Abstract
MULTIGEN technology (T. Vinayagamoorthy, U.S. patent 6,197,510, March 2001) is a modification of conventional sequencing technology that generates a single electropherogram consisting of short nucleotide sequences from a mixture of known DNA targets. The target sequences may be present on the same or different nucleic acid molecules. For example, when two DNA targets are sequenced, the first and second sequencing primers are annealed to their respective target sequences, and then a polymerase causes chain extension by the addition of new deoxyribose nucleotides. Since the electrophoretic separation depends on the relative molecular weights of the truncated molecules, the molecular weight of the second sequencing primer was specifically designed to be higher than the combined molecular weight of the first sequencing primer plus the molecular weight of the largest truncated molecule generated from the first target sequence. Thus, the series of truncated molecules produced by the second sequencing primer will have higher molecular weights than those produced by the first sequencing primer. Hence, the truncated molecules produced by these two sequencing primers can be effectively separated in a single lane by standard gel electrophoresis in a single electropherogram without any overlapping of the nucleotide sequences. By using sequencing primers with progressively higher molecular weights, multiple short DNA sequences from a variety of targets can be determined simultaneously. We describe here the basic concept of MULTIGEN technology and three applications: detection of sexually transmitted pathogens (Neisseria gonorrhoeae, Chlamydia trachomatis, and Ureaplasma urealyticum), detection of contaminants in meat samples (coliforms, fecal coliforms, and Escherichia coli O157:H7), and detection of single-nucleotide polymorphisms in the human N-acetyltransferase (NAT1) gene (S. Fronhoffs et al., Carcinogenesis 22:1405-1412, 2001).
DNA-based technologies such as the use of PCR and DNA probes (16, 17) have led to a wide range of genomic identification methods (1, 2, 3, 8, 12, 13, 20, 21, 32, 38, 40). However, the ultimate method for identifying a DNA target and, hence, a specific organism is considered to be the determination of a signature nucleotide sequence. The conventional chain dideoxynucleotide termination sequencing method (28) is considered the “gold standard” for determining nucleotide sequences, but it can process only one target at a time. This processing inefficiency of conventional sequencing increases the cost of screening for multiple pathogens and has therefore limited the use of this approach in routine diagnostic testing. Although attempts have been made to sequence multiple targets simultaneously (4, 5, 7, 15, 24, 35), none could produce an electropherogram consisting of distinct nucleotide sequences from multiple targets. MULTIGEN (34) overcomes this problem, and we describe here its basic concepts and present three examples of applications for routine diagnostic testing.
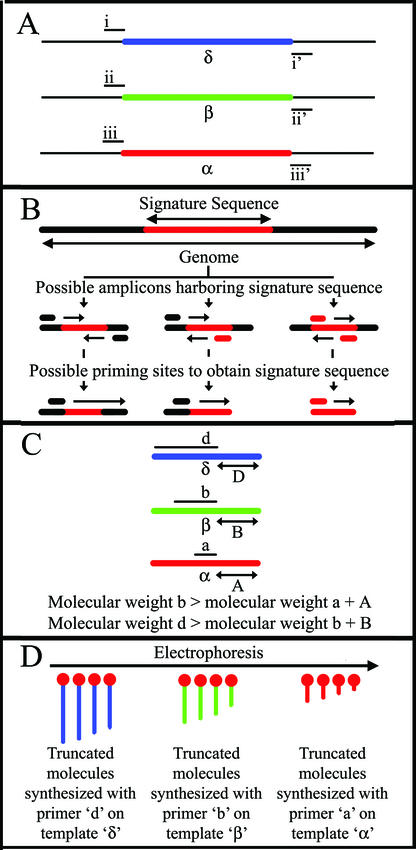
Figure 1A presents a schematic view of MULTIGEN technology, showing the determination of DNA sequences from three genomic regions: α, β, and δ. Each of the three regions is amplified with corresponding pairs of primers: region δ is amplified with primers i and i′, region β is amplified with primers ii and ii′, and region α is amplified with primers iii and iii′. These specific genomic regions can be amplified simultaneously by the multiple PCR (MPCR). As shown in Fig. 1B, there are various options for designing the PCR primers around the segment of DNA that is of interest. Furthermore, the sequencing primer site can be located at different sites around the amplicon. Once amplicons from the α, β, and δ regions are amplified, the desired regions of the amplicons can be sequenced. The determination of nucleotide sequences of known DNA targets is primarily used for microbial identification and genetic variations such as single-nucleotide polymorphisms (SNPs) wherein only short segments of the target DNA are sequenced. In order to restrict the length of the sequencing segments without a readthrough of the amplicon, a short stretch at the 3′ end of the amplicon is sequenced. Thus, the generated DNA sequence will consist of the DNA sequence of the downstream PCR primer plus a variable number of nucleotides from the target DNA sequence beyond the 3′ end of the downstream PCR primer. Sequencing primer a is used to sequence segment A on amplicon α, sequencing primer b is used to sequence segment B on amplicon β, and sequencing primer d is used to sequence segment D on amplicon δ (Fig. 1C). In order to analyze the truncated molecules generated from the different amplicon targets in a single lane, sequencing primers of various molecular weights are used. The sequencing primers are modified such that the molecular weight of sequencing primer b is greater than the combined molecular weights of sequencing primer a and segment A. The molecular weight of sequencing primer d is greater than the combined molecular weights of sequencing primer b and segment B. The sequences generated on the same gel from all three amplicons are shown schematically in Fig. 1D. The truncated molecules generated by the lower-molecular-weight sequencing primers will migrate first, followed by those generated by the larger sequencing primers. This prevents any overlap in the migration of the various series of truncated DNA molecules. They are then detected and recorded when they cross the path of a scanner in an automated sequencer. Therefore, by using a mixture of different sequencing primers of progressively increasing molecular weights, different sets of truncated DNA molecules specific to each DNA target can be generated and detected simultaneously in a single lane by using a slab gel or in the same capillary tube by using capillary electrophoresis.
FIG. 1.
Schematic representation of novel features of MULTIGEN technology. (A) Generation of amplicons α, β, and δ from three different targets by MPCRs; (B) possible PCR primer sites for generating target amplicons and possible sites for sequencing primers; (C) relative molecular sizes and position of sequencing primers a, b, and d to the targets A, B, and D; (D) relative electrophoretic migration of truncated DNA molecular species produced from primer extension of targets A, B, and D.
MATERIALS AND METHODS
Materials.
Escherichia coli O157:H7 was supplied by the Department of Food Science, Faculty of Agriculture, University of Saskatchewan, Saskatoon, Saskatchewan, Canada. PCR-based detection kits for three high-risk subtypes of the human papillomavirus (HPV)—HPV18, HPV31, and HPV33—and three sexually transmitted pathogens (Neisseria gonorrhoeae, Chlamydia trachomatis, and Ureaplasma urealyticum) were purchased from Maxim Biotech. These included primers and template DNA of the target organisms (Table 1). All PCR and sequencing primers (except the primers for HPV subtypes) were designed by Bio-ID Diagnostic, Inc., Saskatoon, Saskatchewan, Canada, by using Oligo 6.0 software (Life Sciences Software Resources) and synthesized by Sigma-Aldrich (Canada). Human DNA was purchased from Boehringer Mannheim GmbH. Primer modification can involve the attachment of a macromolecule with an appropriate molecular weight. One such modification includes coupling the primer to the macromolecule via a bifunctional linker molecule. The sequencing primer is first synthesized with a C6 amino modification at the 5′ end and is then coupled to a carboxyl group on the linker molecule. The resulting complex is purified and coupled to the macromolecule via its single amino group.
TABLE 1.
PCR primers and corresponding amplicons
| Application | Target genome | PCR primersa | Amplicon size (bp) |
|---|---|---|---|
| HPV | HPV18 | F, 5′-CACACCACAATACTATGGCGCGCT-3′ | 360 |
| R, 5′-CTGCTGGATTCAACGGTTTCTGGC-3′ | |||
| HPV33 | F, 5′-GCAGTAAGGTACTGCACGACTATG-3′ | 413 | |
| R, 5′-CGACCCGAAATATTATGAAATCGT-3′ | |||
| HPV31 | F, 5′-TAAGCTCGGCATTGGAAATACCCT-3′ | 350 | |
| R, 5′-CCTTCCTCCTATGTTGTGGAATCG-3′ | |||
| Meat | Stx2 | F, 5′-CATATATCAGTGCCCGGTGTGA-3′ | 158 |
| R, 5′-GCATTTCCACTAAACTCCATTA-3′ | |||
| LacZ | F, 5′-AGGCATGATGCGACGCTTGT-3′ | 275 | |
| R, 5′-TCTGTTATTGGCGCGGGTAG-3′ | |||
| LamB | F, 5′-TCTGGTCCTGGTGCCGGTCT-3′ | 306 | |
| R, 5′-TCAGGACACTCTGAGTATGT-3′ | |||
| STDs | U. urealyticum | F, 5′-CAGGAGCAATTAACTTCGCTGAAG-3′ | 219 |
| R, 5′-CAGTACTGAGAATATCGAAACGACGTC-3′ | |||
| N. gonorrhoeae | F, 5′-CTCTGCTTCGGCTCTCTGCTG-3′ | 298 | |
| R, 5′-GAAGACCTTCGAGCAGACATCAC-3′ | |||
| C. trachomatis | F, 5′-GCAAGATATCGAGTATGCGTTGTTAGG-3′ | 364 | |
| R, 5′-TTCATTGTACTCATTAAACGAGCGG-3′ | |||
| NAT1 gene | Amplicon 1 | F, 5′-ATTCTCCTGCCTACATCAGAAG-3′ | 354 |
| R, 5′-AGGAAAACAAAACGAAAGC-3′ | |||
| Amplicon 2 | F, 5′-TTGATCAAGTTGTGAGAAGA-3′ | 318 | |
| R, 5′-CTAGATACCAGAATCCATTCTCT-3′ |
Orientation: F, forward; R, reverse.
Preparation of total DNA.
Cultures of E. coli O157:H7 were grown overnight in Luria-Bertani broth at 37°C in a shaker water bath. Total DNA from bacterial culture, as well as human genomic DNA from buccal swabs obtained from human volunteers, was extracted by using a QIAmp DNA Minikit (Qiagen). The purity and yield of the DNA was determined by using a spectrophotometer (Ultrospec 3000; Pharmacia Biotech, Cambridge, United Kingdom).
MPCR.
MPCR of target amplicons was performed in a 50-μl volume containing 5 μl of 10× buffer (Bio-ID Diagnostic, Inc., and Maxim Biotech). The corresponding thermocycling protocol is shown in Table 2. The amplified multiple targets were purified by using PSIclone HTS (Princeton Separations, Princeton, N.J.). Then, 5 μl of purified MPCR reaction mixture was separated on 2% agarose electrophoresis, stained with 0.1% ethidium bromide, and visualized under UV light at 254 nm on a transilluminator (FBTIV-88; Fisher Scientific) and photographed with a Polaroid photo documentation camera (FBPDC-34; Fisher Scientific).
TABLE 2.
Conditions for PCR and MPCRa
| Application | Template/50μl | Primer (amt [pmol])/50-μl reactione | Thermocycler profile |
|---|---|---|---|
| HPV | |||
| MPCR | 2.4 × 109 copies of HPV18 | HPV18 (10) | 95° C/3-min denature |
| 2.4 × 109 copies of HPV33 | HPV33 (10) | (95°C/1 min, 55°C/1 min, 72°C/1 min), 35 cycles 72°C/10-min final extension; 4°C/hold | |
| 2.4 × 109 copies of HPV31 | HPV31 (10) | ||
| Cycle sequencing | 4.8 × 1010 total ampliconsb | HPV18 (3.2) | Step up]c (96°C/10 s, 50°C/5 s, 60°C/4 min), cycles: 25;4°C/hold |
| HPV33 (3.2) | |||
| HPV31 (1) | |||
| Meat | |||
| MPCR | 9.7 × 109 copies of genomic DNA | LacZ (5) | 95°C/3-min denature |
| LamB (5) | (95°C/1 min, 56.5°C/1 min, 72°C/1 min), 35 cycles 72°C/10-min final extension; 4°C/hold | ||
| Stx2 (10) | |||
| Cycle sequencing | 2.7 × 1010 total ampliconsb | LacZ (1.6) | (96°C/10 s, 50°C/5 s, 60°C/4 min), 25 cycles; 4°C/hold |
| LamB (1.6) | |||
| Stx2 (1.6) | |||
| STD | |||
| MPCR | 2.4 × 109 copies of UU plasmid | UU (5) | 95°C/3-min denature |
| 2.4 × 109 copies of NG plasmid | NG (5) | (95°C/1 min, 56.5°C/1 min, 72°C/1 min), 35 cycles 72°C/10-min final extension; 4°C/hold | |
| 2.4 × 109 copies of CT plasmid | CT (5) | ||
| Cycle sequencing | 2.2 × 1010 total ampliconsb | UU (1.6) | (96°C/10 s, 50°C/5 s, 60°C/4 min), 25 cycles; 4°C/hold |
| NG (1.6) | |||
| CT (1.6) | |||
| NAT1 | |||
| MPCR | 9.7 × 104 copies of genomic DNA | Amp1 (10) | 95°C/3-min denature |
| Amp2 (10) | (95°C/1 min, 54°C/1 min, 72°C/1 min), 35 cycles; 72°C/10-min final extension; 4°C/hold | ||
| Cycle sequencing | 1.8 × 1010 total ampliconsb | Amp1 (1) | Step upd (96°C/10 s, 55°C/10 s, 60°C/4 min), 25 cycles; 4°C/hold |
| Amp2 (1) |
UU primer, U. urealyticum primer; NG, N. gonorrhoeae primer; CT, C. trachomatis primer; Amp1, amplicon 1; Amp2, amplicon 2.
Total amplicon calculation was based on the average size of the target amplicons.
HPV33 primers were added initially; HPV18 primers were added after eight cycles, and then HPV31 primers were added after 20 cycles.
Amplicon2 primers were run for 12 cycles, and then amplicon 1 primers were added for the remaining cycles.
Cycle sequencing was performed with a 20-μl reaction volume.
Cycle sequencing and capillary electrophoresis.
The respective amplicons were sequenced by using corresponding sequencing primers (Table 3) by cycle sequencing by using the ABI Prism BigDye terminator cycle sequencing ready reaction kit (version 3.0; PE Applied Biosystems) on a GeneAmp 2400 thermocycler (PE Applied Biosystems). Unincorporated dye terminators were removed by using Centricep chromatography columns (Princeton Separations). The samples were then dried in a speedvac (DNA 120; ThermoSavant) and resuspended in 20 μl of ABI Prism template suppression reagent. Samples were analyzed by capillary electrophoresis by using the ABI Prism genetic analyzer 310. The 47-by-50-μm uncoated capillary was filled with performance-optimized polymer 6 (acrylamide-urea polymer) and heated to 50°C. Next, 20 μl of the sequencing mixture was transferred to an Eppendorf tube. Samples were drawn into the capillary by an electrokinetic injection at 2 kV for 75 s. The electrophoresis was carried out at 15 kV for 36 min.
TABLE 3.
Sequencing primers
| Application | Target genome | Sequencing primera |
|---|---|---|
| HPV | HPV18 | 5′-Δ-AATTTATTAATAAGGTGCCTGCGGTGCCAG-3′ |
| HPV33 | 5′-Δ-AAAAAAAACGACATGTGGATTTAAACAAAC-3′ | |
| HPV31 | 5′-Δ-TATAACGTGTCAAAGACCGTTGTGTCCAGA-3′ | |
| Meat | Stx2 | 5′-Δ-TCGTCACTCACTGGTTTCATCATA-3′ |
| LacZ | 5′-Δ-CCACGACGTTTGGTGGAATGTCTTTTGTGA-3′ | |
| LamB | 5′-Δ-GCGCATCGAAAGACGGCTGGTTATTCACTG-3′ | |
| STDs | U. urealyticum | 5′-Δ-AACGAAGACAAAGAACGTAAAGTTGCTTAT-3′ |
| N. gonorrhoeae | 5′-Δ-CGTTTGTTGCTCTATGCTGGCGGCTTCGGT-3′ | |
| C. trachomatis | 5′-Δ-CTGAAGAAAATTTGAGCAATTTCATTTTCC-3′ | |
| NAT1 gene | Amplicon 1 | 5′-Δ-GAGAGATTCCAACTGGTATC-3′ |
| Amplicon 2 | 5′-Δ-TTAATTTCTGGGAAGGATCAGCCTCAGGTG-3′ |
Δ, Modification to increase the primer molecular weight.
RESULTS
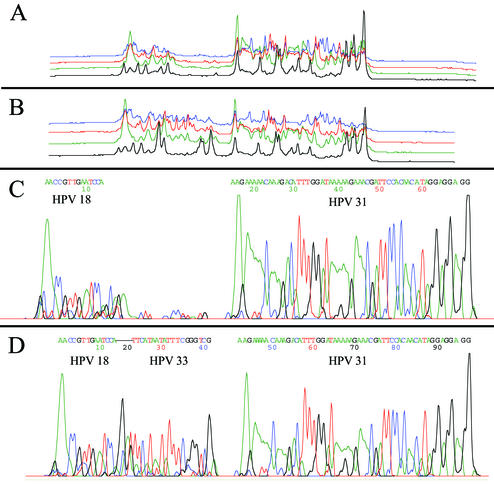
To illustrate the basic concept and technical feasibility of MULTIGEN technology, experiments were performed with three HPV subtypes (HPV18, HPV31, and HPV33) to show that accurate identification of such closely related genomes could be achieved. The basic experimental model involved producing specific amplicons from plasmid clones carrying the complete genomes of HPV subtypes and then simultaneous cycle sequencing of the 3′-terminal end of the pooled amplicons by using sequencing primers of various molecular weights. The DNA sequences of the elecropherograms thus produced were BLAST searched (National Institutes of Health [NIH]) to identify homology between these respective loci of the target sequences and those in GenBank (NIH). Initially, we generated three amplicons of 360, 350, and 413 bp from three separate plasmid clones carrying target DNA segments (E6 region) HPV18, HPV31, and HPV33, respectively (10, 26, 29). Two combinations of these amplicons were used as templates to simultaneously sequence the 3′ end of the amplicons. The first experiment was designed to show that MULTIGEN could generate and read sequences simultaneously from two separate DNA targets (Table 4). We used two sequencing primers (HPV18 [molecular weight, 13,899] and HPV31 [molecular weight, 28,276]). The electropherogram produced two separate sequences consisting of 14 and 55 nucleotides, respectively. As determined by BLAST search, the sequences showed 100% homology with HPV18 and HPV31, respectively. As predicted, the electropherogram had a distinct “nonsignal” region between the two sequences (Fig. 2A and C). In our next experiment, the number of targets was increased to three subtypes. Amplicons from all three HPV templates were simultaneously sequenced by using sequencing primers HPV18 (molecular weight, 13,899), HPV33 (molecular weight, 19,108), and HPV31 (molecular weight, 28,276). The electropherogram produced had three distinct sequences of 14, 19, and 55 nucleotides accurately identifying HPV18, HPV33, and HPV31, respectively (Fig. 2B and D).
TABLE 4.
Species-specific nucleotide sequences and corresponding sequencing primers
| Primer (mol wt) | Expected electropherograma for:
|
||
|---|---|---|---|
| Locus and target sequence 1 | Locus and target sequence 2 | Locus and target sequence 3 | |
| HPV18 (13,899) | HPV18 (436-450) and 5′-AACCGTTGAATCCA-3′ | ||
| HPV31 (28,276) | HVP31 (445-486) and 5′-AAGAAAAACAA AGACATTTGGATAAAAAGAAACGAT TCCACAACATAGGAGGAGG-3′ | ||
| HPV18 (13,899) | HPV18 (436-450) and 5′-AACCGTTGAATCCA-3′ | ||
| HPV33 (19,108) | HPV33 (481-500) and 5′-TTCATAATATTTCGGGTCG-3′ | ||
| HPV31 (28,276) | HVP31 (445-486) and 5′-AAGAAAAACAA AGACATTTGGATAAAAAGAAACGAT TCCACAACATAGGAGGAGG-3′ | ||
| UU (9,280) | UU-UreB gene (257-284) and 5′-GGACGTCGTTTCGATATTCTCAGTACTG-3′ | ||
| NG (18,383) | NG-CppB gene (3506-3530) 5′-GCGTGATGTCTGCTCGAAGGTCTTC-3′ | ||
| CT (28,048) | CT-pCTT1 plasmid (453-475) and 5′-GCTCG TTTAATGAGTACAATGAA-3′ | ||
| Stx2 (7,253) | Stx2 gene (492-521) and 5′-TCTGGCGTTAATGGAGTTTAGTGGAAATGC-3′ | ||
| LacZ (18,509) | Lac operon (6216-6192) and 5′-CGATACTACCCGCGCCAATAACAGA-3′ | ||
| LamB (27,921) | LamB gene (824-847) and 5′-CTGAACATAC TCAGAGTGTCCTGA-3′ | ||
| NAT1 Amp1 (6,141) | −340 mutation and 5′-TTATAAAC(G/A)T-3′ | ||
| NAT1 Amp2 (30,958) | 445 and 549 mutations and 5′-CCTTGT(G/A) TCTTCC-3′; 5′-GTTTGAC(G/A)GA-3′ | ||
See definitions in Table 2, footnote a.
The nucleotide range is given in parentheses.
FIG. 2.
Electropherogram showing electrophoretic separation and sequences from multiple targets. (A) Electropherogram of raw data showing two distinct regions of fluorescence signals representing two stretches of sequences, (B) electropherogram of raw data showing distinct regions of fluorescence signals representing three stretches of sequences; (C) electropherogram of analyzed data showing two sequences—a 15-base nucleotide sequence generated by sequencing primer HPV18, followed immediately by a nonsignal, which is then followed by a 41-nucleotide sequence generated by sequencing primer HPV31; (D) electropherogram analyzed data showing three sequences—a 13-base nucleotide sequence generated by sequencing primer HPV18, followed immediately by a 20-nucleotide sequence generated by sequencing primer HPV33, followed immediately by a 41-nucleotide sequence generated by sequencing primer HPV31.
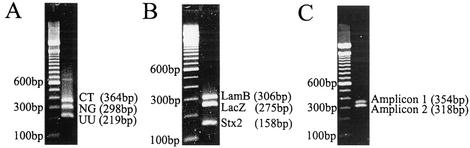
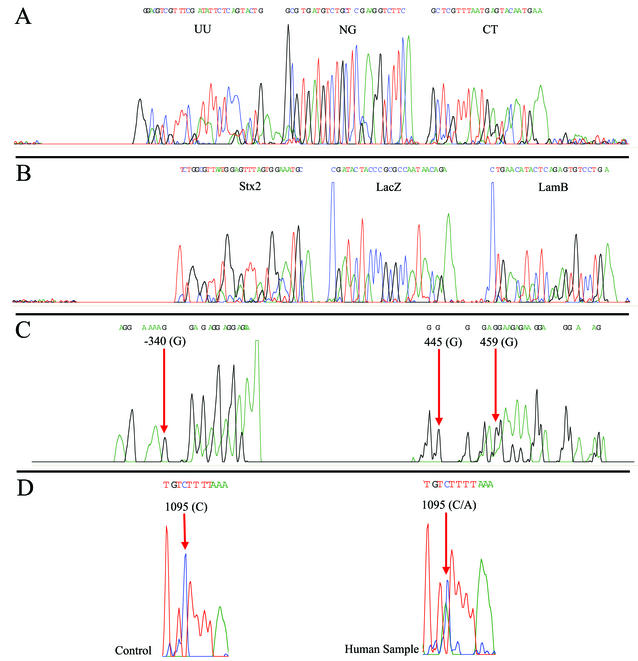
To show the utility of MULTIGEN technology in routine diagnostic testing, three applications were selected. In the first experiment we showed successful detection of three pathogens—N. gonorrhoeae, C. trachomatis, and U. urealyticum—that are associated with sexually transmitted diseases (STDs). Signature DNA segments from all three STD targets were amplified by using MPCR, generating three amplicons of 298, 364, and 219 bp, respectively (Fig. 3A). The amplicons were simultaneously cycle sequenced by using specific sequencing primers. Figure 4A shows a single electropherogram consisting of 28 nucleotides of U. urealyticum, followed by 25 nucleotides of N. gonorrhoeae, and then 23 nucleotides C. trachomatis. A BLAST search of the nucleotide sequence of the electropherogram detected 100% homology with specific DNA segments in the ureB gene (19), the CppB gene (39), and the cryptic plasmid (31). In subsequent experiments we spiked these samples with DNA from HPV and were still able to obtain clean signature sequences for all three targets with no interference. In our second experiment, signature DNA segments from the β-galactosidase (lacZ) gene (14) representing coliforms, the lambda receptor (lamB) gene (6) representing fecal coliforms, and the Shiga toxin (stx2) gene (27) representing E. coli O157:H7 were simultaneously amplified generating three amplicons lamB (306 bp), lacZ (275 bp), and stx2 (158 bp; Fig. 3B) from the same E.coli isolate. These amplicons were simultaneously sequenced by using specific sequencing primers. Figure 4B shows a single electropherogram exhibiting all three signature nucleotide sequences of an stx2 sequence of 30 nucleotides, followed by a lacZ sequence of 25 nucleotides and a lamB sequence of 24 nucleotides. A BLAST search of this electropherogram exhibited 100% homology with respective target gene sequences. To illustrate the specificity of MULTIGEN technology, 50 ng of the genomic DNA from the E. coli isolate was spiked with genomic DNA from common water protozoans: 2.5 ng of Cryptosporidium parvum and 2.5 ng of Giardia lamblia. Even with 10% contaminants we were able to generate an electropherogram with all three signature nucleotide sequences for the lacZ, lamB, and stx2 gene segments. The sensitivity of MULTIGEN technology was estimated at two stages. The first was a sequencing step. Template titration shows that the optimal amount of double-stranded DNA per target is ca. 2 ng (12 fmol = 7.4 × 109 copies) of the target amplicon. The second stage was an MPCR step. We were able to generate all three-signature nucleotide sequences for the lacZ, lamB, and stx2 gene segments from 100 pg (0.032 fmol = 1.9 × 107 copies) of the genomic the DNA. In our third experiment, as an example of the ability of MULTIGEN technology to identify human genetic markers, two segments of the human NAT1 (9) gene were simultaneously amplified from human genomic DNA by using MPCR primers producing two amplicons of 354 and 318 bp (Fig. 3C). These two amplicons were sequenced simultaneously by using modified sequencing primers. The electropherogram of the single-nucleotide polymorphism (G/A) at locus −345 on the first segment and of the loci at 445 and 459 in the second segment are shown in Fig. 4C. Haploid genotyping was carried out independently on the third segment (620 bp) of the NAT1 gene for the C/A SNP at locus 1095. The homozygous C/C and the heterozygous C/A are depicted in Fig. 4D. All of these experiments were repeated a number of times, some of them as many as 25 times, with identical results obtained with each and every repetition.
FIG. 3.
Agarose gel electrophoresis showing amplicons from MPCRs. (A) Pathogens associated with STDs: C. trachomatis (CT; 364 bp), N. gonorrhoeae (NG; CppB gene, 298 bp), and U. urealyticum (UU; ureB gene, 219 bp). (B) Bacterial contaminants associated with meat: fecal coliforms (lamB gene, 306 bp), coliforms (lacZ gene, 275 bp), and E. coli O157: H7 (stx2 gene, 158 bp). (C) Human N-acetyltransferase (NAT1) gene amplicon one (354 bp) and amplicon two (318 bp).
FIG. 4.
Electropherogram of analyzed data showing U. urealyticum (UU; 28 nucleotides), N. gonorrhoeae (NG; 25 nucleotides), and C. trachomatis (CT; 23 nucleotides) (A); stx2 gene (30 nucleotides), lacZ gene (25 nucleotides), and lamB gene (24 nucleotides) (B); two segments of human N-acetyltransferase single nucleotide polymorphism showing (G/A) SNP at locus −340 on the first segment and at 445 and 459 in the second segment (C); and homozygous wild type (control) showing cystine at locus 1095 with the test (human) sample showing heterozygocity (cystine/adenine).
DISCUSSION
The determination of nucleic acid identity involves the binding of an oligonucleotide to a specific segment of the target DNA. Some of the available methods (e.g., dot blot, microarray, etc.) stop at this stage, where detection is determined by relative fluorescent signals from the bound and labeled oligonucleotide probes, whereas others (e.g., PCR, ligase chain reaction, etc.) are processed further by polymerase-mediated reactions but are still assessed by the single fluorescent signal they produce (e.g., the size of amplicon or the release of signal quenching in real-time PCR). Features that distinguish MULTIGEN technology are that (i) MULTIGEN goes beyond mere target amplification and produces a distinctive and unique target sequence; that (ii) the technology involves three specific oligonucleotide primers per target, similar to nested PCR; and that (iii) as MULTIGEN generates n number of signals using all four nucleotides in a specific sequence, it increases the specificity of detection by at least 4n times relative to probe- and PCR-based methods.
In order to avoid only the sequence of the downstream PCR primer showing up on the electropherogram, the sequencing primers are designed such that there are at least a few target specific nucleotides in between +1 of the annealing site and that of the 3′ downstream PCR primer. The truncated molecules generated during MULTIGEN cycle sequencing are less than 100 nucleotides long and therefore generate signals that are sharp, avoiding the broad signals associated with long truncated molecular species. We report three test models that demonstrate the potential place of MULTIGEN technology in routine diagnostic testing.
C. trachomatis, N. gonorrhea, and U. urealyticum are common causes of STDs in humans. Conventionally, these organisms are identified by culture and/or serological methods (25, 33). Compared to other DNA-based methods that identify only two of these organisms (i.e., C. trachomatis and/or N. gonorrhoeae) such as PCR (39), strand displacement (33), ligase chain reaction (33), nucleic acid-based amplification (22), and ramification amplification (42), we show that a single MULTIGEN test menu of STDs could include all of the important pathogens in routine clinical practices, significantly enhancing pathogen detection and thereby the level of care for patients with STDs.
Testing of food products includes tests for indicator organisms such as coliforms, fecal coliforms (18a), and toxin-producing pathogenic organisms such as E. coli O157:H7. Conventional testing includes the culture method (plate count) to determine bacterial load and acid and gas production in special growth medium (MacConkey broth or indicator media) (18), followed by confirmation by serological methods. In order to provide a test with better specificity, we show that MULTIGEN can detect the presence of coliforms, fecal coliforms, and E. coli O157:H7 simultaneously. The speed and cost-effectiveness of MULTIGEN testing offer a significant potential contribution toward more effective inventory control in agribusiness.
The correlation of SNPs with human diseases (30) has generated an interest in determining haploid genotypes, which include the determination of nucleotides at specific loci on both alleles. Although the determination of a nucleotide could be achieved by using single-nucleotide primer extension (23, 41), target-specific nucleotide probes (20), or single-nucleotide sequence analysis (11, 36), only MULTIGEN can provide both the nucleotide and the specific locus by identifying nucleotides on either side of the SNP locus simultaneously at a number of SNP sites.
We have applied MULTIGEN technology here to determine the nucleotide sequence from three segments (the lacZ gene, lamB gene, and stx2 gene) of the same microbial genome, i.e., E. coli O157:H7. This capability has applications in three areas. (i) The first is when DNA segments with desired traits are inserted in plasmid vectors to produce pharmaceuticals such as insulin via the β-galactosidase fusion protein (37). MULTIGEN technology ensures the specific orientation of the insert in the plasmid host for proper expression and that all of the essential elements of gene expression such e.g., promoters are intact. (ii) The second application is in the posttranscriptional modification of eukaryotic RNA, leading to phenotypic abnormalities such as β thalassemia (37). Simultaneously sequencing a number of splicing regions of mRNA would ensure a proper template for translation. (iii) Finally, MULTIGEN technology can be applied to the detection of a chimeric genome carrying a number of “foreign” DNA segments that could be used as biological-threat agents.
In summary, we illustrate here the scientific basis for simultaneously obtaining multiple nucleotide sequences from viral, bacterial, and human genomic loci; the ability to subtype microbes, emphasizing immediate applications in clinical testing and the food industry; human haplogenotyping; and the potential for determining genetic elements of “chimeric” genomes that could be used as biological-threat agents. MULTIGEN technology adds a novel technical modification to the proven scientific principles of conventional sequencing and the gel electrophoresis process. MULTIGEN combines the desired characteristics of high sensitivity, high specificity, and cost-effectiveness that are paramount for critical medical decision making in routine high-volume diagnostic settings and allows the development of tests for the detection of virtually any combination of target sequences in any type of sample that contains nucleic acid material.
Acknowledgments
We acknowledge Anette Kerviche of the Cancer Research Center, Saskatoon, Canada, for technical assistance with the HPV electropherogram, and the Department of Food Science, University of Saskatchewan, for providing bacterial cultures.
REFERENCES
- 1.Afshari, C. A., E. F. Nuwaysir, and J. C. Barrett. 1999. Application of complementary DNA microarray technology to carcinogen identification, toxicology, and drug safety evaluation. Cancer Res. 59:4759-4760. [PubMed] [Google Scholar]
- 2.Aono, T., K. Kondo, H. Miyoshi, K. Tanaka-Taya, M. Kondo, Y. Osugi, J. Hara, S. Okada, and K. Yamanishi. 1998. Monitoring of human cytomegalovirus infections in pediatric bone marrow transplant recipients by nucleic acid sequence-based amplification. J. Infect. Dis. 178:1244-1249. [DOI] [PubMed] [Google Scholar]
- 3.Bonora, S., M. C. Gutierrez, G. Di-Perri, F. Brunello, B. Allegranzi, M. Ligozzi, R. Fontana, E. Concia, and V. Vincent. 1999. Comparative evaluation of ligation-mediated PCR and spoligotyping as screening methods for genotyping of Mycobacterium tuberculios strains. J. Clin. Microbiol. 37:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee, M. 1991. Enzymatic multiplex DNA sequencing.Nucleic Acids Res. 19:3301-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church, G. M., and S. Kieffer-Higgins. 1988. Multiplex DNA sequencing. Science 240:185-188. [DOI] [PubMed] [Google Scholar]
- 6.Clement, J. M., and M. Hofnung. 1981. Gene sequence of the lambda receptor, an outer membrane protein of Escherichia coli K-12. Cell 27(Pt. 2):507-514. [DOI] [PubMed] [Google Scholar]
- 7.Creasey, A., L., Jr., D'Angio, T. S. Dunne, C. Kissinger, T. O'Keeffe, H. Perry-O'Keefe, L. S. Moran, M. Roskey, I. Schildkraut, and L. E. Sears. 1991. Application of a novel chemiluminescence-based DNA detection method to single-vector and multiplex DNA sequencing. BioTechniques 11:102-109. [PubMed] [Google Scholar]
- 8.Erlich, A. H. 1992. PCR technology: principles and application. Oxford University Press, New York, N.Y.
- 9.Fronhoffs, S., T. Bruning, E. Ortiz-Pallardo, P. Brode, B. Koch, V. Harth, A. Sachinidis, H. M. Bolt, C. Herberhold, H. Vetter, and Y. Ko. 2001. Real-time PCR analysis of the N-acetyltransferase NAT allele *3,*4,*10,*11,*14 and *17 polymorphism in squamous cell cancer of head and neck. Carcinogenesis 22:1405-1412. [DOI] [PubMed] [Google Scholar]
- 10.Goldsborough, M. D., D. DiSilvestre, G. F. Temple, and A. T. Lorincz. 1989. Nucleotide sequence of human papillomavirus type 31: a cervical neoplasia-associated virus. Virology 171:306-311. [DOI] [PubMed] [Google Scholar]
- 11.Griffin, T. J, and L. M. Smith. 2000. Single nucleotide polymorphism analysis by MALDI-TOF mass spectrometry. Trends Biotechnol. 18:77-84. [DOI] [PubMed] [Google Scholar]
- 12.Hames, B. D., and S. J. Higgins. 1995. Gene probe 1. Oxford University Press, New York, N.Y.
- 13.Hebart, H., D. Gamer, J. Loeffler, C. Mueller, C. Sinzger, G. Jahn, P. Bader, T. Klingebiel, J. Kanz, and H. Einsele. 1998. Evaluation of murex CMV DNA hybrid capture assay for detection and quantitation of cytomegalovirus infection in patients following allogeneic stem cell transplantation. J. Clin. Microbiol. 36:1333-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hediger, M. A., D. F. Johnson, P. Nierlich, and I. Zabin. 1985. DNA sequence of the lactose operon: the LacA gene and the transcriptional termination region. Proc. Natl. Acad. Sci. USA 82:6414-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbell, E. 2001. Multiplex sequencing by hybridization. J. Comput. Biol. 8:141-149. [DOI] [PubMed] [Google Scholar]
- 16.Innis, M. A., D. H. Gelfand, J. J. Sninsky, and T. J. White. 1990. PCR protocols. Academic Press, Inc., New York. N.Y.
- 17.Keller, G. H., and M. M. Manak. 1989. DNA probes. MacMillan Publishers, Ltd., London, United Kingdom.
- 18.Koneman, W. E., S. D. Allen, V. R. Dowell, M. W. Janda, H. M. Sommers, and W. C. Winn. 1988. Diagnostic microbiology. Lippincott Co., New York, N.Y.
- 18a.Kelly, M. T., D. J. Brenner, and J. J. Farmer III. 1985. In E. H. Lennette, A. Balows, W. J. Hausler, and H. J. Shadomy (ed.), Manual of clinical microbiology, 4th ed. American Society for Microbiology, Washington, D.C.
- 19.Kong, F., G. James, Z. Ma, S. Gordon, W. Bin, and G. L. Gilbert. 1999. Phylogenetic analysis of Ureaplasma urealyticum: support for the establishment of a new species, Ureaplasma parvum. Int. J. Syst. Bacteriol. 49(Pt. 4):1879-1889. [DOI] [PubMed] [Google Scholar]
- 20.Kwiatkowski, R. W., V. Lyamichev, M. de Arruda, and B. Neri. 1999. Clinical, genetic, and pharmacogenetic applications of the Invader assay. Mol. Diagn. 4:353-364. [DOI] [PubMed] [Google Scholar]
- 21.Lizardi, P. M., X. Huang, Z. Zhu, P. Bray-Ward, D. C. Thomas, and D. C. Ward. 1998. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 19:225-232. [DOI] [PubMed] [Google Scholar]
- 22.Mahony, J. B., X. Song, S. Chong, M. Faught, T. Salonga, and J. Kapala. 2001. Evaluation of the NucliSens basic kit for detection of Chlamydia trachomatis and Neisseria gonorrheae in genital tract specimens using nucleic acid sequence-based amplification of 16S rRNA. J. Clin. Microbiol. 39:1429-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makridakis, N. M., and J. K. Reichardt. 2001. Multiplex automated primer extension analysis: simultaneous genotyping of several polymorphisms. BioTechniques 31:1374-1380. [DOI] [PubMed] [Google Scholar]
- 24.Monstein, H. J., Y. Johansson, and J. Jonasson. 2000. Detection of vancomycin resistance genes combined with typing of enterococci by means of multiplex PCR amplification and multiple primer DNA sequencing. APMIS 108:67-73. [DOI] [PubMed] [Google Scholar]
- 25.Morse, S. A. and S. K. Sarafian. 1985. Sexually transmitted diseases, p. 863-876. In E. H. Lennette, A. Balows, W. J. Hausler, and H. J. Shadomy (ed.), Manual of clinical microbiology, 4th ed. American Society for Microbiology, Washington, D.C.
- 26.Pao, C. C., C. Y. Lin, J. S. Maa, C. H. Lai, S. Y. Wu, and Y. K. Soong. 1990. Detection of human papilloma viruses in cervicovaginal cells using polymerase chain reaction. J. Infect. Dis. 161:113-115. [DOI] [PubMed] [Google Scholar]
- 27.Ramachandran, V., M. A Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2001. The common Shiga toxin 2-containing Escherichia coli serotypes and human isolates of the same serotypes possess a Stx2d toxin type. J. Clin. Microbiol. 39:1932-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seedorf, K., T. Oltersdorf, G. Krammer, and W. Rowekamp. 1987. Identification of early proteins of human papilloma virus type 16 (HPV16) and HPV18 in cervical carcinoma cells. EMBO J. 6:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shastry, B. S. 2002. SNP alleles in human disease and evolution. J. Hum. Genet. 47:561-566. [DOI] [PubMed] [Google Scholar]
- 31.Sriprakash, K. S., and E. S. Macavoy. 1987. Characterization and sequence of a plasmid from the trachoma biovar of Chlamydia trachomatis. Plasmid 18:205-214. [DOI] [PubMed] [Google Scholar]
- 32.Tanke, H. J., J. Wiegant, R. P. van Gijlswijk, V. Bezrookove, H. Pattenier, R. J. Heetebrij, E. G. Talman, A. K. Raap, and J. Vrolijk. 1999. New strategy for multi-colour fluorescence in-situ hybridization—COBRA: combined binary ratio labeling. Eur. J. Hum.Genet. 7:2-11. [DOI] [PubMed] [Google Scholar]
- 33.Van Dyck, E., M. Leven, L. Pattyn, V. Damme, and M. Laga. 2001. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by enzyme immunoassay, culture, and three nucleic acid amplification tests. J. Clin. Microbiol. 39:1751-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinayagamoorthy, T. March. 2001. Multi-loci genome analysis. U.S. patent 6,197,510.
- 35.Wang, G., M. S. Rahman, M. Z. Humayun, and D. E. Taylor. 1999. Multiplex sequence analysis demonstrates the competitive growth advantage of the A-to-G mutants of clarithromycin-resistant Helicobacter pylori. Antimicrob. Agents Chemother. 43:683-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasson, J., G. Skolnick, L. Love-Gregory, and M. A. Permutt. 2002. Assessing allele frequencies of single nucleotide polymorphism in DNA pools by pyrosequencing technonology. BioTechniques 32:1144-1152. [DOI] [PubMed] [Google Scholar]
- 37.Watson, D. J., M. Gilman, J. Witkowski, and M. Zoller. 1997. Recombinant DNA, 2nd ed.W. H. Freeman & Co., New York, N.Y.
- 38.Winkler, M. A., J. Uher, and S. Cepa. 1999. Direct analysis and identification of Helicobacter and Campylobacter species by MALDI-TOF mass spectrometry. Anal. Chem. 71:3416-3419. [DOI] [PubMed] [Google Scholar]
- 39.Wong, K. C., B. S. Ho, S. I. Egglestone, and W. H. Lewis. 1995. Duplex PCR system for simultaneous detection of Neisseria gonorrhoeae and Chlamydia trachomatis in clinical specimens. J. Clin. Pathol. 48:101-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yager, T. D., L. Baron, R. Batra, A. Bouevitch, D. Chan, K. Chan, S. Darasch, R. Gilchrist, A. Izmailov, J. M. Lacroix, K. Marchelleta, J. Renfrew, D. Rush-low, E. Steinbach, C. Ton, P. Waterhouse, H. Zaleski, J. M. Dunn, and J. Stevens. 1999. High performance DNA sequencing, and the detection of mutations and polymorphisms, on the Clipper sequencer. Electrophoresis 20:1280-1300. [DOI] [PubMed] [Google Scholar]
- 41.Ye, F., M. S. Li, J. D. Taylor, Q. Nguyen, H. M. Colton, W. M. Casey, M. Wagner, M. P. Weiner, and J. Chen. 2001. Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Hum. Mutat. 17:305-316. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, W., M. Cohenford, B. Lentricha, H. D. Isenberg, E. Simon, H. Li, H. Yi, and D. Y. Zhang. 2002. Detection of Chlamydia trachomatis by isothermal ramification amplification method: a feasibility study. J. Clin. Microbiol. 40:128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]