously described (46) to the V1 region of gp120, because the remainder of the envelope protein from variants derived at this time point was virtually identical. All potential N-linked glycosylation sites were conserved between the two Q23 chimeras, although secondary structure analysis predicted greater surface exposure of V1 in Q23A4 (data not shown).
FIG. 1.
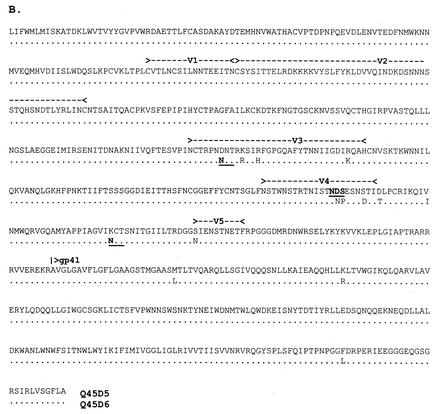
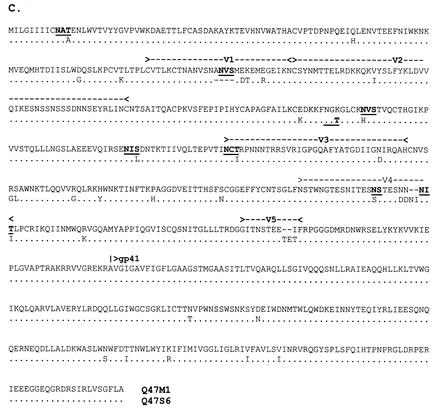
Alignments of the full-length envelope glycoproteins used in chimera construction. (A) Q23; (B) Q45; (C) Q47. Amino acids are given in single-letter codes. Gaps (-) are included to maintain the alignment. Amino acid identity between the two sequences is indicated with a dot (.). Potential N-linked glycosylation sites that differ between the chimera pairs are underlined and in boldface. Locations of variable loops and the gp120/gp41 cleavage site are indicated above the sequence.
Chimeras Q47 M1 and Q47S6 also differed in the V1 region, resulting in an additional potential N-linked glycosylation site in Q47 M1 (Fig. 1C). The Q47 chimeras were the most divergent of the variant pairs, and the 41 amino acid positions that differed between these two viruses were distributed throughout the extracellular envelope-coding region. Eight of the substitutions affected potential N-linked glycosylation sites, seven of which conferred additional potential glycosylation sites to Q47 M1. Variant Q47S6 was 99.7% homologous to the group consensus sequence, which was derived from nine sequences that ranged from 94.2 to 99.7% identity to the consensus. Variant Q47 M1 was 98.4% homologous to the group consensus sequence, which was derived from 10 sequences that ranged in identity to the consensus from 94.2 to 98.6%.
In contrast to Q23 and Q47, there was extensive diversity in the envelope genes from the Q45 virus population that often was represented by an amino acid substitution that was unique to a single sequence. However, diversity that distinguished variant groups was concentrated in the V3 and V4 regions. The envelope sequences chosen to represent the diversity from Q45 were identical in the region N terminal to V3 but differed at four positions in V3, which involved one potential N-linked glycosylation site, and in V4 and the CD4 binding domain (Fig. 1B). Chimera Q45D5 was 98.7% identical to the consensus sequence of the variant pool that it represented (range, 98.1 to 98.8%), and Q45D6 was 99.3% identical to the consensus sequence for the group of variants that it represented (range, 93.9 to 99.3%).
Plasmids containing each chimera were transfected into 293T cells, and infectious stocks were generated in PHA-stimulated PBMC. None of the viruses caused any visible cytopathology in primary cells or in MT-2 cells.
Replication properties of clade A HIV-1 envelope chimeras.
We evaluated each chimera for the ability to infect both resting and stimulated PBMC at several virus concentrations. Features of the donor cells affected replication of all chimeras (Fig. 2). Because of the pronounced donor effect, a complete set of experiments, which included replication kinetics at two or three different virus concentrations in stimulated and resting cells and competition assays between chimeras from the same infected host (described below), was replicated in cells from four or five different donors. Cells were evaluated for expression of CD4, CCR5, CXCR4, CD45 isotype, and CD25 prior to infection. Although donor cells in which viruses replicated to lower levels expressed low levels of CCR5, not all donor cells with low levels of CCR5 were nonpermissive (data not shown). Thus, there was no correlation between permissive or resistant cell phenotype and any of the markers evaluated.
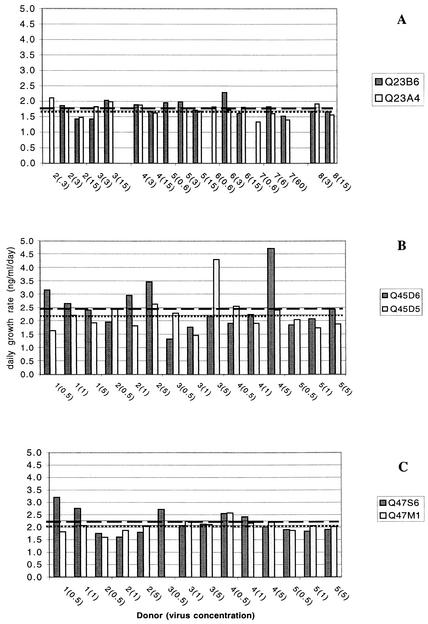
FIG. 2.
Summary of clade A HIV-1 chimera daily growth rates in stimulated cells from different donors. (A) Q23; (B) Q45; (C) Q47. The daily growth rate was determined from the slope of log p24gag versus day plots. The numbers on the x axis indicate an individual donor, and the virus concentrations used to initiate infection is given in parentheses. Data from donor 1 are not shown in panel A because Q23B6 was not included in those experiments. A different set of donor cells was used for each of the virus pairs (e.g., donor 3 in Q23 experiments is not the same as donor 3 in experiments using Q45 or Q47). The mean daily growth rate is shown as the dashed line for Q23B6 (1.76), Q45D6 (2.48), and Q47S6 (2.19) and a dotted line for Q23A4 (1.73), Q45D5 (2.22), and Q47 M1 (2.05).
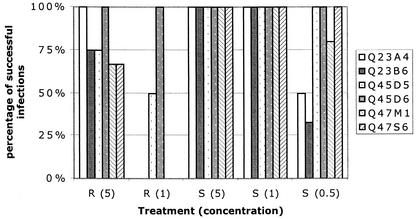
Q45D6 was the only clade A chimera that initiated infection in all donors under all experimental conditions (Fig. 3). Although Q45D5 consistently produced less p24gag than Q45D6, it was able to infect stimulated donor cells at all input virus concentrations. Q45D5 only established infection in resting cells in three of four experiments when the inoculum contained 5 ng of p24gag per ml and in two of four experiments at an infecting concentration of 1 ng of p24gag per ml, suggesting that this chimera required cell stimulation for efficient infection. Both Q47S6 and Q47 M1 could infect stimulated cells at low levels of virus inoculum (i.e., 0.5 ng of p24 per ml), although Q47 M1 was not able to infect PHA blasts from one donor at this concentration. Neither of the Q23 chimeras consistently infected stimulated cells at low virus inoculum levels. Consistent with previous results (46), Q23A4 was more efficient than Q23B6 in infecting resting cells, although none of the Q23 or Q47 chimeras could infect resting cells if the level of inoculum did not exceed 1 ng per ml.
FIG. 3.
Replication success of clade A HIV-1 chimeras in stimulated or resting cells in individual cultures. A virus was determined to successfully infect a culture under specified conditions if p24gag was detectable at day 12 PI by antigen ELISA. R, resting cells; S, cells stimulated by PHA. The number in parentheses is the total amount of viral p24gag in the inoculum in nanograms per milliliter. Q23 chimeras are included in the indicated treatment categories, although the actual amount of virus in the categories indicated by S (0.5), S (1), and S (5) was 0.3 to 0.6, 3, and greater than 10 ng/ml, respectively.
The nonparametric Wilcoxon signed ranks test was used to test the null hypothesis that growth rate (Fig. 2) and initial amount of virus (data not shown) were not different between the two chimeras from an individual that were grown in the same donor cells under the same experimental conditions. There were no statistical differences between growth rates of viruses grown in PHA blasts. Because daily growth rates of variant pairs were not significantly different, we determined virus production at day 0 from the y intercept of the log-growth curves as an estimate of the number of cells initially infected with each virus. There was also no significant difference between predicted day 0 virus production of variants from the same individual. Thus, in stimulated cells, there was no statistical support to merit rejection of the null hypothesis that growth rate and initial virus production were the same between paired viruses.
The growth rate of a virus in stimulated cells was compared to growth in resting cells from the same donors. Because Q47 and Q23 chimeras did not always replicate in resting cells, there were insufficient data for this paired analysis. Thus, for these comparisons, growth rate data for each of the chimera pairs in stimulated cells was pooled and evaluated against the pooled data for the respective virus pair in resting cells. Q45 chimeras replicated well in all conditions, and thus the analysis was conducted on the individual virus in resting or stimulated cells. Virus replication in resting cells was significantly slower than in stimulated cells for Q45 and Q47 chimeras (P = 0.04) but not for the Q23 chimeras. However, there was no difference in either the rate of replication or the predicted initial virus production between virus pairs in resting cells.
Fitness of clade A HIV-1 envelope chimeras in mixed infections.
Replication properties of chimeras in individual infections of primary cultures were not significantly different. However, relative fitness of variants can best be tested in mixed infections for which efficiency of using shared resources can be evaluated. Competition assays were conducted by infecting both stimulated and resting PBMC with a mixture of the paired chimeras at different proportions and amounts of total virus. Experiments were conducted in cells from the same five donors that were used for replication kinetic studies of individual variants. A virus was considered able to establish infection if p24gag production from that variant was detectable at day 12 of the experiment, even if the proportional amount of virus was less than that in the inoculum. The relative proportion of each variant was determined by quantitative HTA.
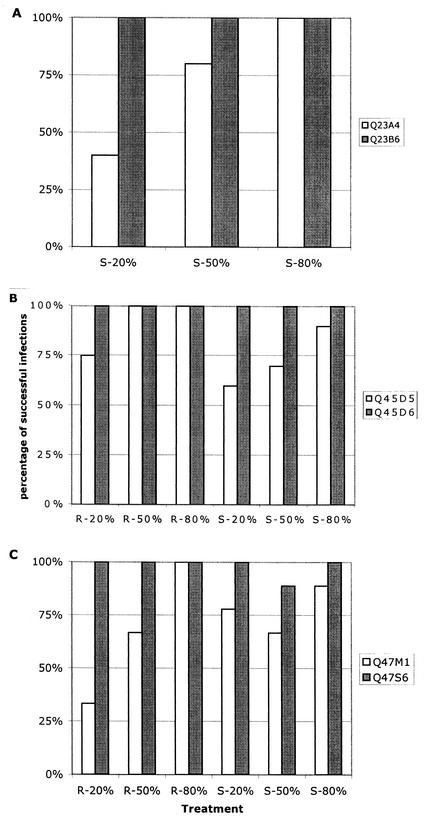
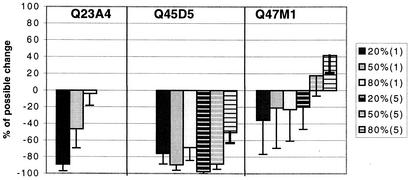
For each of the three pairs of chimeras, one of the variants was consistently successful in establishing infection regardless of its proportional representation in the inoculum (Fig. 4). Competition experiments for Q23 variants were only conducted in stimulated cells because Q23B6 infection of resting cells was inconsistent. In addition, a total virus inoculum of 3 ng of p24gag per ml was used for Q23 chimera competitions because volumes required for higher concentrations of this low-titer virus were excessive and infection was inconsistent at lower concentrations. In PHA blasts, both Q23 variants could establish infection if they constituted 80% of the infecting virus population, but Q23A4 was not able to consistently establish infection if it constituted 50% or less of the inoculum. In all experiments in which Q23A4 was detectable at day 12, the proportional representation was less than that of the input inoculum, except when it made up 80% of the infecting inoculum (Fig. 5). Q45 and Q47 chimeras were competed at two different amounts of total virus inoculum (1 and 5 ng/ml) in stimulated cells and at 5 ng of total virus per ml in resting cells. Q45D6 dominated competitions under all conditions tested. However, despite the lower overall yields of Q45D5 in individual infections, this chimera was successful in establishing and maintaining a small proportion of the virus population in 64% of all competition experiments in both resting and stimulated cells when present at only 20% of the inoculum. Despite its ability to persist in mixed cultures, Q45D5 always declined relative to the proportional representation in the inoculum (Fig. 5). Of the Q47 chimeras, Q47S6 successfully established infection in both resting and stimulated cells in all but one of the competition experiments. Q47 M1 was inconsistent in establishing infection in competition with Q47S6 even if it represented 80% of the inoculum. Although Q47S6 was the more successful variant in terms of consistently establishing infection under different conditions, Q47 M1 was able to maintain or increase in relative proportion compared to the inoculum ratio in many experiments (Fig. 5). The large variance associated with the proportional change of Q47 M1 in mixed culture suggests that relative fitness of the Q47 variants is a function of the donor cell environment but that Q47S6 may be less sensitive to these differences.
FIG. 4.
Replication success of clade A HIV-1 chimeras in stimulated or resting donor cells in the presence of a competitor. (A) Q23; (B) Q45; (C) Q47. The percentage of successful replications was determined by dividing the number of experiments in which the viral RNA of a chimera could be identified in the supernatant fraction at day 12 by the total number of experiments conducted under each of the indicated conditions. The proportional representation of the chimera in the inoculum is shown on the x axis. S, experiment conducted in PHA-stimulated cells; R, resting cells. The data displayed for stimulated-cell competition studies in panel A were conducted at 3 ng/ml total virus, whereas data in panels B and C include experiments in which infection was initiated with a total virus concentration of both 1 and 5 ng/ml.
FIG. 5.
Change in proportional representation of lower-fitness chimeras during the course of competition experiments. The percent possible change was determined by quantitative HTA from the proportion of the chimera in the inoculum and in the supernatant at day 12. A chimera that represented 20% of the total inoculum could decrease to 0% or increase to 100%. Thus, a virus that constituted 60% of the total virus recovered at day 12 achieved 50% of its potential increase, or if it was not represented at day 12, it achieved 100% of its possible decline. Black, gray, and white bars indicate that the virus was present at 20, 50, and 80% of the inoculum, respectively. The numbers in parentheses in the key indicate the total amount of virus inoculum, with solid bars indicating 1 ng/ml and striped bars indicating 5 ng/ml of total virus.
These data indicated that the clade A variants from an individual had different abilities to establish and maintain infection in mixed cultures, despite the fact that there were no significant differences in replication kinetics of viruses grown alone. We compared growth rates of each variant grown alone and in competition to determine if the presence of a competitor affected replication parameters of a virus. None of the differences in growth rates were significant (data not shown). There was a significant increase (P = 0.043) in the initial virus production of Q45D5 in mixed culture compared to day 0 production when grown alone. This suggests that the threshold for successful infection was higher for Q45D5 in competition than when grown alone. The growth rates and initial virus production of Q47 M1 alone and in competition were virtually identical, and whereas the growth rate of Q47S6 was lower in competition than in individual cultures, this difference was not significant. Thus, there was no indication that the replication rate of a variant was altered in mixed culture compared to the rate in individual culture. However, the greater fitness of one member of each variant pair suggested that the viruses differed in their ability to establish infection and spread in primary cells.
Coreceptor utilization of clade A HIV-1 envelope chimeras.
Data from competition studies could be explained by differential use of coreceptors by the variant pairs, which would allow access to different subsets of primary cells in culture. To determine if this was the case, we investigated the coreceptor requirement of each chimera by using a panel of cells engineered to express one of eight chemokine receptors in conjunction with CD4 (GHOST cells) (39). All of the variants utilized CCR5 exclusively (Table 2). However, the ability of chimeras to infect the GHOST cells varied. In each experiment, the number of cells infected with Q23A4 was about half the number of cells infected with Q23B6 and Q47 M1 infected approximately one-third as many cells as Q47S6. This was an interesting observation because in both cases the virus that was dominant in competition assays also infected more CCR5-expressing GHOST cells. However, the number of GHOST cells infected with equal amounts of the two Q45 chimeras was the same.
TABLE 2.
Coreceptor use and 2D7 neutralization summarya
| Virus | Coreceptor | % Controlb (SE) |
|---|---|---|
| Q23A4 | CCR5 | 52.6 (6.7) |
| Q23B6 | CCR5 | 58.4 (4.7) |
| Q45D5 | CCR5 | 36.2 (8.9) |
| Q45D6 | CCR5 | 37.0 (6.5) |
| Q47M1 | CCR5 | 40.7 (3.8)c |
| Q47S6 | CCR5 | 94.8 (6.3) |
Ratios of infected cells are as follows: A4/B6, 0.54; D5/D6, 1.00, M1/S6, 0.38. The number of cells infected with the chimeras is significantly different for A4/B6 and M1/S6 (Student's two-tailed t test, P < 0.01.). The number of infected CCRS-GHOST cells was determined by flow cytometry for each of the clade A chimeras in eight replicate experiments. The data are displayed as the ratio of the mean number of infected cells for each virus pair.
The percent control is the number of cells infected in the presence of 2D7 (10 μg/ml) divided by the number of cells infected in the presence of the control antibody. Mean percent control and standard error were determined from four replicate experiments.
The percent of cells infected with Q47M1 is significantly different from the percent of cells infected with Q47S6 in the presence of 2D7 (10 μg/ml) relative to respective controls (Student's two-tailed t test, P < 0.01).
Neutralization of clade A HIV-1 envelope chimeras.
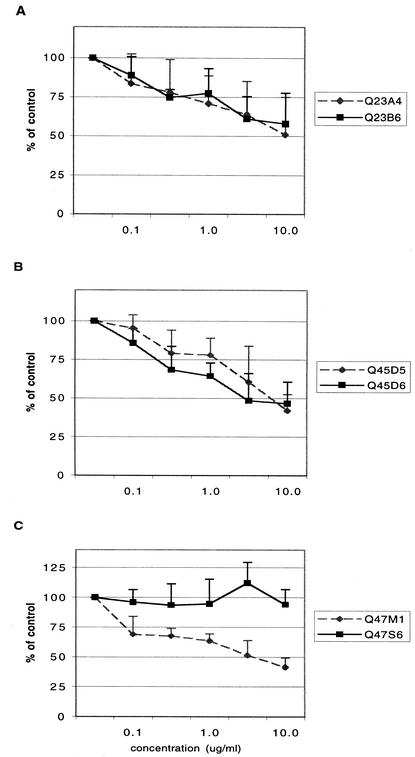
Although all of the chimeras exclusively utilized CCR5, there was clearly a difference between the ability of the Q23 and Q47 chimeras in to infect CCR5 GHOST cells. If greater fitness and more efficient infection of CCR5 GHOST cells by some chimeras were associated with different requirements for CD4-CCR5-gp120 interaction, then we expected that variant pairs would be differentially neutralized by antibody to this chemokine receptor or to the CD4 binding site on gp120. GHOST cells expressing CCR5 and CD4 were preincubated with different concentrations of antibody 2D7 and infected with each of the virus chimeras (Fig. 6 and Table 2). Q23 variants were resistant to 2D7 neutralization, and the highest concentration of 2D7 used (10 μg/ml) did not result in a 50% reduction of infection for either chimera. Although Q45 variants were more sensitive to 2D7 neutralization, there was no difference in relative sensitivity to anti-CCR5 neutralization of these chimeras. However, 2D7 differentially inhibited the Q47 chimeras. Chimera Q47S6, which dominated competition experiments and which had an enhanced ability to infect CCR5 GHOST cells, was resistant to neutralization with this antibody, while 10 μg/ml of 2D7 reduced the number of Q47 M1-infected cells by 60%.
FIG. 6.
Neutralization of clade A chimeras by 2D7. Antibody to CCR5 was incubated with CCR5 expressing GHOST cells prior to virus infection, and infection was initiated with 10 ng of p24gag of each chimera per ml. The number of infected cells was determined by flow cytometry. Data are presented as the percentage of cells infected in the presence of 2D7 compared to cells treated with an isotype control antibody and represent the results of four replicate experiments. Concentrations of 2D7 are given in micrograms per milliliter.
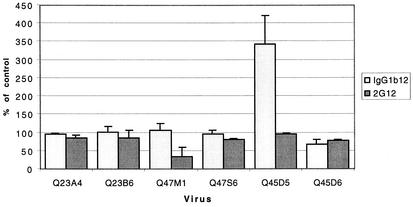
These data indicated that although both Q47 chimeras use CCR5 as a coreceptor, CCR5 contact with gp120 differed between these two variants. However, both Q23 and Q45 chimeras had similar neutralization profiles with antibody 2D7, although Q23 chimeras were more resistant than Q45 chimeras to neutralization. We further investigated potential differences in neutralization properties that might distinguish paired chimeras using antibodies IgGb12 and 2G12, which are specific for gp120 and are reported to have cross-clade neutralizing ability at concentrations between 2 and 10 μg/ml (58). Antibodies at concentrations of 10 μg/ml were preincubated with each variant for 30 min prior to infection of GHOST cells (Fig. 7). Again, Q23 viruses were resistant to neutralization by both antibodies at this concentration. Both of the Q47 chimeras were inhibited by 2G12, which recognizes a carbohydrate epitope on the outer face of gp120 (50), but Q47 M1 was more sensitive than Q47S6 to inhibition with this antibody. It is of interest that Q47 M1 has seven more N-linked glycosylation sites than Q47S6, which might account for the increased sensitivity to neutralization by antibody 2G12. Notably, antibody IgG1b12, which binds to the CD4 binding site (8), significantly enhanced the infectivity of Q45D5. In contrast, the number of cells infected with chimera Q45D6 was reduced by 34%, and this was, therefore, the only clade A chimera that was inhibited by IgG1b12 at the concentrations used. Thus, in two of the three cases examined, related variants clearly differed with respect to receptor or coreceptor binding.
FIG. 7.
Neutralization of clade A chimeras by 2G12 and IgG1b12. Virus (10 ng of p24gag per ml) and antibodies (10 μg/ml) were incubated together for 30 min and then applied to CCR5-expressing GHOST cells. The number of infected cells was determined by flow cytometry. Data are presented as the percentage of cells infected in the presence of anti-gp120 antibody compared to cells treated with an isotype control antibody and represent the results of two replicate experiments (three replicates for Q45 chimeras).
DISCUSSION
Viral diversity is a hallmark of infection with HIV-1, although viral heterogeneity typically is limited near the time of infection because of a bottleneck imposed on the transmitted virus population. However, it has previously been reported (31, 45) that women infected with clade A HIV-1 harbored a diverse population of viral variants at the time of infection and seroconversion. It is not clear whether these variants are phenotypically similar or if the variant pool contains viruses with biologically distinct properties. This is a significant question because colonization of a new host by a biologically diverse virus population versus a genetically homogeneous virus population may create different challenges for early intervention strategies and may alter the progression of disease. Thus, in the present study we tested the hypothesis that clade A HIV-1 envelope glycoproteins that represented different variant groups in the heterogeneous virus population near the time of infection conferred unique properties on a virion.
We first evaluated the genetic diversity of env in three individuals at one to three time points during the first year of infection. Pairwise analysis of all sequences indicated that virus sequences formed two clusters, with intergroup genetic diversity threefold greater than intragroup diversity. The sequence within each group that most closely resembled the consensus sequence for each variant cluster was selected for chimera construction. We were not able to determine if other variants within a group had phenotypes similar to that of the one chosen for chimera construction because, despite numerous attempts to produce additional chimeras, only those described were infectious.
Chimeric viruses were identical in all viral genes except the extracellular portion of gp160, which consisted of the entire coding region of gp120 and the extracellular and membrane-spanning region of gp41. Thus, any observed differences in viral properties could be attributed solely to this region of env. We anticipated that if differences existed, they would involve interaction of gp120 with receptor or coreceptor and hence would influence viral entry, although processing, assembly, and budding could also be affected. Because our hypothesis tested whether phenotypic differences occurred between representatives of the virus population within an infected host, all experiments and analyses compared chimeras containing envelope genes from the same individual.
We evaluated the replication of viruses in individual cultures of stimulated and resting primary cells. These experiments were confounded by the pronounced differences in virus replication in cells from different donors (see Fig. 2). Although there were cells in which the two chimeras replicated equally well or poorly, cells in other cases were more permissive for one of the variants than the other. Thus, the variance was high, and statistically significant differences in growth rates were not detected. Because virus production varied between chimeras in paired experiments and replication rates were not significantly different, we also examined the y intercepts of the log-transformed growth curves to determine if one variant was able to initiate infection of a larger number of cells. However, there was also no statistical support for a difference in the initial number of cells infected by each of the chimeras. Thus, we had no support for differences in replicative ability of virus chimeras based on kinetic experiments in individual culture.
The relative fitness of variant viruses is best determined by allowing viruses to compete for resources in the same environment. If, for example, variants replicate in different cell subsets, then the presence of a competing virus will have little effect on the replication kinetics of each virus. On the other hand, if variants infect the same cell subsets, they must compete for a limiting resource. A virus that can utilize existing resources more efficiently than its competitor will leave more progeny. In mixed infections of primary cells, one variant in each pair—Q23B6, Q45D6, and Q47S6—successfully established infection and in most cases outgrew the competing chimera regardless of the activation state of cells, total amount of virus, and proportional representation in the virus inoculum. The daily growth rate and initial virus production of dominant chimeras grown alone or in mixed culture was the same, suggesting that there was no interaction between variants in mixed culture, although replication variability in different donor cells may have precluded the detection of small differences. Similarly, there were no detectable differences between growth rates of the less-fit chimeras—Q23A4, Q45D5, and Q47 M1—in mixed or individual cultures. However, the number of cells initially infected with Q45D5 was significantly increased in competition compared to individual cultures, suggesting that the threshold number of cells required for this chimera to sustain infection was greater in mixed than in individual cultures. Thus, results of competition studies indicated that representative envelope variants from each of three infected individuals have different fitness in vitro, but there was no indication that the presence of a competing virus affected replication properties of any chimera.
The greater in vitro fitness of chimeras Q23B6, Q45D6, and Q47S6 could be due to either an expanded host cell range, which would allow the chimeras to replicate in a wider variety of cell types, or more efficient use of the same cell subsets. HIV-1 requires CD4 as a receptor and one of several chemokine receptors as a coreceptor for virus entry (1, 20, 22). The distribution of chemokine receptors varies on CD4-expressing cells, and expression levels of a receptor fluctuate with the state of cell activation and the chemical environment. Therefore, we utilized cell lines expressing a fixed level of both CD4 and one of the eight chemokine receptors identified as coreceptors for HIV-1 to determine if chimeras had different coreceptor requirements. Our data indicated that all of the chimeras utilized CCR5 exclusively. However, in coreceptor determination experiments, dominant chimera Q23B6 infected almost 50% more cells than Q23A4. Similarly, Q47S6, which also dominated all competition experiments, infected over 60% more CCR5-expressing GHOST cells than did Q47 M1. Thus, it is possible that either binding or entry of Q23A4 and Q47 M1 was restricted by a more stringent requirement for the relative density or configuration of receptor and coreceptor on the cell surface. If dominant chimeras Q23B6 and Q47S6 had less rigorous requirements for receptor-coreceptor configuration, then in competition assays Q23A4 and Q47M1 could be excluded from potential target cells both at the time of initial infection and during expansion in culture. The notable difference between Q47 chimeras in susceptibility to neutralization with antibody to CCR5 provided supporting evidence that the binding site for CCR5 on the gp120 of the Q47 chimeras was different.
It is clear that coreceptors play a significant role in HIV-1 pathogenesis, because genetic differences in chemokine receptors or ligands are present in the human population that can confer an individual with increased susceptibility or resistance to infection (9, 23, 24, 33, 40, 43). Furthermore, it has been shown that in addition to the type of chemokine receptor expressed on a cell, the density of chemokine receptor relative to CD4 is also an important determinant of susceptibility of target cells to HIV-1 infection (20, 56). The expression of chemokine receptors is controlled by a variety of cytokines (2, 5, 10, 49), indicating that the inflammatory milieu of a potential target cell may be as significant a determinant of infection susceptibility as the genetic features of either the host or the virus. Additionally, the dynamic cellular environment in women newly infected with clade A HIV-1 may help sustain the phenotypic diversity of the virus population by providing multiple niches for viruses of different relative fitness levels.
Coreceptor density cannot be the only factor dictating the differential fitness of clade A chimeras evaluated in this study. Q45D6 was conspicuously the most competent of the clade A chimeras for replicating to high titers in all donor cells under any experimental condition, so it was not surprising that it emerged as the dominant chimera in competition with Q45D5. However, in this case there was no indication that chimeras differed in their interaction with CCR5 because both Q45 variants infected the same number of GHOST cells and both were neutralized effectively and to the same extent with 2D7. Instead, an epitope in the CD4-binding site, which is recognized by antibody IgG1b12, distinguished Q45D5 and Q45D6. These data suggest that Q45 variants differ in their interaction with CD4 or in a postreceptor binding conformation that either enhances or diminishes virus entry. Differences in relative levels of CD4 expression can influence the susceptibility of a cell to infection, as exemplified by the resistance of primate cells to HIV-1 infection due to low levels of cell surface CD4 (3). Furthermore, CD4 has been shown to be the principal determinant of efficient viral binding, endocytosis, and proteolysis if coreceptor levels exceed a minimum density (27). Because the density and ratio of these surface receptors vary between tissues (41), differences in expression levels of either receptor or coreceptor may expedite niche adaptation or compartmentalization of variants to different tissues, which has been described previously for HIV-1 in subjects Q23, Q45, and Q47 (47).
Although we used antibodies to probe for disparity between the closely related gp120 chimeras, the differences in neutralization sensitivity are a significant finding that merits further investigation in primary cells. The markedly different profiles of Q47 chimeras to inhibition with 2D7 demonstrate both that the binding site for CCR5 is different on the two chimeras and that therapeutic intervention with this antibody would not be equally effective against all members of the variant pool in this individual. Similarly, IgG1b12 is widely accepted as a potent neutralizing antibody and although it exhibited inhibitory activity towards Q45D6, it significantly enhanced infection of Q45D5 at the same concentration. Thus, our data support the hypothesis that variants coexisting within women infected with clade A HIV-1 do have distinct phenotypes, which could subsequently influence tissue distribution and therapeutic efficacy.
Acknowledgments
We thank Joan Kreiss, Harold Martin, Jr., and their collaborators at the Ganjoni Municipal Clinic and Coast Provincial General Hospital (Mombasa, Kenya) for sample collection, Julie Overbaugh for providing plasma samples, John Graham for statistical assistance, and Rita Luther for her assistance with Q23 chimera construction.
This work was supported by Public Health Service grant AI44609.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Bailer, R. T., B. Lee, and L. J. Montaner. 2000. IL-13 and TNF-α inhibit dual-tropic HIV-1 in primary macrophages by reduction of surface expression of CD4, chemokine receptors CCR5, CXCR4 and post-entry viral gene expression. Eur. J. Immunol. 30:1340-1349. [DOI] [PubMed] [Google Scholar]
- 3.Bannert, N., D. Schenten, S. Craig, and J. Sodroski. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 74:10984-10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbas, C. F., III, E. Bjorling, F. Chiodi, N. Dunlop, D. Cababa, T. M. Jones, S. L. Zebedee, M. A. Persson, P. L. Nara, E. Norrby, et al. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. USA 89:9339-9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleul, C. C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 94:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 7.Burton, D. R., C. F. Barbas III, M. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 9.Carrington, M., M. Dean, M. P. Martin, and S. J. O'Brien. 1999. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum. Mol. Genet. 8:1939-1945. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, R. G., J. L. Riley, B. L. Levine, P. J. Blair, D. C. St. Louis, and C. H. June. 1998. The role of co-stimulation in regulation of chemokine receptor expression and HIV-1 infection in primary T lymphocytes. Semin. Immunol. 10:195-202. [DOI] [PubMed] [Google Scholar]
- 11.Cho, M. W., M. K. Lee, M. C. Carney, J. F. Berson, R. W. Doms, and M. A. Martin. 1998. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J. Virol. 72:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford, J. M., P. L. Earl, B. Moss, K. A. Reimann, M. S. Wyand, K. H. Manson, M. Bilska, J. T. Zhou, C. D. Pauza, P. W. Parren, D. R. Burton, J. G. Sodroski, N. L. Letvin, and D. C. Montefiori. 1999. Characterization of primary isolate-like variants of simian-human immunodeficiency virus. J. Virol. 73:10199-10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Torre, J. C., and J. J. Holland. 1990. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J. Virol. 64:6278-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delwart, E. L., J. I. Mullins, P. Gupta, G. H. Learn, Jr., M. Holodniy, D. Katzenstein, B. D. Walker, and M. K. Singh. 1998. Human immunodeficiency virus type 1 populations in blood and semen. J. Virol. 72:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delwart, E. L., H. Pan, H. W. Sheppard, D. Wolpert, A. U. Neumann, B. Korber, and J. I. Mullins. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J. Virol. 71:7498-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 17.de Roda Husman, A. M., R. P. van Rij, H. Blaak, S. Broersen, and H. Schuitemaker. 1999. Adaptation to promiscuous usage of chemokine receptors is not a prerequisite for human immunodeficiency virus type 1 disease progression. J. Infect. Dis. 180:1106-1115. [DOI] [PubMed] [Google Scholar]
- 18.Domingo, E., C. Escarmis, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859-864. [DOI] [PubMed] [Google Scholar]
- 19.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 20.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276:229-237. [DOI] [PubMed] [Google Scholar]
- 21.Etemad-Moghadam, B., Y. Sun, E. K. Nicholson, G. B. Karlsson, D. Schenten, and J. Sodroski. 1999. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immunodeficiency virus passaged in vivo. J. Virol. 73:8873-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng, Y., C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez, E., M. Bamshad, N. Sato, S. Mummidi, R. Dhanda, G. Catano, S. Cabrera, M. McBride, X. H. Cao, G. Merrill, P. O'Connell, D. W. Bowden, B. I. Freedman, S. A. Anderson, E. A. Walter, J. S. Evans, K. T. Stephan, R. A. Clark, S. Tyagi, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 1999. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc. Natl. Acad. Sci. USA 96:12004-12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez, E., R. Dhanda, M. Bamshad, S. Mummidi, R. Geevarghese, G. Catano, S. A. Anderson, E. A. Walter, K. T. Stephan, M. F. Hammer, A. Mangano, L. Sen, R. A. Clark, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 2001. Global survey of genetic variation in CCR5, RANTES, and MIP-1α: impact on the epidemiology of the HIV-1 pandemic. Proc. Natl. Acad. Sci. USA 98:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, Q., A. P. Barry, Z. Wang, S. M. Connolly, S. C. Peiper, and M. L. Greenberg. 2000. Evolution of the human immunodeficiency virus type 1 envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J. Virol. 74:11858-11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klenerman, P., S. Rowland-Jones, S. McAdam, J. Edwards, S. Daenke, D. Lalloo, B. Koppe, W. Rosenberg, D. Boyd, A. Edwards, et al. 1994. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature 369:403-407. [DOI] [PubMed] [Google Scholar]
- 27.Kozak, S. L., S. E. Kuhmann, E. J. Platt, and D. Kabat. 1999. Roles of CD4 and coreceptors in binding, endocytosis, and proteolysis of gp120 envelope glycoproteins derived from human immunodeficiency virus type 1. J. Biol. Chem. 274:23499-23507. [DOI] [PubMed] [Google Scholar]
- 28.Lahm, H. W., and S. Stein. 1985. Characterization of recombinant human interleukin-2 with micromethods. J. Chromatogr. 326:357-361. [DOI] [PubMed] [Google Scholar]
- 29.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 1959. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, S. L., T. Schacker, L. Musey, D. Shriner, M. J. McElrath, L. Corey, and J. I. Mullins. 1997. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J. Virol. 71:4284-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 32.Lukashov, V. V., C. L. Kuiken, and J. Goudsmit. 1995. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J. Virol. 69:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangano, A., E. Gonzalez, R. Dhanda, G. Catano, M. Bamshad, A. Bock, R. Duggirala, K. Williams, S. Mummidi, R. A. Clark, S. S. Ahuja, M. J. Dolan, R. Bologna, L. Sen, and S. K. Ahuja. 2001. Concordance between the CC chemokine receptor 5 genetic determinants that alter risks of transmission and disease progression in children exposed perinatally to human immunodeficiency virus. J. Infect. Dis. 183:1574-1585. [DOI] [PubMed] [Google Scholar]
- 34.Markham, R. B., W. C. Wang, A. E. Weisstein, Z. Wang, A. Munoz, A. Templeton, J. Margolick, D. Vlahov, T. Quinn, H. Farzadegan, and X. F. Yu. 1998. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc. Natl. Acad. Sci. USA 95:12568-12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald, R. A., D. L. Mayers, R. C.-Y. Chung, K. F. Wagner, S. Ratto-Kim, D. L. Birx, and N. L. Michael. 1997. Evolution of human immunodeficiency virus type 1 env sequence variation in patients with diverse rates of disease progression and T-cell function. J. Virol. 71:1871-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGrath, K. M., N. G. Hoffman, W. Resch, J. A. Nelson, and R. Swanstrom. 2001. Using HIV-1 sequence variability to explore virus biology. Virus Res. 76:137-160. [DOI] [PubMed] [Google Scholar]
- 38.Merat, R., H. Raoul, T. Leste-Lasserre, P. Sonigo, and G. Pancino. 1999. Variable constraints on the principal immunodominant domain of the transmembrane glycoprotein of human immunodeficiency virus type 1. J. Virol. 73:5698-5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mummidi, S., M. Bamshad, S. S. Ahuja, E. Gonzalez, P. M. Feuillet, K. Begum, M. C. Galvis, V. Kostecki, A. J. Valente, K. K. Murthy, L. Haro, M. J. Dolan, J. S. Allan, and S. K. Ahuja. 2000. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J. Biol. Chem. 275:18946-18961. [DOI] [PubMed] [Google Scholar]
- 41.Nokta, M. A., X. D. Li, J. Nichols, M. Mallen, A. Pou, D. Asmuth, and R. B. Pollard. 2001. Chemokine/CD4 receptor density ratios correlate with HIV replication in lymph node and peripheral blood of HIV-infected individuals. AIDS 15:161-169. [DOI] [PubMed] [Google Scholar]
- 42.Novella, I. S., D. K. Clarke, J. Quer, E. A. Duarte, C. H. Lee, S. C. Weaver, S. F. Elena, A. Moya, E. Domingo, and J. J. Holland. 1995. Extreme fitness differences in mammalian and insect hosts after continuous replication of vesicular stomatitis virus in sandfly cells. J. Virol. 69:6805-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Brien, S. J., and J. P. Moore. 2000. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 177:99-111. [DOI] [PubMed] [Google Scholar]
- 44.Park, E. J., and G. V. Quinnan, Jr. 1999. Both neutralization resistance and high infectivity phenotypes are caused by mutations of interacting residues in the human immunodeficiency virus type 1 gp41 leucine zipper and the gp120 receptor- and coreceptor-binding domains. J. Virol. 73:5707-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poss, M., H. Martin, J. Kreiss, P. Nyange, K. Mandaliya, B. Chohan, and J. Overbaugh. 1995. Diversity in virus populations from genital mucosa and peripheral blood in women recently infected with HIV. J. Virol. 69:8118-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poss, M., and J. Overbaugh. 1999. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J. Virol. 73:5255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poss, M., A. G. Rodrigo, J. J. Gosink, G. H. Learn, D. D. Panteleeff, H. L. Martin, Jr., J. Bwayo, J. K. Kreiss, and J. Overbaugh. 1998. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J. Virol. 72:8240-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roben, P., J. Moore, M. Thali, J. Sodroski, C. Barbas III, and D. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowland-Jones, S., and R. Tan. 1997. Control of HIV co-receptor expression: implications for pathogenesis and treatment. Trends Microbiol. 5:300-302, 302-303. [DOI] [PubMed]
- 50.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shioda, T., S. Oka, X. Xin, H. Liu, R. Harukuni, A. Kurotani, M. Fukushima, M. K. Hasan, T. Shiino, Y. Takebe, A. Iwamoto, and Y. Nagai. 1997. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J. Virol. 71:4871-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmons, G., J. D. Reeves, S. Hibbitts, J. T. Stine, P. W. Gray, A. E. Proudfoot, and P. R. Clapham. 2000. Co-receptor use by HIV and inhibition of HIV infection by chemokine receptor ligands. Immunol. Rev. 177:112-126. [DOI] [PubMed] [Google Scholar]
- 54.Singh, A., and R. G. Collman. 2000. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J. Virol. 74:10229-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth, R. J., Y. Yi, A. Singh, and R. G. Collman. 1998. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J. Virol. 72:4478-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tokunaga, K., M. L. Greenberg, M. A. Morse, R. I. Cumming, H. K. Lyerly, and B. R. Cullen. 2001. Molecular basis for cell tropism of cxcr4-dependent human immunodeficiency virus type 1 isolates. J. Virol. 75:6776-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verrier, F., A. Nadas, M. K. Gorny, and S. Zolla-Pazner. 2001. Additive effects characterize the interaction of antibodies involved in neutralization of the primary dualtropic human immunodeficiency virus type 1 isolate 89.6. J. Virol. 75:9177-9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, B., R. B. Lal, D. E. Dwyer, M. Miranda-Saksena, R. Boadle, A. L. Cunningham, and N. K. Saksena. 2000. Molecular and biological interactions between two HIV-1 strains from a coinfected patient reveal the first evidence in favor of viral synergism. Virology 274:105-119. [DOI] [PubMed] [Google Scholar]
- 60.Weaver, S. C., A. C. Brault, W. Kang, and J. J. Holland. 1999. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J. Virol. 73:4316-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 62.Wu, L., W. A. Paxton, N. Kassam, N. Ruffing, J. B. Rottman, N. Sullivan, H. Choe, J. Sodroski, W. Newman, R. A. Koup, and C. R. Mackay. 1997. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med. 185:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, L. Q., P. MacKenzie, A. Cleland, E. C. Holmes, A. J. Leigh Brown, and P. Simmonds. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y. J., T. Dragic, Y. Cao, L. Kostrikis, D. S. Kwon, D. R. Littman, V. N. KewalRamani, and J. P. Moore. 1998. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 72:9337-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]