Abstract
A screening method for methicillin-resistant Staphylococcus aureus (MRSA) by using selective broth and real-time PCR (broth-PCR) was developed and evaluated. The samples (n = 304) were cultured in the broth overnight, followed by nuc gene detection by real-time PCR. nuc-negative samples were further checked for the presence of nuc amplification inhibitors by a PCR internal inhibitor assay. nuc-positive samples and nuc-negative samples with PCR inhibitors were cultured onto plates and processed further. The diagnostic values for this MRSA screening method were 93.3% sensitivity, 89.6% specificity, 31.8% positive predictive value, and 99.6% negative predictive value. The application of the broth-PCR method will be able to report most of the negative samples (258 of 289 [89.3%]) on the next morning and can save as much as 84.9% (258 of 304) of the labor and cost spent on processing the nuc-negative specimens on plates. In the study, all the samples were processed in parallel by the broth enrichment method and the plating method for comparison. To identify MRSA, the isolated oxacillin-resistant S. aureus strains were tested by a duplex real-time PCR targeting the mecA gene and the nuc gene. A collection of MRSA, methicillin-susceptible Staphylococcus aureus, methicillin-resistant Staphylococcus epidermidis, and methicillin-susceptible Staphylococcus epidermidis strains and a panel of standard strains of 11 bacterial species other than S. aureus were also tested by this method, which was proved to be a valuable tool for MRSA identification in a routine microbiological laboratory.
Staphylococcus aureus is one of the most significant human pathogens, causing both nosocomial and community-acquired infections. Its main habitats are the nasal membranes and the skin of humans and warm-blooded animals. S. aureus can cause a range of infectious diseases from mild conditions, such as skin and soft tissue infections, to severe, life-threatening debilitation (14, 23). Strains of methicillin-resistant S. aureus (MRSA) were first detected in the early 1960s, shortly after methicillin came into clinical use. Resistance to methicillin is mediated by the presence of penicillin-binding protein 2a (PBP-2a), encoded by the mecA gene (4). No available β-lactam binds effectively to PBP-2a, and staphylococci resistant to methicillin or oxacillin should be generally regarded as resistant to all β-lactams (13). Since the end of the 1970s, the occurrence of MRSA has increased steadily. Molecular epidemiological studies have shown that a limited number of MRSA strains have spread by clonal dissemination between different hospitals, cities, countries, and even continents and are now the cause of hospital infections worldwide (5, 15). MRSA strains are usually introduced into an institution by an infected or colonized patient or by a colonized health care worker. Thus, epidemiological surveys and control measures are particularly important for MRSA. Rapid screening followed by accurate and timely identification of MRSA becomes an elemental procedure in preventive measures.
In the present study, a MRSA screening method using MRSA-selective broth and real-time PCR, and a duplex real-time PCR assay for rapid identification of MRSA strains, were developed and evaluated.
MATERIALS AND METHODS
Clinical samples.
Three hundred four consecutive clinical samples sent to our laboratory for MRSA screening were investigated. The samples were from wounds or abscesses (35.9%), the anterior nares (27.0%), the perineum (19.7%), urine (7.9%), catheter insertion sites (3%), skin and soft tissues (0.7%), sputum (0.3%), the trachea (0.3%), and other sites (5.2%). Most specimens were sampled by swabs.
Bacterial strains.
Culture collection strains tested in the study included Enterococcus faecalis ATCC 29212, Enterococcus faecium ATCC 19434, Staphylococcus cohni ATCC 29974, Staphylococcus saprophyticus ATCC 15305, Staphylococcus xylosus ATCC 29971, Streptococcus pneumoniae ATCC 6305, Streptococcus pyogenes ATCC 19615, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 13883, Proteus mirabilis ATCC 29245, Pseudomonas aeruginosa ATCC 27853, MRSA strains CCUG 46147 (a homogeneously, highly resistant strain) and CCUG 31966 (a heterogeneously, weakly resistant strain), methicillin-susceptible S. aureus (MSSA) strain ATCC 29213, methicillin-resistant Staphylococcus epidermidis (MRSE) strain ATCC 29887, and methicillin-susceptible Staphylocoocus epidermidis (MSSE) strains ATCC 29886 and ATCC 12228. A collection of 19 representative clinical MRSA isolates, which had different pulsed-field gel electrophoresis banding patterns, and the 15 MRSA strains isolated in this study were also tested by a duplex real-time PCR for detecting the mecA and nuc genes.
Selective media and culture conditions.
For evaluation and for comparison with the new MRSA screening method (broth-PCR method), strains were processed in parallel by the broth enrichment method and the conventional plating method during the study.
The samples were first plated onto two types of agar plates: a blood agar plate and a mannitol salt agar (MSA) plate with 1 μg of oxacillin/ml (19). The plates were incubated at 35°C for 24 to 48 h. After being streaked onto the agar plates, the samples were inoculated in the MRSA-selective broth. The MRSA broth being used to enrich MRSA in clinical specimens was composed of Iso-Sensitest broth (Oxoid), 2.3% NaCl, 1 μg of aztreonam (Bristol-Myers Squibb)/ml, and 2 μg of oxacillin (Sigma)/ml. The inoculated broth was incubated at 30°C.
In the conventional plating method, S. aureus was isolated and identified by a standard procedure (11). Isolates that were oxacillin resistant in an oxacillin disk diffusion test were further verified by mecA and nuc duplex PCR.
In the broth enrichment method, the bacterial growth was indicated by turbidity. The turbid broth (100 μl), or the broth after a maximal 5-day incubation, was spread onto each of two agar plates, blood agar and MSA agar, and was further investigated as for the plating method.
In the broth-PCR method, all the samples were tested by real-time PCR for nuc in the overnight-cultivated broth, whether the broth turned turbid or not; then the nuc-negative samples were checked by a PCR internal inhibitor assay. In order to evaluate this new method, all the samples were further processed when the broth turned turbid or after a maximal 5-day incubation as described for the broth enrichment method.
Detection of nuc gene from broth after overnight cultivation. (i) DNA extraction.
An aliquot (100 μl) of broth was centrifuged at 20,800 × g for 2 min. The supernatant was carefully removed, and the pellet was suspended in 100 μl of MilliQ water (i.e., water purified by reverse osmosis and filtration). The suspension was then heated at 95°C for 15 min. After centrifugation for 1 min at 20,800 × g to sediment the debris, the clear supernatant was ready to be used as template DNA in PCR.
(ii) Real-time PCR.
The real-time PCR assay was carried out with the LightCycler system (Roche). Primers NUC1 (5′-GCG ATT GAT GGT GAT ACG GTT-3′) and NUC2 (5′-AGC CAA GCC TTG ACG AAC TAA AGC-3′), directed to the nuc gene, an S. aureus-specific marker, were used (2). Amplification reactions were performed in a volume of 20 μl containing 2 μl of DNA template, 4 mM MgCl2, 0.25 μM each primer, and 2 μl of 10× LightCycler FastStart DNA Master SYBR Green I mixture (Roche). Following an initial denaturation at 95°C for 10 min to activate the FastStart Taq DNA polymerase, the 35-cycle amplification program consisted of heating at 20°C/s to 95°C with a 0-s hold, cooling at 20°C/s to 55°C with a 5-s hold, and heating at 20°C/s to 72°C with an 8-s hold. Then the one-cycle melting curve program consisted of heating at 20°C/s to 95°C with a 0-s hold, cooling at 20°C/s to 58°C with a 60-s hold, and heating at 0.1°C/s to 95°C with a 0-s hold. Finally, the experiment protocol ended with one cycle of cooling at 20°C/s to 35°C with a 30-s hold. The fluorescence channel was set at F1 (530 nm).
(iii) Data analysis.
The identity of the PCR product from a sample can be confirmed by performing a melting curve analysis comparing its melting temperature (Tm) with the Tm of the product from the positive control. In the study, the samples with Tms within the range of the Tm of the positive control's product ± 0.5°C were regarded as nuc positive.
(iv) Sensitivity assay.
The MRSA strains CCUG 46147 and CCUG 31966 in serial 10-fold dilutions were inoculated into the MRSA broth and incubated at 30°C. nuc PCR was performed at time zero (just after inoculation), day 1 (after overnight cultivation), day 3 (after a 3-day cultivation), and day 6 (after a 6-day cultivation).
(v) PCR internal inhibitor assay.
For nuc-negative samples, a PCR-inhibitor assay was performed to check if nuc amplification inhibitors were present in the extracted DNA. A minimal amount of purified nuc-positive DNA (5 pg), which was equivalent to the detection limit (overnight cultivation) of the broth-PCR, was added to the amplification mixture. Other PCR parameters were exactly the same as those of the nuc PCR used in the broth-PCR method. The sample was considered to contain a nuc amplification inhibitor if the nuc gene could not be amplified in the PCR inhibitor assay.
Identification of MRSA by duplex real-time PCR.
A pure bacterial culture was used in the duplex real-time PCR assay.
(i) DNA extraction.
A single colony was picked and suspended in 100 μl of MilliQ water. The suspension was then heated at 95°C for 15 min. After centrifugation for 1 min at 20,800 × g to sediment the debris, the clear supernatant was ready to be used as template DNA in PCR.
(ii) Duplex real-time PCR.
The duplex real-time PCR was run by use of the LightCycler system (Roche). Primers MECA1 (5′-GCA ATC GCT AAA GAA CTA AG-3′) and MECA2 (5′-GGG ACC AAC ATA ACC TAA TA-3′) and primers NUC1 and NUC2, targeting the mecA gene and the nuc gene, respectively, were used (2, 19). Amplification mixtures contained 2 μl of DNA template, 3 mM MgCl2, 1 μM (each) MECA1 and MECA2, 0.25 μM (each) NUC1 and NUC2, and 2 μl of 10× LightCycler FastStart DNA Master SYBR Green I mixture (Roche) in a final volume of 20 μl. The cycling program was the same as that used for detection of nuc except for 32 cycles of amplification.
(iii) Data analysis.
Melting curve analysis was performed to determine which specific gene(s) had been detected from the samples. Strains with Tms within the range of the positive control's Tm (mecA) ± 0.8°C and within the range of the positive control's Tm (nuc) ± 0.5°C were regarded as mecA and nuc positive, respectively.
RESULTS
MRSA screening.
A total of 15 MRSA strains were detected by all three screening methods. The sample types and the detection of each of the 15 strains by the different screening methods are shown in Table 1.
TABLE 1.
The 15 MRSA strains detected by different screening methods and their sample types in the study
| Sample or strain (n = 15) | Sample type | Resultsa with:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Broth-PCR (nuc, overnight culture) | Broth enrichment method
|
Plating method
|
||||||||
| Turbidity
|
MRSA detected | Blood agar plate
|
MSA plate
|
|||||||
| Overnight culture | 2-day culture | Overnight culture | 2-day culture | Overnight culture | 2-day culture | |||||
| 1 | Wound or abscess | + | − | + | + | + | ND | − | − | |
| 2 | Anterior nares | + | + | ND | + | + | ND | + | ND | |
| 3 | Wound or abscess | + | + | ND | + | + | ND | + | ND | |
| 4 | Anterior nares | + | + | ND | + | + | ND | + | ND | |
| 5 | Wound or abscess | + | + | ND | + | + | ND | + | ND | |
| 6 | Anterior nares | + | − | + | + | + | ND | − | + | |
| 7 | Perineum | + | − | + | + | + | ND | − | + | |
| 8 | Urine | + | − | + | + | + | ND | − | + | |
| 9 | Sputum | + | − | + | + | − | − | − | + | |
| 10 | Perineum | + | + | ND | + | + | ND | + | ND | |
| 11 | Anterior nares | + | + | ND | + | + | ND | + | ND | |
| 12 | Wound or abscess | + | − | + | + | − | − | − | − | |
| 13 | Perineum | − | − | + | − | − | − | − | + | |
| 14 | Vestibulum nasi | + | + | ND | + | + | ND | − | + | |
| 15 | Urine | + | + | ND | + | + | ND | − | + | |
| No. of positive cases among the 15 MRSA samples | 14 | 8 | 7 | 14 | 12 | 0 | 6 | 7 | ||
+, positive; −, negative; ND, not determined.
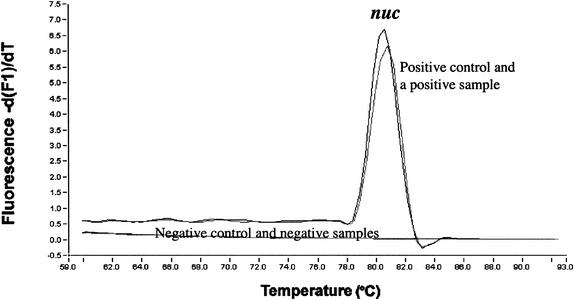
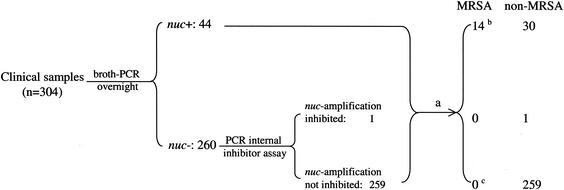
By use of the broth-PCR method, among the 304 clinical samples investigated, 44 samples (14.5%) were found to be nuc positive after overnight cultivation in the MRSA broth (Fig. 1 and 2). Among the 260 nuc-negative samples, one sample was proved to contain a nuc PCR inhibitor. Among the 15 MRSA strains detected in the study, 14 strains were derived from nuc-positive samples and one was from a nuc-negative sample (sample 13) in which no nuc PCR inhibitor was detected. The strain (strain 13) missed in the broth-PCR assay was not detected by the broth enrichment method or the blood-agar plate method either, but it was found on the MSA plate, with only 2 colonies, after a 2-day cultivation (Table 1). The diagnostic values of the broth-PCR method were as follows: sensitivity, 93.3% (14 of 15); specificity, 89.6% (259 of 289); positive predictive value, 31.8% (14 of 44); and negative predictive value, 99.6% (259 of 260).
FIG. 1.
Tm curves in the broth-PCR for detection of the nuc gene (MRSA screening).
FIG. 2.
Flowchart and results of the broth-PCR method in this study. a, the broth, when it turned turbid or after a maximal 5-day incubation, was spread to the agar plates and processed in a conventional way. b, one (sample 12) of the MRSA strains was not detected by the direct plating method. c, an MRSA strain was isolated from sample 13 on the MSA plate by the direct plating method.
Among the 15 samples in which MRSA was found, turbid broth was observed in 8 samples after a 1-day incubation and in all 15 samples after incubation for 2 days (Table 1). However, in one case (sample 13), MRSA was not isolated from the enriched broth; instead, a coagulase-negative staphylococcus (CoNS) was isolated. The predominant bacteria isolated from the turbid broth in this study were CoNS (64% of the cases), while MRSA and MSSA accounted for only 11 and 4%, respectively. The diagnostic values of the broth-enrichment method, after 2 days of incubation, were as follows: sensitivity, 93.3% (14 of 15); specificity, 61.2% (177 of 289); positive predictive value, 11.1% (14 of 126); and negative predictive value, 99.4% (177 of 178).
By the plating method, when the two types of agar plates were taken together, 14 of the 15 MRSA strains were detected after incubation for 2 days, with a sensitivity (93.3%) as high as those of the other two methods, but the specificity was poor (10.1% [31 of 289]). MRSA was detected from sample 13, but not from sample 12 (Table 1), by this method.
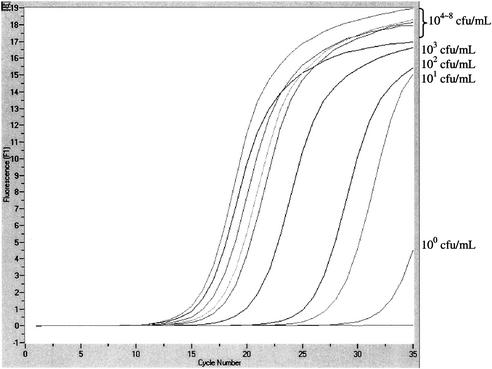
The sensitivity assay showed that the detection limit of the broth-PCR for nuc at the time of inoculation, time zero, was 104 to 105 CFU/ml of inoculum in broth. After overnight incubation, the detection limit was improved to 100 to 101 CFU/ml of inoculum in broth (Fig. 3). After 3 and 6 days of incubation, the detection limits were 100 to 101 and 100 to 102 CFU/ml of inoculum in broth, respectively.
FIG. 3.
Sensitivity of the real-time PCR assay for detection of the nuc gene from the overnight-cultured broth.
MRSA identification.
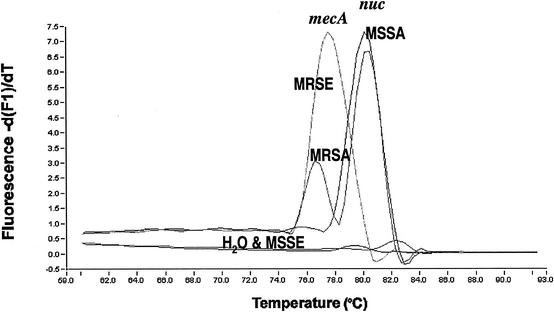
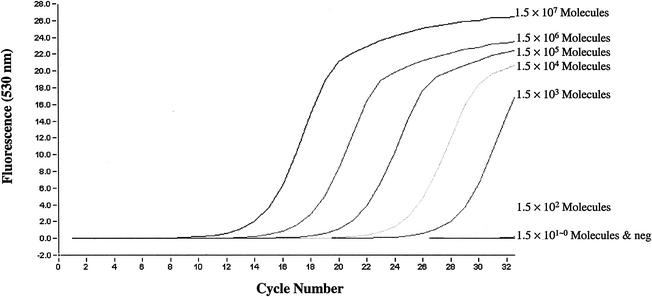
All MRSA strains tested in the study presented two peaks in the melting curve analysis; one peak was specific for the mecA gene with a Tm of 77.50 to 79.00°C, and one was specific for the nuc gene with a Tm of 79.90 to 80.60°C. MSSA strains had only a nuc peak, MRSE strains had only a mecA peak, and MSSE strains had no peak (Fig. 4). The mecA and nuc genes could be detected, by use of LightCycler real-time PCR, with an amount of DNA template as small as 1.5 × 102 genomic molecules (Fig. 5).
FIG. 4.
Tm curves for MRSA, MSSA, MRSE, and MSSE in mecA and nuc duplex real-time PCR (MRSA identification).
FIG. 5.
Sensitivity of the mecA and nuc duplex real-time PCR assay, determined through serial dilutions of the template DNA extracted from MRSA strain CCUG 31966.
The specificity of the real-time PCR assay was further determined with a panel of 11 gram-negative and gram-positive standard strains of species other than S. aureus. No cross-reactivity was observed.
DISCUSSION
MRSA is now one of the most important nosocomial pathogens worldwide. The prevalence of MRSA, however, varies markedly by country. The prevalence of MRSA in northern European countries is low; this is assumed to be due at least in part to the prompt implementation of aggressive infection control measures (1, 6, 21, 22). Screening high-risk patients and health care workers for MRSA is one of the control measures. Several studies have found that such screening programs are cost-effective (3, 10, 12, 16).
The conventional culture methods are time- and labor-consuming, and the diagnostic values are not as good as those of the new MRSA screening method. Especially because the number of samples for MRSA screening has increased dramatically in recent years, a more efficient method is needed to meet the clinical requirements. In the present study, we report a rapid and sensitive method, using MRSA-selective broth and real-time PCR, to screen for the presence of MRSA. This procedure started with enrichment of bacteria in a selective broth that favors the growth of MRSA, followed by detection of an S. aureus-specific gene (nuc) via real-time PCR. The nuc-negative samples were further processed by a PCR inhibitor assay to check for nuc amplification-inhibitory substances. nuc-negative samples without nuc PCR inhibitors were regarded as MRSA negative. Compared to the broth enrichment method and the conventional plating method, which required incubation for 2 days, the overnight broth-PCR assay achieved the same sensitivity, 93.3%, and a higher specificity (89.6% versus 61.2 and 10.7%, respectively), positive predictive value (31.8% versus 11.1 and 5.1%, respectively), and negative predictive value (99.6% versus 99.4 and 96.9%, respectively).
The gene coding for methicillin resistance (mecA) was not targeted in this screening assay (broth-PCR), because the DNA template used at this step was directly extracted from the broth, which was a mixed culture. In these mixed cultures, CoNS were the predominant bacteria. It is known that methicillin resistance is frequent among CoNS on a global scale (8, 9, 17). At our hospital the incidence of methicillin resistance in CoNS is 45%. Therefore, the addition of mecA in the screening method would not really increase the specificity and positive predictive value of the method, but it would give rise to false-positive results in cases such as those involving methicillin-resistant CoNS combined with MSSA.
The sensitivity assay showed that the nuc gene could be identified with an inoculum as low as 1 to 10 CFU/ml in the broth after overnight cultivation. Prolonged incubation did not produce higher sensitivity. The study indicated that overnight was a suitable interval for incubation before proceeding to the following PCR assay.
Since the sample used for nuc PCR was from the enriched broth, it is possible that PCR-inhibitory substances were present in the sample. To determine whether nuc amplification inhibitors were present in the nuc-negative samples, a PCR internal inhibitor assay was performed as a complementary test to the broth-PCR method. The compositions of clinical samples are usually complicated and display great variation, which could range from mixed culture to bacteria-free, so a possible way to check for PCR internal inhibitors is to add a minimal amount of positive DNA to the PCR system. In this study, the PCR inhibitor assay was aimed at determining if nuc amplification was inhibited or disturbed in those nuc-negative samples, so the direct method was to add the nuc-positive DNA to the PCR system. One advantage of using the nuc-positive DNA was that all the other parameters in the PCR inhibitor assay, including the criteria in data analysis, were exactly the same as those in the nuc PCR, so the inhibitory status of the nuc PCR could be truly reflected through the PCR internal inhibitor assay. A negative result in the PCR inhibitor assay indicated that nuc amplification was inhibited or disturbed by the inhibitory substances in the sample DNA. Although only one sample was found to inhibit nuc amplification in this study, and it was proved to be a non-MRSA sample (Fig. 2), we still think it necessary to include this assay in the broth-PCR method in order to reduce the number of false nuc-negative cases.
With the application of this method, most of the negative samples (258 of 289 [89.3%]) can be identified the next morning after sampling, so the time to obtain the negative result can be reduced to 16 to 18 h. Meanwhile, as much as 84.9% (258 of 304) of the labor and cost spent on processing nuc-negative specimens on plates can be saved, which is especially cost-effective in countries such as Sweden, where the prevalence of MRSA remains low. This new MRSA-screening method has now been applied at our routine laboratory. The introduction of this new method is of significance in clinics. According to our policy, patients who are highly suspected to be colonized with MRSA, such as patients hospitalized abroad, are isolated in a single room until the cultures become MRSA negative. An earlier MRSA-negative report makes it possible for patients to go to open wards earlier, saving much expense.
One explanation for the missed MRSA strain in the broth-PCR assay and broth enrichment assay is that the inoculum of the bacteria in the broth was lower than the detection limits of the assays. PCR inhibitors were not detected from the extracted DNA of this sample (sample 13), and no S. aureus was isolated from the enriched broth.
Identification of MRSA by real-time PCR has been reported previously (7, 18, 20). In the study by Tan et al. (20), the detection of MRSA was performed with two separate PCRs, for an S. aureus-specific gene (Sa442) and the mecA gene, respectively, by using a cyanine 5-labeled probe and SYBR Green I. In the studies of Reischl et al. (18) and Grisold et al. (7), the PCR assay was carried out with two pairs of hybridization probes labeled with different dyes. Here we report a real-time PCR assay which applies SYBR Green I as the only fluorescent agent for detecting mecA and nuc, simultaneously, in one PCR. By using the conditions described in the present study, both mecA and nuc could be easily detected with a DNA template in a range of 1.5 × 102 to 1.5 × 107 genomic molecules in the reaction mixture (Fig. 5). The DNA extracted from a single colony, by using our simple and rapid boiling procedure (described in Materials and Methods), usually has a concentration of 1.5 × 106 to 6.0 × 106 molecules/μl. Thus, the duplex PCR with SYBR Green I described in this study is sensitive enough to be used as a routine diagnostic method. The specificity of this method has been tested with a panel of standard strains of species other than S. aureus, and no cross-reactivity was observed. Furthermore, by the combination of the simple template DNA preparation with the rapid thermocycling of the LightCycler system, results are available within 1 h.
In conclusion, the broth-PCR method is an efficient MRSA-screening assay. The duplex real-time PCR for rapid identification of MRSA is a valuable tool in a routine microbiological laboratory.
Acknowledgments
We are grateful to Agneta Ahlin and Gerd Fessé for skillful technical assistance.
REFERENCES
- 1.Boyce, J. M. 2002. Understanding and controlling methicillin-resistant Staphylococcus aureus infections. Infect. Control Hosp. Epidemiol. 23:485-487. [DOI] [PubMed] [Google Scholar]
- 2.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaix, C., I. Durand-Zaleski, C. Alberti, and C. Brun-Buisson. 1999. Control of endemic methicillin-resistant Staphylococcus aureus: a cost-benefit analysis in an intensive care unit. JAMA 282:1745-1751. [DOI] [PubMed] [Google Scholar]
- 4.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fluit, A. C., C. L. C. Wielders, J. Verhoef, and F.-J. Schmitz. 2001. Epidemiology and susceptibility of 3,051 Staphylococcus aureus isolates from 25 university hospitals participating in the European SENTRY study. J. Clin. Microbiol. 39:3727-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grisold, A. J., E. Leitner, G. Mühlbauer, E. Marth, and H. H. Kessler. 2002. Detection of methicillin-resistant Staphylococcus aureus and simultaneous confirmation by automated nucleic acid extraction and real-time PCR. J. Clin. Microbiol. 40:2392-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarlov, J. O. 1999. Phenotypic characteristics of coagulase-negative staphylococci: typing and antibiotic susceptibility. APMIS 91(Suppl.):1-42. [PubMed] [Google Scholar]
- 9.Jarlov, J. O., and N. Hoiby. 1998. Coagulase-negative staphylococci in a major Danish university hospital: diversity in antibiotic susceptibility between wards. APMIS 106:411-416. [DOI] [PubMed] [Google Scholar]
- 10.Karchmer, T. B., L. J. Durbin, B. M. Simonton, and B. M. Farr. 2002. Cost-effectiveness of active surveillance cultures and contact/droplet precautions for control of methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 51:126-132. [DOI] [PubMed] [Google Scholar]
- 11.Kloos, W. E., and T. L. Bannerman. 1999. Staphylococcus and Micrococcus, p. 264-282. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 12.Kunori, T., B. Cookson, J. A. Roberts, S. Stone, and C. Kibbler. 2002. Cost-effectiveness of different MRSA screening methods. J. Hosp. Infect. 51:189-200. [DOI] [PubMed] [Google Scholar]
- 13.Livermore, D. M., and J. D. Williams. 1996. β-Lactams: mode of action and mechanisms of bacterial resistance, p. 502-578. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, Md.
- 14.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 15.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 16.Papia, G., M. Louie, A. Tralla, C. Johnson, V. Collins, and A. E. Simor. 1999. Screening high-risk patients for methicillin-resistant Staphylococcus aureus on admission to the hospital: is it cost effective? Infect. Control Hosp. Epidemiol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 17.Petinaki, E., F. Kontos, V. Miriagou, M. Maniati, F. Hatzi, A. N. Maniatis, and The Bacterial Resistance Study Group. 2001. Survey of methicillin-resistant coagulase-negative staphylococci in the hospitals of central Greece. Int. J. Antimicrob. Agents 18:563-566. [DOI] [PubMed] [Google Scholar]
- 18.Reischl, U., H. J. Linde, M. Metz, B. Leppmeier, and N. Lehn. 2000. Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence PCR. J. Clin. Microbiol. 38:2429-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth, R. W., G. Kahlmeter, B. Olsson-Liljequist, and B. Hoffman. 2001. Methods for identifying methicillin resistance in Staphylococcus aureus. J. Hosp. Infect. 48:103-107. [DOI] [PubMed] [Google Scholar]
- 20.Tan, T. Y., S. Corden, R. Barnes, and B. Cookson. 2001. Rapid identification of methicillin-resistant Staphylococcus aureus from positive blood cultures by real-time fluorescence PCR. J. Clin. Microbiol. 39:4529-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhoef, J., D. Beaujean, H. Blok, A. Baars, A. Meyler, C. van der Werken, and A. Weersink. 1999. A Dutch approach to methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 18:461-466. [DOI] [PubMed] [Google Scholar]
- 22.Voss, A., D. Milatovic, C. Wallrauch-Schwarz, V. T. Rosdahl, and I. Braveny. 1994. Methicillin-resistant Staphylococcus aureus (MRSA) in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 13:50-55. [DOI] [PubMed] [Google Scholar]
- 23.Waldvogel, F. A. 2000. Gram-positive cocci, p. 2069-2092. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, Pa.