Abstract
A new mycobacteriophage-based technique (PhageTek MB) was compared with standard culture and staining techniques for diagnosis of pulmonary tuberculosis. A total of 2,048 respiratory specimens from 1,466 patients collected from February 2000 to March 2001 were studied by both (i) conventional methods (direct microscopic examination [auramine-rhodamine fluorochrome], and culture in BacT/ALERT 3D and solid media) and (ii) the PhageTek MB assay. This phenotypic test utilizes specific mycobacteriophages to detect the presence of live Mycobacterium tuberculosis complex organisms within a decontaminated clinical sample. Overall, 205 (10%) specimens were positive for mycobacteria (134 patients): 144 (70.2%) M. tuberculosis isolates and 61 (29.8%) nontuberculous mycobacterium isolates (30 Mycobacterium kansasii, 12 Mycobacterium xenopi, 9 Mycobacterium gordonae, 7 Mycobacterium avium complex, 2 Mycobacterium chelonae, and 1 Mycobacterium fortuitum isolate). PhageTek MB was more likely to give a positive result with specimens in which high numbers of acid-fast bacilli were observed on the smear. The sensitivity, specificity, and positive and negative predictive values of this mycobacteriophage-based technique versus culture for M. tuberculosis were 58.3, 99.1, 83.2, and 96.9%, respectively. PhageTek MB is a rapid (48-h), specific, safe, and easy-to-perform test. According to the prevalence of the disease in the population studied, the test would require improved sensitivity in order to be used as a screening test for routine diagnosis of respiratory tuberculosis in our setting.
Tuberculosis continues to be a global health problem, with more than 8 million new cases and 2 million deaths each year (24). The spread of human immunodeficiency virus (HIV) infection and the breakdown in health services have contributed to a dramatic rise in the incidence of tuberculosis. The great majority of the new tuberculosis cases (95%) and tuberculosis deaths (98%) are in developing regions such as Southeast Asia, sub-Saharan Africa, Latin America, and Eastern Europe (15, 20). One of the principles of tuberculosis control is rapid and accurate diagnosis of infected patients in order to allow prompt initiation of antibiotic therapy and to prevent transmission. Although the conventional procedures are irreplaceable diagnostic tools, detection of acid-fast bacilli by microscopy shows poor sensitivity, and solid culture methods can take as long as 8 weeks (liquid culture methods are faster but unaffordable for laboratories in low-income areas). In addition, coinfection with HIV has changed the clinical presentation of tuberculosis and reduced the sensitivity of classical microbiology methods (5, 9, 17). Therefore, in developing countries, it is especially important to have an inexpensive and rapid test for tuberculosis identification so that infected individuals can be isolated and treated immediately (10).
Mycobacteriophages constitute a potentially useful approach for detecting viable Mycobacterium tuberculosis bacilli as well as for susceptibility studies (22). Several mycobacteriophages have been reported to be highly specific for all M. tuberculosis complex species (11, 13, 25). These mycobacteriophages have been important in the development of phage-based technology, as shown in some studies (7, 14, 28). Their relatively rapid replication (which compensates for the otherwise slow growth of their hosts), the simplicity of the methodology, and the relatively inexpensive equipment required make the use of mycobacteriophages a suitable tool for the rapid diagnosis of tuberculosis.
The PhageTek MB assay (Organon Teknika Corporation, Durham, N.C.; manufactured by Biotec Laboratories Ltd., Ipswich, United Kingdom, as a variant of its FASTPlaqueTB test) is a phenotypic assay that uses M. tuberculosis complex-specific mycobacteriophages to report the presence of live M. tuberculosis complex organisms within a sample. The target bacteria in a decontaminated respiratory specimen are rapidly infected by the target-specific bacteriophage. A selective virucide is added which, without affecting the cells, destroys all exogenous phages that have not infected the M. tuberculosis complex bacilli. The phages protected within the M. tuberculosis complex organisms replicate and form clear areas (plaques) in a lawn of rapidly growing host helper cells. Although the number of plaques generated from a given sample is related to the number of viable M. tuberculosis complex cells containing mycobacteriophage, this test is qualitative. Results from specimens can be read by eye after 48 h.
The aim of this study was to evaluate the diagnostic value of the PhageTek MB kit for pulmonary tuberculosis in comparison with standard culture (solid and liquid media) and staining techniques for respiratory specimens. In addition, a specificity study of this mycobacteriophage-based assay was performed with different mycobacterial species.
(This study was presented in part at the 11th European Congress of Clinical Microbiology and Infectious Diseases, Istanbul, Turkey, April 2001 [P. Orús, P. Berlanga, N. Galí, F. Alcaide, S. Blanco, M. A. Benítez, J. Domínguez, P. Pedroso, V. Ausina, and R. Martín, abstr. P1136, p. 235-236].)
MATERIALS AND METHODS
Specimens.
A total of 2,048 respiratory specimens collected from 1,466 patients between February 2000 and March 2001 at the Hospital Universitari de Bellvitge and the Hospital Universitari Germans Trias i Pujol, Barcelona, Spain, were studied. The clinical samples included 1,443 (70.5%) sputum specimens, 525 (25.6%) bronchial or tracheal aspirate specimens, 79 (3.9%) bronchoalveolar lavage specimens, and 1 (0.04%) transthoracic aspirate specimen.
Specimen processing, microscopy, and culture.
All samples were processed within 24 h of specimen collection. The specimens were processed according to the conventional N-acetyl-l-cysteine-NaOH digestion-decontamination procedure (16). After they had been processed and concentrated by centrifugation, the final sediments were suspended in 2 ml of phosphate buffer (pH 6.8). This suspension was then used for the preparation of smears for acid-fast staining with auramine-rhodamine fluorochrome and for inoculation into two media: an MB/BacT bottle (bioMérieux sa, Marcy l'Etoile, France) and a Löwenstein-Jensen (LJ) slant (MAIM, Barcelona, Spain) used as a solid medium. The MB/BacT antibiotic supplement (amphotericin B, azlocillin, nalidixic acid, polymyxin B, trimethoprim, and vancomycin) was added for culture of nonsterile specimens. An equal volume (0.5 ml) of the processed specimens was inoculated at the same time in each medium studied. All cultures were incubated at 35 to 37°C for as long as 6 weeks. The MB/BacT bottles were registered and placed in the BacT/ALERT 3D instrument (bioMérieux sa) for incubation and continuous monitoring as recommended by the manufacturer. The LJ medium was incubated under 5% CO2 and was examined for colonies on the slant once a week.
Mycobacterial species identification.
Mycobacterial isolates were identified by conventional biochemical and culture tests, PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the hsp65 gene (2, 23), and DNA probes (AccuProbe; GenProbe Inc., San Diego, Calif.). These gene probes were selectively applied to each positive culture to identify M. tuberculosis complex, Mycobacterium avium complex, Mycobacterium kansasii, and Mycobacterium gordonae on the basis of the pigmentation of the pellet and morphological characteristics by microscopic examination.
Mycobacteriophage-based assay (PhageTek MB).
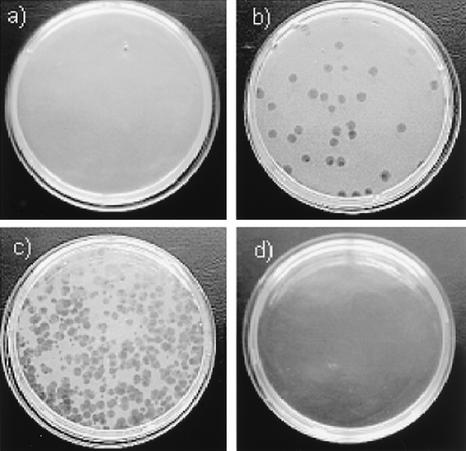
The mycobacteriophage-based assay was performed and interpreted according to the manufacturer's recommendations by using the reagents supplied in the PhageTek MB kit. Briefly, 1 ml of the decontaminated specimens was washed and incubated overnight at 37°C in Middlebrook 7H9 medium with 10% oleic acid-albumin-dextrose-catalase (OADC). One hundred microliters of phages was then added, and the mixture was incubated at 37°C for 1 h. Following incubation, extracellular phage were inactivated by addition of 100 μl of virucidal solution. After careful mixing to ensure that all the phage were exposed to the virucide, the sample was incubated at room temperature for 5 min, followed by addition of 5 ml of Middlebrook 7H9 medium with 10% OADC and 1 ml of a suspension of rapidly growing host helper cells (Mycobacterium smegmatis) in which the released mycobacteriophage could infect and continue amplification. Finally, the mixture was plated with 5 ml of molten agar. The plates were allowed to set and were incubated for 18 to 24 h at 37°C. Positive and negative controls were included with each batch of specimens. Released phage (from target M. tuberculosis) were detected as zones of lysis (plaques) in a lawn of helper cells. Plaques are representative of viable M. tuberculosis complex bacilli present in the sample. Results were interpreted as follows: 0 to 19 plaques, negative; ≥20 plaques, positive (Fig. 1).
FIG. 1.
Interpretation of results of the PhageTek MB assay. (a) Negative; no plaques. (b) Positive; countable plaques, numbering 20 or more. (c) Positive; confluent lysis. (d) Positive; complete lysis. This image was kindly provided by Biotec Laboratories Ltd.
Assay specificity.
The analytical specificity of the PhageTek MB assay was assessed by testing mycobacterial strains isolated from clinical specimens (1 Mycobacterium abscessus, 1 M. avium complex, 1 Mycobacterium gastri, 1 M. gordonae, 1 Mycobacterium intracellulare, 7 M. kansasii, and 2 Mycobacterium xenopi isolates), 5 reference strains (1 M. avium [ATCC 25291], 1 M. gastri [IP140340005], 2 M. kansasii [ATCC 12478 and ATCC 35755], and 1 Mycobacterium mucogenicum [ATCC 49650] strain), and 2 isolates from environmental samples (both M. kansasii). All the strains had been identified by standard methods previously. M. kansasii strains had also been studied previously according to subtype, determined by a reverse hybridization line-probe assay (LiPA Mycobacteria Test; Innogenetics, Zwijnaarde, Belgium) or by AccuProbe and PCR-RFLP of the hsp65 gene (2, 23, 26). Fresh cultures (3 to 6 weeks old) from solid media were used as sources of mycobacterial strains. The mycobacterial growths were transferred to sterile screw-cap glass tubes containing liquid culture medium (Middlebrook 7H9 medium with 10% OADC) and six to eight glass beads. The suspensions were homogenized by using a vortex mixer for 15 to 20 s. Large clumps were allowed to settle by letting the suspensions stand for at least 5 min. The supernatants were transferred into sterile tubes, and the turbidity was adjusted to 0.5 McFarland standard. Several dilutions of the standardized suspensions were tested following the assay procedure. Colony counts were performed by plating duplicate 0.1-ml volumes of dilutions of the suspensions onto Middlebrook 7H11 medium and incubating for as long as 6 weeks at 37°C. Final colony counts ranged from 107 to 102 CFU/ml.
Statistical analysis.
Sensitivities, specificities, and positive and negative predictive values were calculated by standard methods (12).
RESULTS
A total of 205 (10%) specimens from 134 patients were positive for mycobacteria. The distribution of mycobacteria by species isolated was as follows: 144 (70.2%) M. tuberculosis complex isolates and 61 (29.8%) nontuberculous mycobacteria (NTM) (30 M. kansasii, 12 M. xenopi, 9 M. gordonae, 7 M. avium complex, 2 Mycobacterium chelonae, and 1 Mycobacterium fortuitum isolate). Of the overall specimens from which mycobacteria could be isolated, 122 (59.5%) were smear positive (105 M. tuberculosis and 17 NTM isolates) and 83 (40.5%) were smear negative (39 M. tuberculosis and 44 NTM isolates).
Table 1 compares PhageTek MB results with culture results, while Table 2 presents PhageTek MB results for M. tuberculosis culture-positive specimens according to smear results. In comparison with culture, the sensitivity, specificity, and positive and negative predictive values of auramine smear microscopy in the detection of M. tuberculosis were 72.9, 98.7, 80.8, and 98%, respectively. PhageTek MB was more likely to give positive results for specimens in which high numbers of acid-fast bacilli were observed on the smear (Table 2). Quantification results of culture on solid media were available for 27 out of the 144 M. tuberculosis culture-positive specimens (Table 3). The highest number of PhageTek MB-positive specimens was found among samples from which the recovery of M. tuberculosis bacilli was higher (>30 colonies isolated/slant). The overall sensitivity, specificity, and positive and negative predictive values for detection of M. tuberculosis by PhageTek MB, compared with culture results, were 58.3, 99.1, 83.2, and 96.9%, respectively (Table 4). Seventy-seven discrepant results between PhageTek MB and culture were observed: 60 specimens from 47 patients with M. tuberculosis isolates were negative by PhageTek MB but positive by culture, and 17 specimens from 17 patients were positive by PhageTek MB but negative by culture. Of the 47 patients with false-negative PhageTek MB results (26 of which were smear-positive results), 17 patients were undergoing tuberculosis treatment. In addition, of the 17 patients with false-positive PhageTek MB results (6 of which were smear positive), 8 patients had cultures positive for M. kansasii and 9 patients had no clinical or radiological criteria for diagnosis of tuberculosis or any other mycobacterial infection.
TABLE 1.
Comparison of PhageTek MB results with culture results
| PhageTek MB result | Total no. of specimens | No. of cultures with the indicated result in the following medium:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MB/BacT
|
LJ medium
|
MB/BacT plus LJ medium
|
||||||||
| M. tuberculosis positive | NTM positive | Negative | M. tuberculosis positive | NTM positive | Negative | M. tuberculosis positive | NTM positive | Negative | ||
| Positive | 101 | 84 | 8 | 9 | 79 | 8 | 14 | 84 | 8a | 9b |
| Negative | 1,947 | 54 | 48 | 1,845 | 55 | 32 | 1,860 | 60c | 53 | 1,834 |
| Total | 2,048 | 138 | 56 | 1,854 | 134 | 40 | 1,874 | 144 | 61 | 1,843 |
Eight patients with M. kansasii (6 smear positive).
Nine patients without diagnosis of tuberculosis or NTM.
Forty-seven patients (17 under treatment).
TABLE 2.
PhageTek MB results for culture-positive M. tuberculosis specimens according to smear result
| Smear resulta | No. of specimens with the following PhageTek MB result:
|
Total no. of specimens | |
|---|---|---|---|
| Positive | Negative | ||
| Negative | 5 | 34 | 39 |
| + | 23 | 13 | 36 |
| ++ | 23 | 9 | 32 |
| +++ | 33 | 4 | 37 |
| Total | 84 | 60 | 144 |
Negative, no acid-fast bacilli observed; +, 1 to 9 bacilli/10 fields; ++, 1 to 9 bacilli/field; +++, >9 bacilli/field.
TABLE 3.
Comparison of PhageTek MB results with solid-culture quantification results
| PhageTek MB result (no. of specimens) | No. of specimens with the following culture resulta:
|
||
|---|---|---|---|
| + | ++ | +++ | |
| Positive (15) | 0 | 3 | 12 |
| Negative (12) | 6 | 4 | 2 |
| Total (27) | 6 | 7 | 14 |
+, 1 to 10 colonies isolated; ++, 11 to 30 colonies isolated; +++, >30 colonies isolated.
TABLE 4.
Detection of M. tuberculosis complex by PhageTek MB in smear-positive and smear-negative specimens
| Smear result (no. of specimens) | No. of specimens
|
PhageTek MB performancea
|
||||||
|---|---|---|---|---|---|---|---|---|
|
M. tuberculosis culture positive (144)
|
M. tuberculosis culture negative or NTM (1,904)
|
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
| Phage positive | Phage negative | Phage positive | Phage negative | |||||
| Smear positive (130)b | 79 | 26 | 6c | 19d | 75.2 | 76 | 92.9 | 42.2 |
| Smear negative (1,918) | 5 | 34 | 11e | 1,868 | 12.8 | 99.4 | 31.3 | 98.2 |
| Total (2,048) | 84 | 60f | 17 | 1,887 | 58.3 | 99.1 | 83.2 | 96.9 |
PPV, positive predictive value; NPV, negative predictive value.
Comprising 105 specimens with M. tuberculosis, 16 with M. kansasii, 1 with M. gordonae, and 8 culture-negative samples.
Six patients with M. kansasii.
Eight specimens from 6 patients (4 with tuberculosis under treatment), 10 samples with M. kansasii, and 1 specimen with M. gordonae.
Two patients with M. kansasii and 9 patients without diagnosis of tuberculosis.
Forty-seven patients (17 under treatment).
Analysis of assay specificity using different mycobacterial strains revealed that all the reference and clinical strains of M. kansasii and M. gastri studied were positive by PhageTek MB when the highest CFU counts were tested (Table 5). M. avium complex and M. fortuitum were also PhageTek MB positive in testing of the highest bacillus counts at 1 × 106 and 2 × 106 CFU/ml, respectively. The remaining strains (M. abscessus, M. avium, M. gordonae, M. intracellulare, M. mucogenicum, and M. xenopi), including the two environmental M. kansasii strains, were PhageTek MB negative (data not shown).
TABLE 5.
PhageTek MB results according to the strain and type of M. kansasii and M. gastri
| Strain and typea | Source | No. of bacilli (CFU/ml) | No. of plaques | PhageTek result |
|---|---|---|---|---|
| M. kansasii I | Reference strain (ATCC 12478) | 1.5 × 107 | Total lysis | Positive |
| 1.5 × 105 | Confluent lysis | Positive | ||
| 1.5 × 103 | 198 | Positive | ||
| M. kansasii I | Reference strain (ATCC 35775) | 1 × 106 | Confluent lysis | Positive |
| 1 × 104 | 102 | Positive | ||
| 1 × 102 | 0 | Negative | ||
| M. kansasii I | Clinical isolate | 1.4 × 106 | Total lysis | Positive |
| 1.4 × 105 | Confluent lysis | Positive | ||
| 1.4 × 104 | 276 | Positive | ||
| M. kansasii I | Clinical isolate | 1.1 × 107 | Total lysis | Positive |
| 1.1 × 105 | >300 | Positive | ||
| 1.1 × 103 | 5 | Negative | ||
| M. kansasii II | Clinical isolate | 1.3 × 107 | Confluent lysis | Positive |
| 1.3 × 105 | >300 | Positive | ||
| 1.3 × 103 | 23 | Positive | ||
| M. kansasii III | Clinical isolate | 1.8 × 107 | Confluent lysis | Positive |
| 1.8 × 105 | 246 | Positive | ||
| 1.8 × 103 | 15 | Negative | ||
| M. kansasii IV | Clinical isolate | 1.8 × 107 | Confluent lysis | Positive |
| 1.8 × 105 | 186 | Positive | ||
| 1.8 × 103 | 9 | Negative | ||
| M. kansasii V | Clinical isolate | 2 × 107 | Total lysis | Positive |
| 2 × 105 | Confluent lysis | Positive | ||
| 2 × 103 | 268 | Positive | ||
| M. kansasii VI | Clinical isolate | 1.8 × 107 | Total lysis | Positive |
| 1.8 × 105 | Confluent lysis | Positive | ||
| 1.8 × 103 | 192 | Positive | ||
| M. kansasii III, IV, Vb | Environmental | 3.5 × 105 | 0 | Negative |
| M. kansasii III, IV, Vb | Environmental | 5.5 × 104 | 0 | Negative |
| M. gastri | Reference strain (IP 140340005) | 4.8 × 107 | Confluent lysis | Positive |
| 4.8 × 105 | 25 | Positive | ||
| 4.8 × 103 | 0 | Negative | ||
| M. gastrib | Clinical isolate | 1.8 × 107 | Confluent lysis | Positive |
| 1.8 × 105 | 176 | Positive | ||
| 1.8 × 103 | 4 | Negative |
DISCUSSION
Since 1947 more than 250 mycobacteriophage types have been identified (13). Nevertheless, the potential clinical application of mycobacteriophages for rapid diagnosis and drug susceptibility testing of M. tuberculosis has only recently been proposed (4, 6, 14, 21, 27, 28). The sensitivity of mycobacteriophage-based techniques relies mainly on the biological amplification of bacteriophage particles. In this way, our study showed a high sensitivity of PhageTek MB with smear-positive specimens as well as with those samples from which a high number of M. tuberculosis bacilli were isolated. However, although these techniques are able to detect fewer than 10 CFU of bacilli in a 100-μl sample (data not shown), the PhageTek MB showed poor sensitivity with smear-negative specimens and with those specimens from which a low number of colonies were recovered on solid media.
Although this study was performed in a high-NTM-prevalence population, the specificity of the test was found to be acceptable when clinical specimens were evaluated. Nevertheless, the mycobacteriophage-based test provided nine positive results for patients without a microbiological or clinical diagnosis of tuberculosis (smear-negative, culture-negative specimens from patients not under therapy affecting the viability of bacilli). A possible explanation could be insufficient virucide activity leading to plaque formation from infected helper cells, not from M. tuberculosis bacilli previously present in the specimen.
The positive results found with highly concentrated suspensions of M. kansasii, M. gastri, and M. avium complex in the assay specificity evaluation may not have important clinical implications in developing countries, where the NTM prevalence is substantially lower than in our area (8).
Only a few studies have assessed the diagnostic utility of the phage technology by using clinical samples (1, 3, 18, 19). Although the results of those studies have shown a high sensitivity and specificity of the test, some important issues should be taken into consideration. Two trials conducted in Cape Town, South Africa (1), and Karachi, Pakistan (19), compared the mycobacteriophage-based test with microscopic examination and a solid medium (LJ), while a combination of solid and liquid media (MB/BacT) was used in the present study. Furthermore, the LJ slants were inoculated with 0.1 and 0.2 ml of the treated specimen suspensions in the Cape Town and Karachi trials, respectively, instead of 0.5 ml of suspension, the amount used in our evaluation. These factors, as is well known, improve the recovery rates of mycobacteria, which could have important implications for the sensitivity and positive predictive values achieved by the mycobacteriophage-based technique in comparison with culture findings. Even so, the overall sensitivity of the mycobacteriophage-based test in the present study (58.3%) is lower than that found (70%) in the Cape Town trial (1); if one considers only the smear positive specimens (data not specified in that evaluation), then the sensitivity is similar (75.2%). On the other hand, in one study performed in Mexico (3), the luciferase reporter mycobacteriophages detected 92% of the mycobacterial isolates recovered from sputum samples (all positive acid-fast bacillus smear specimens) and were able to distinguish M. tuberculosis complex from other mycobacteria by using p-nitro-α-acetylamino-β-hydroxy propiophenone. Our evaluation, unlike the one conducted in Mexico (3), included smear-positive and smear-negative samples, and not only sputum samples but also other kinds of specimens, which gave us the opportunity of assessing the clinical performance of the phage-based test under nonoptimal conditions. In addition, specimens from patients on tuberculosis treatment were also included in the present study, whereas only new tuberculosis suspects were included in both the Cape Town and Karachi trials.
The mycobacteriophage-based assay showed low sensitivity in our study, making it unsuitable as a screening test in our area. However, this methodology could be implemented in low-income countries where access to health services is poor and the patients' samples may present higher bacillus counts due to more-extensive disease. In addition, because this assay is based on the principle of detecting viable bacilli, it has the potential to be used as a technique to evaluate the response to antituberculosis drug therapy. Four patients under tuberculosis treatment included in our study were smear positive and culture and PhageTek MB negative, indicating the importance of the viability of the bacilli within the specimen. Further investigations are required in order to assess the real utility of the phage-based technique in antituberculosis therapy monitoring.
In the last annual report on global tuberculosis control published by the World Health Organization (29), reaching 70% case detection by 2005 is presented as one of the most important aims. This mycobacteriophage-based test could be considered a suitable and affordable tool to aid in improving case detection in developing areas, because it is a rapid method that does not require any specialized or automated equipment. Furthermore, the reagents needed to perform the assay do not represent a major expense for laboratories in resource-poor areas.
Acknowledgments
We are grateful to M. Tutusaus, M. Juncal, E. Martínez, M. Pérez, and P. Pedroso for technical assistance.
We also thank Organon Teknika Corporation for providing us with the PhageTek MB kits.
REFERENCES
- 1.Albert, H., A. Heydenrych, R. Brookes, R. J. Mole, B. Harley, E. Subotsky, R. Henry, and V. Azevedo. 2002. Performance of a rapid phage-based test, FASTPlaqueTBTM, to diagnose pulmonary tuberculosis from sputum specimens in South Africa. Int. J. Tuberc. Lung Dis. 6:529-537. [DOI] [PubMed] [Google Scholar]
- 2.Alcaide, F., I. Richter, C. Bernasconi, B. Springer, C. Hagenau, R. Schulze-Röbbecke, E. Tortoli, R. Martín, E. C. Böttger, and A. Telenti. 1997. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J. Clin. Microbiol. 35:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banaiee, N., M. Bobadilla-Del-Valle, S. Bardarov, Jr., P. F. Riska, P. M. Small, A. Ponce-De-Leon, W. R. Jacobs, Jr., G. F. Hatfull, and J. Sifuentes-Osornio. 2001. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J. Clin. Microbiol. 39:3883-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carriere, C., P. F. Riska, O. Zimhony, J. Kriakov, S. Bardarov, J. Burns, J. Chan, and W. R. Jacobs, Jr. 1997. Conditionally replicating luciferase reporter phages: improved sensitivity for rapid detection and assessment of drug susceptibility of Mycobacterium tuberculosis. J. Clin. Microbiol. 35:3232-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colebunders, R., and I. Bastian. 2000. A review of the diagnosis and treatment of smear-negative pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 4:97-107. [PubMed] [Google Scholar]
- 6.Eltringham, I. J., F. A. Drobniewski, J. A. Mangan, P. D. Butcher, and S. M. Wilson. 1999. Evaluation of reverse transcription-PCR and a bacteriophage-based assay for rapid phenotypic detection of rifampin resistance in clinical isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:3524-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eltringham, I. J., S. M. Wilson, and F. A. Drobniewski. 1999. Evaluation of a bacteriophage-based assay (phage amplified biologically assay) as a rapid screen for resistance to isoniazid, ethambutol, streptomycin, pyrazinamide, and ciprofloxacin among clinical isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:3528-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falloon, J. 2000. Pulmonary manifestations of human inmunodeficiency virus infection, p. 1415-1426. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 5th ed. Churchill Livingstone, New York, N.Y.
- 10.Foulds, H., and R. O'Brien. 1998. New tools for the diagnosis of tuberculosis: the perspective of developing countries. Int. J. Tuberc. Lung Dis. 2:778-783. [PubMed] [Google Scholar]
- 11.Froman, S., D. W. Will, and E. Bogen. 1954. Bacteriophage active against virulent Mycobacterium tuberculosis. I. Isolation and activity. Am. J. Public Health 44:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galen, R. S., and S. R. Gambino. 1975. Beyond normality: the predictive value and efficiency of medical diagnosis. John Wiley & Sons, New York, N.Y.
- 13.Gardner, G. M., and R. S. Weiser. 1947. A bacteriophage for Mycobacterium smegmatis. Proc. Soc. Exp. Biol. Med. 66:205-206. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs, W. R., Jr., R. G. Barletta, R. Udani, J. Chan, G. Kalkut, G. Sosne, T. Kieser, G. J. Sarkis, G. F. Hatfull, and B. R. Bloom. 1993. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science 260:819-822. [DOI] [PubMed] [Google Scholar]
- 15.Kochi, A. 1991. The global tuberculosis situation and the new control strategy of the World Health Organization. Tubercle 72:1-6. [DOI] [PubMed] [Google Scholar]
- 16.Kubica, G. P., W. E. Dye, M. L. Cohn, and G. Middlebrook. 1963. Sputum digestion and decontamination with N-acetyl-l-cysteine-sodium hydroxide for culture of mycobacteria. Am. Rev. Respir. Dis. 87:775-779. [DOI] [PubMed] [Google Scholar]
- 17.Long, R. 2001. Smear-negative pulmonary tuberculosis in industrialized countries. Chest 120:330-334. [DOI] [PubMed] [Google Scholar]
- 18.Mole, R. J., and T. W. O. Maskell. 2001. Phage as a diagnostic—the use of phage in TB diagnosis. J. Chem. Technol. Biotechnol. 76:683-688. [Google Scholar]
- 19.Muzaffar, R., S. Batool, F. Aziz, A. Naqvi, and A. Rizvi. 2002. Evaluation of the FASTPlaqueTB assay for direct detection of Mycobacterium tuberculosis in sputum specimens. Int. J. Tuberc. Lung Dis. 6:635-640. [PubMed] [Google Scholar]
- 20.Narain, J. P., M. C. Raviglione, and A. Kochi. 1992. HIV-associated tuberculosis in developing countries: epidemiology and strategies for prevention. Tuber. Lung Dis. 73:311-321. [DOI] [PubMed] [Google Scholar]
- 21.Pearson, R. E., S. Jurgensen, G. J. Sarkis, G. F. Hatfull, and W. R. Jacobs, Jr. 1996. Construction of D29 shuttle phasmids and luciferase reporter phages for detection of mycobacteria. Gene 183:129-136. [DOI] [PubMed] [Google Scholar]
- 22.Perkins, M. D. 2000. New diagnostic tools for tuberculosis. Int. J. Tuberc. Lung Dis. 4:S182-S188. [PubMed] [Google Scholar]
- 23.Picardeau, M., G. Prod'Hom, L. Raskine, M. P. LePennec, and V. Vincent. 1997. Genotypic characterization of five subspecies of Mycobacterium kansasii. J. Clin. Microbiol. 35:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raviglione, M. C., D. E. Snider, Jr., and A. Kochi. 1995. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA 273:220-226. [PubMed] [Google Scholar]
- 25.Redmond, W. B., and J. C. Cater. 1960. A bacteriophage specific for Mycobacterium tuberculosis varieties hominis and bovis. Am. Rev. Respir. Dis. 82:781-786. [DOI] [PubMed] [Google Scholar]
- 26.Richter, E., S. Niemann, S. Rüsch-Gerdes, and S. Hoffner. 1999. Identification of Mycobacterium kansasii by using a DNA probe (AccuProbe) and molecular techniques. J. Clin. Microbiol. 37:964-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riska, P. F., W. R. Jacobs, Jr., B. R. Bloom, J. McKitrick, and J. Chan. 1997. Specific identification of Mycobacterium tuberculosis with the luciferase reporter mycobacteriophage: use of p-nitro-α-acetylamino-β-hydroxy propiophenone. J. Clin. Microbiol. 35:3225-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson, S. M., Z. al-Suwaidi, R. McNerney, J. Porter, and F. A. Drobniewski. 1997. Evaluation of a new rapid bacteriophage-based method for the drug susceptibility testing of Mycobacterium tuberculosis. Nat. Med. 3:465-468. [DOI] [PubMed] [Google Scholar]
- 29.World Health Organization. 2002. Global tuberculosis control: surveillance, planning, financing. WHO report 2002. WHO/CDS/TB/2002.295. World Health Organization, Geneva, Switzerland.