Abstract
Computer-assisted analysis of pulsed-field gel electrophoresis (PFGE) libraries can facilitate comparisons of fragment patterns present on multiple gels. We evaluated the ability of the Advanced Analysis (version 4.01) and Database (version 1.12) modules of the Phoretix gel analysis software package (Nonlinear USA, Inc., Durham, N.C.) to accurately match DNA fragment patterns. Two gels containing 38 lanes of SmaI-digested Enterococcus faecalis OG1RF DNA were analyzed to assess the impact of (i) varying the lane position of the standards, (ii) using gel plugs made at different times, and (iii) normalizing the fragment patterns by using molecular weight (MW) algorithms versus retardation factor (Rf) algorithms. Two sets of PFGE libraries (one containing SmaI restriction patterns from 62 Enterococcus faecium isolates and the other containing SmaI restriction patterns of 89 Staphylococcus aureus isolates) were analyzed to assess the impact of varying the matching tolerance algorithm (designated as the vector box setting [VBS]) in the Phoretix software. Varying the lane position of standards on a gel and using gel plugs made on different days resulted in different VBSs, although it was not possible to judge whether those differences were statistically significant. Normalization of E. faecalis OG1RF fragment patterns by Rf and MW methodology yielded no statistically significant differences in variability between the same fragment on different lanes. Suboptimal VBSs decreased the specificity with which related isolates were grouped together in dendrograms. The optimal VBS for analysis of PFGE fragment patterns from E. faecalis isolates differed from that for S. aureus isolates and sometimes was not that recommended by the manufacturer. Thus, computer-assisted analysis of PFGE patterns seemed to compensate for the intra- and intergel variation evaluated in the present study, and optimizing the software for the species to be tested was a critical preliminary step before further PFGE library analysis.
Pulsed-field gel electrophoresis (PFGE) is the typing method of choice for enterococci and staphylococci due to its high discriminatory power for these organism groups (8, 19). However, comparisons of fragment patterns present on multiple gels from large sets of isolates are technically difficult (4, 21). Many variables, such as the concentration of DNA in the agarose plugs, the amount of agarose in the gel, the electrophoresis voltage, the gel temperature, and the buffer strength, contribute to intra- and intergel variation and complicate comparisons of fragment patterns on multiple gels and in PFGE libraries (2, 4). (PFGE libraries consist of multiple gels that contain fragment patterns of a single bacterial species.) Much of the interlaboratory variation reported for PFGE results can be attributed to individual user technique, different types of PFGE instrumentation, and different protocols used for testing (4, 21). However, variation can also be attributed to fragment position normalization and matching tolerance algorithms used for pattern interpretation (3, 21). Random lane and fragment distortions occur within PFGE libraries (2, 4). These distortions can impact the matching of isolates by PFGE analysis software. Slight variation in the preparation of gel plugs is another source of intragel variation that can lead to fragment or lane distortions. The lanes selected to contain the molecular weight (MW) or retardation factor (Rf) standards are also critical for analysis, since the first and last lanes of a gel tend to be more distorted than the interior lanes (2, 7). These varying sources of intra- and intergel variation can affect matching tolerance algorithms in PFGE analysis software packages and therefore can distort PFGE matching results.
Computer-assisted analysis of PFGE data allows investigators to create searchable databases of fragment patterns where similarity calculations and cluster analyses can easily be performed (18). Comparisons of software packages to date have given some insight into their ability to identify outbreak-related strains (7) but have not directly addressed the question of compensating for the intra- and intergel variation within PFGE libraries. The present study assessed the impact of three potential sources of intra- and intergel variation of fragment banding patterns: (i) the location of the standards in the gel, (ii) the age of the agarose gel plugs containing the sample DNA, and (iii) the normalization of fragment position by Rf versus MW methods. In addition, the impact of varying matching tolerance algorithms was assessed by using Enterococcus faecium and Staphylococcus aureus PFGE libraries.
MATERIALS AND METHODS
Study design.
Two gels containing 38 lanes of SmaI-digested Enterococcus faecalis OG1RF DNA (13) were analyzed to assess the impact of intra- and intergel variation. Varying the matching tolerance algorithms for both E. faecium (62 isolates) and S. aureus (89 isolates) PFGE libraries demonstrated how different settings impacted macrorestriction fragment pattern matching. This process helped determine an optimized matching tolerance setting for each PFGE library. Organisms were identified by standard biochemical procedures (5, 9). Antimicrobial susceptibility testing was performed by broth microdilution, and the resulting MICs were converted into categorical interpretations according to NCCLS guidelines (14, 15).
PFGE protocol for E. faecalis and E. faecium isolates.
The isolates were inoculated into 1 ml of Trypticase soy broth (Remel, Lenexa, Kans.) and incubated at 37°C overnight. The turbidity of the broth was adjusted to an optical density of 1.00 at 540 nm with TE buffer (10 mM Tris-HCl [Sigma, St. Louis, Mo.] and 1 mM EDTA [Sigma] at pH 7.5). Cells were washed twice in TE buffer and resuspended in 300 μl of buffer before the addition of 300 μl of melted 2% SeaPlaque agarose (BioWhittaker Molecular Applications, Rockland, Maine; final agarose concentration, 1%). Plugs were prepared in reusable plug molds (Bio-Rad Laboratories, Hercules, Calif.). The bacterial cells in the plugs were lysed overnight in 20 μl of mutanolysin (stock concentration, 10,000 U/ml; Sigma) per 1 ml of EC lysis buffer (6 mM Tris-HCl, 1 M NaCl, 100 mM EDTA, 0.5% Brij-58 [Sigma], 0.2% sodium deoxycholate [Sigma], 0.5% Sarkosyl [Sigma]). Deproteination of the samples was performed by incubating each plug in 0.1 mg of proteinase K (Life Technologies, Inc., Rockville, Md.)/ml of Sarkosyl-EDTA-Tris buffer (10 mM Tris-HCl, 0.1 M EDTA, 1% Sarkosyl) at pH 6.8 for 4 to 6 h in a 55°C water bath. After deproteination, the plugs were washed three times in TE buffer. Slices of the plugs were cut and incubated with the restriction endonuclease SmaI (New England BioLabs, Inc., Beverly, Mass.) for 2 to 4 h in a dry shaker at 25°C. Digested plug slices were inserted into 20-well 1% chromosomal grade agarose (Bio-Rad Laboratories) gels prepared with 0.5× Tris-borate-EDTA buffer (Life Technologies, Inc.). Gels were run on a CHEF DR-III apparatus (Bio-Rad Laboratories) at 14°C for 20 h at 6 V/cm with an initial pulse time of 1 s and a final pulse time of 20 s. Gels were stained in ethidium bromide, destained in fresh distilled water, and photographed. Digital images of the gels were saved as TIFF files by using the Foto/Analyst Archiver (Fotodyne, Inc., Hartland, Wis.) for subsequent gel analysis. Manual banding pattern interpretation was based upon published criteria (20). Computer-assisted analysis was performed with the Advanced Analysis (version 4.01) and Database (version 1.12) modules of Phoretix gel analysis software (Nonlinear USA, Inc., Durham, N.C.). (The modules together are currently named Phoretix 1D Pro.)
PFGE protocol for S. aureus isolates.
S. aureus isolates were inoculated into 5 ml of brain heart infusion broth (BD BioSciences, Sparks, Md.) and incubated without shaking at 37°C overnight. The cultures were adjusted with sterile brain heart infusion broth to an optical density of 1.3 by using a Microscan turbidity meter (Dade MicroScan, Sacramento, Calif.). A total of 200 μl of each cell suspension was transferred to microcentrifuge tubes and centrifuged at 13,000 rpm for 5 min. The supernatant was aspirated and cells were resuspended in 300 μl of 0.01 M TE buffer (pH 8.0). To each tube, 4 μl of conventional lysostaphin (stock solution of 1 mg/ml in 20 mM sodium acetate; Sigma) and 300 μl of 1.8% SeaKem Gold agarose (BioWhittaker Molecular Applications) were added, with mixing after the addition of each reagent. Plugs were prepared in nondisposable plug molds. After they solidified, the plugs were transferred to tubes containing 3 ml of EC lysis buffer and then incubated for 4 h at 37°C. After lysis, the plugs were washed four times for 30 min with 4 ml of TE buffer. Slices of the plugs were cut and incubated with SmaI for 4 h at 25°C. Digested plug slices were placed directly onto a 30-well comb that was subsequently placed securely into the gel-casting platform. One percent SeaKem Gold agarose (equilibrated to 55°C) made with 0.5× Tris-borate-EDTA buffer was poured into the gel-casting platform around the comb and allowed to solidify at room temperature for 45 min. Gels were run on a CHEF DR-II (Bio-Rad Laboratories) apparatus at 14°C for 21 h at 200 V with an initial pulse time of 5 s and a final pulse time of 40 s. Gels were stained with an ethidium bromide solution, destained in fresh distilled water, and photographed. Polaroid pictures of the gels were saved digitally as TIFF files for subsequent gel analysis. Manual banding pattern interpretation and computer-assisted analysis were performed as previously described for the E. faecium isolates (20).
Sources of intra- and intergel variation investigated.
The following sources of variability were evaluated to determine their influence on the ability of PFGE analysis software packages to accurately match 32 identical, non-standard-lane pulsed-field patterns on two PFGE gels: (i) variation of standard lane position, (ii) gel plugs made on different days, and (iii) normalization of fragment position by MW and Rf methods. In the software, MW normalization of a fragment position occurred by estimating unknown fragment values from the E. faecalis OG1RF MW standard fragment values by using a log curve-fitting algorithm. The log regression method was used because it appeared to provide the best fit of the E. faecalis OG1RF strain fragment positions run by PFGE compared to other curve type choices in the software (i.e., cubic spline, first-order Lagrange, linear, linear log, and quadratic). Conversely, Rf normalization of fragment position was a linear function of the pixel position that fragments occupied within a lane. Thus, for the Rf method, unknown fragment positions were not estimated based on the “fit” of a log curve; rather, they represented the location of the peak pixel intensity associated with each identified fragment. All statistical comparisons of MW and Rf normalization results were performed by using SAS version 6.12 (SAS Institute, Inc., Cary, N.C.). For each fragment present in the 32 lanes examined, the mean, range, variance, and coefficient of variation (CV; standard deviation divided by the mean) (17) were calculated. CVs, the within-fragment variability normalized by MW (CVMW) and Rf (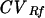 ), were compared for each fragment by using the Z-test. A P value of ≤0.05 defined significant associations.
), were compared for each fragment by using the Z-test. A P value of ≤0.05 defined significant associations.
Optimization of matching tolerance studies.
The effects of varying the matching tolerance algorithm were assessed by using PFGE libraries of E. faecium (n = 62; 57 isolates were vancomycin resistant) and S. aureus (n = 89; 88 isolates were oxacillin resistant) macrorestriction patterns analyzed by Rf. Optimization of matching tolerance algorithms was achieved by manipulation of the vector box setting (VBS), which is the maximum difference in Rf values between two fragments that the PFGE analysis software defined to match. For both the E. faecium and S. aureus matching tolerance optimization studies, the visually similar group (VSG) was defined as fragment patterns that were within six fragment differences from an arbitrarily chosen reference fragment pattern that was visually similar to more than one fragment pattern in each respective PFGE library. At different VBS settings, unweighted pair group method using arithmetic averages (UPGMA) dendrograms were generated from Dice coefficients for each library. At each VBS, the match similarity on the dendrograms that included all members of the VSG was determined. All isolates included in this subset were defined as the dendrogram group. Within each dendrogram group, the total number of isolates, the number of non-VSG isolates, and the specificity (%) were determined. The sensitivity was 100% by definition since all VSG isolates were included in each dendrogram group. The specificity was defined according to the following relationship: (true negatives/[false positives + true negatives] × 100). False positives were non-VSG isolates located within the dendrogram group. True negatives were non-VSG isolates located outside of the dendrogram group. The dendrogram group with the highest specificity was designated the “optimized” VBS. In a similar manner, the optimized matching tolerance for the E. faecium library analyzed by MW was determined, and results were compared to the Rf analysis.
RESULTS
Analysis of E. faecalis gels.
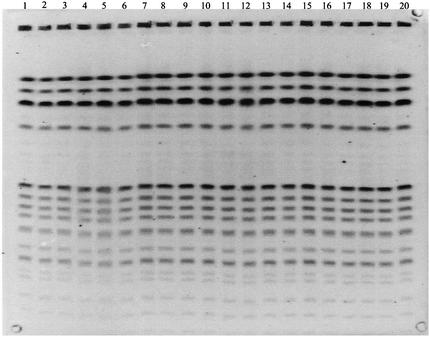
Two PFGE gels containing a total of 38 lanes with SmaI-digested E. faecalis OG1RF DNA (run on two different days with the same CHEF apparatus) were analyzed by using Phoretix software. Figure 1 shows one of the two gel images (gel 2) containing 20 of the 38 lanes. Visual inspection showed distortions in lanes 4, 5, and 6 of gel 2. No substantial lane distortions were noted in gel 1.
FIG. 1.
Gel 2: lanes containing SmaI-digested E. faecalis OG1RF DNA. Some lane-to-lane variations, or distortions, exist in gel 2. Distortions are pronounced in lanes 4 to 6.
Location of the standards on the gel.
Both gel 1 and gel 2 were analyzed five times by using Rf normalization of fragment positions. For each time, three different lanes were designated as the electrophoresis standards. These included (i) the first, middle, and last lanes; (ii) the second, middle, and last lanes;, (iii) the third, middle, and last lanes; (iv) the first, middle, and second-to-last lanes; and (v) the first, middle, and third-to-last lanes. The fragment patterns in the remaining 32 lanes in each gel were analyzed as though they were unique isolates to determine the minimum VBS that grouped the largest number of patterns together on the dendrogram. The maximum match similarity was defined as the highest similarity percentage at which all 32 identical fragment patterns matched together within the Phoretix software. The minimum VBS that yielded the maximum match similarity for all 32 lanes was determined for gel 1, gel 2, and gels 1 and 2 together. The range of minimum VBS for all standard lane positions was less for gel 1 (0.750 to 0.875) than for gel 2 (0.700 to 1.075), a result that was perhaps due to the distortions in the fragment positions of lanes 4 and 5 of gel 2. When both gels were matched together and all five sets of experimental lane positions were analyzed, the minimum VBS range was 0.750 to 1.025. The third, middle, and last lanes containing E. faecalis OG1RF DNA appeared to show the smallest minimum VBS (gel 1, 0.750; gel 2, 0.700), whereas the first, middle, and third-to-last lanes of gels 1 and 2 demonstrated the highest minimum VBS (gel 1, 0.875; gel 2, 1.075). It was not possible to judge whether differences that occurred between VBSs were statistically significant.
Age of agarose plug production.
The impact of using agarose plug preparations made on different days on the minimum VBS for both gels was determined. Lot 1 plugs were prepared on a different day than lot 2 plugs. The standard and nonstandard plug combinations used were as follows: lot 1 in both the standard and the nonstandard lanes; lot 1 in the standard lanes and lot 2 in the nonstandard lanes; lot 2 in both the standard lanes and nonstandard lanes; and lot 2 in the standard lanes with lot 1 in the nonstandard lanes. The lanes of each combination were matched together by Rf normalization with the range of minimum VBSs varying between 0.525 and 0.875% (gel 1) and between 0.650 and 1.2% (gel 2). The lowest minimum VBS for gel 1 (0.525%) was achieved by using lot 2 in the standard lanes and lot 1 in the nonstandard lanes. Conversely, the lowest minimum VBS for gel 2 (0.650%) was found by using lot 2 in both the standard and the nonstandard lanes. For all lot 1 and lot 2 standard and nonstandard lanes matched together for both gels, the minimum VBSs were 1.2% and 0.750%, respectively. When gels 1 and 2 were matched to duplicate images of themselves, the minimum VBSs for gels 1 and 2 were 0.975 and 1.0%, respectively.
MW versus Rf normalization.
The PFGE fragment position for each lane of E. faecalis OG1RF DNA in gels 1 and 2 was normalized by MW and Rf methods by using Phoretix software. After the automated band finding feature of the software was used, all of the DNA fragment positions of the 38 E. faecalis OG1RF isolates were confirmed visually to ensure that no artifacts were present on the gel prior to the normalization. During the MW and Rf normalization process, the first, middle, and last lanes of each gel were considered standard lanes and were excluded from further normalization analysis, leaving 32 nonstandard lanes for analysis. For normalization of the MW values, the MWs of fragments in the lanes not designated as standard were calculated by the software using the known MWs of E. faecalis OG1RF DNA fragments (13, 20) in the standard lane positions. The Phoretix software fitted a log curve between the E. faecalis OG1RF MW standard fragment values and used this curve to estimate the MWs of the fragments in the nonstandard lanes. The Rf values of fragments, calculated by the software, represented the migratory distance in pixels that fragments traveled relative to the length of each lane on a gel (Phoretix 1D Advanced manual). For Rf analysis, the middle of the loading well in each lane was standardized as the top of the lane. The software calculated Rf values by assigning Rf lines to each fragment position within the leftmost standard lane position. Once these Rf lines were assigned, linear interpolation of pixel position in the software generated Rf values for fragment positions in nonstandard lane positions with “0” for the top and “1” for the bottom of each lane.
Table 1 summarizes statistics calculated from the intrafragment MW values from all 11 fragments in the 32 nonstandard lanes of gels 1 and 2. The actual MWs are different from the calculated MW mean due to the variation associated with how well the log curve “fit” the known E. faecalis OG1RF MWs. The calculated MW range and variance generally decreased from fragments 1 to 11. CVs ranged between 0.0096 and 0.0230 for fragments 1 to 6 and fragments 10 to 11, but they were ≤0.0070 for fragments 7 to 9.
TABLE 1.
Intrafragment MW normalization analysis of 32 lanes containing E. faecalis OG1RF
| Fragment no. | Actual MW (kb)a | Calculated MW
|
|||
|---|---|---|---|---|---|
| Mean (kb) | Range (kb) | Variance (kb) | CV | ||
| 1 | 625 | 587 | 568-600 | 99.0 | 0.0170 |
| 2 | 370 | 425 | 412-442 | 47.0 | 0.0161 |
| 3 | 300 | 315 | 306-326 | 31.3 | 0.0177 |
| 4 | 240 | 190 | 181-197 | 19.1 | 0.0230 |
| 5 | 139 | 96 | 93-98 | 1.86 | 0.0142 |
| 6 | 112 | 90 | 88-91 | 0.738 | 0.0096 |
| 7 | 97 | 86 | 85-87 | 0.281 | 0.0062 |
| 8 | 85 | 84 | 83-84 | 0.257 | 0.0061 |
| 9 | 72 | 81 | 80-82 | 0.319 | 0.0070 |
| 10 | 50 | 78 | 77-80 | 0.749 | 0.0111 |
| 11 | 34 | 77 | 75-79 | 1.14 | 0.0139 |
Table 2 contains statistics calculated from the intrafragment Rf values from all fragments in the 32 nonstandard lanes. Averaging raw Rf values generated a single set of Rf mean values that were used in subsequent analyses. The observed Rf means and variances were much lower compared to the corresponding MW values due to different units of measure. However, for each fragment the unitless 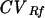 and CVMW values were close to each other and, when compared, were not found to be significantly different (P > 0.1 for fragments 1 to 6 and fragments 10 to 11, P = 0.07 for fragments 7 to 8, and P = 0.08 for fragment 9).
and CVMW values were close to each other and, when compared, were not found to be significantly different (P > 0.1 for fragments 1 to 6 and fragments 10 to 11, P = 0.07 for fragments 7 to 8, and P = 0.08 for fragment 9).
TABLE 2.
Intrafragment Rf normalization analysis of 32 lanes containing E. faecalis OG1RF
| Fragment no. | Calculated Rf values
|
|||
|---|---|---|---|---|
| Mean | Range | Variance | CV | |
| 1 | 0.176 | 0.165-0.190 | 0.000053 | 0.0412 |
| 2 | 0.222 | 0.208-0.236 | 0.000092 | 0.0431 |
| 3 | 0.267 | 0.251-0.283 | 0.000151 | 0.0460 |
| 4 | 0.354 | 0.337-0.372 | 0.000228 | 0.0427 |
| 5 | 0.556 | 0.532-0.585 | 0.000454 | 0.0383 |
| 6 | 0.596 | 0.571-0.626 | 0.000495 | 0.0373 |
| 7 | 0.631 | 0.603-0.662 | 0.000528 | 0.0364 |
| 8 | 0.662 | 0.635-0.696 | 0.000595 | 0.0369 |
| 9 | 0.710 | 0.680-0.744 | 0.000656 | 0.0361 |
| 10 | 0.769 | 0.737-0.807 | 0.000758 | 0.0358 |
| 11 | 0.810 | 0.779-0.848 | 0.000780 | 0.0345 |
Matching tolerance optimization for the E. faecium PFGE library.
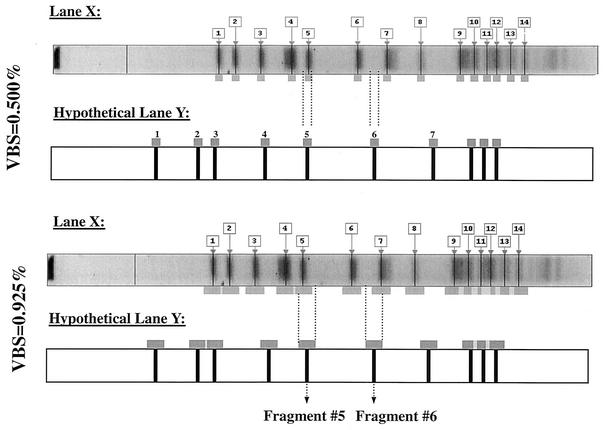
In order to build upon both the variation and MW versus Rf normalization studies, an applied analysis was undertaken to determine organism-specific optimal matching tolerance settings for both E. faecium and S. aureus PFGE libraries containing patterns of clinical isolates. Figure 2 illustrates how a matching tolerance, such as VBS, can impact fragment-to-fragment matching when the value of the VBS increases. At a VBS of 0.500%, fragment 5 of lane X matches fragment 5 of lane Y, but fragment 7 of lane X does not match fragment 6 of lane Y. At a VBS of 0.925%, fragment 7 of lane X does match fragment 6 of lane Y due to fragment overlap caused by increasing VBS.
FIG. 2.
Comparison of fragment-to-fragment matching at two vector box sizes of 0.500 and 0.925%. At a VBS of 0.500%, fragment 5 of lane X will match fragment 5 of lane Y, but fragment 7 of lane X does not match fragment 6 of lane Y. At a VBS of 0.925%, fragment 7 of lane X does match fragment 6 of lane Y due to fragment overlap caused by increasing VBS.
Four gels containing SmaI-digested DNA restriction patterns from 62 E. faecium isolates obtained from Project Intensive Care Antimicrobial Resistance Epidemiology (Project ICARE) (1, 6) (the E. faecium PFGE library) were analyzed by using Phoretix software. All fragment positions within the PFGE library were normalized by using both MW and Rf methods. The mean Rf values generated from the MW versus Rf study for both the E. faecium and S. aureus PFGE libraries were used as standards for Rf normalization of fragment position. A subset of 26 isolates of E. faecium was designated the VSG. The remaining 35 isolates on the four gels comprised the non-VSG. In order to determine how the 26 VSG isolates were grouped among the 35 non-VSG by the Phoretix software, UPGMA dendrograms of the 62 E. faecium isolates were generated from Dice coefficients for 11 different VBSs, including the manufacturer's recommended setting (0.500%) and selected values above and below 0.500%.
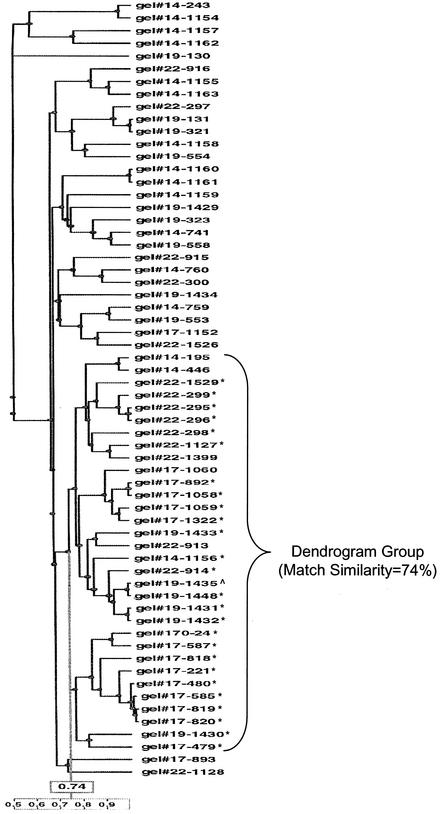
For the E. faecium PFGE library, 5 of the 11 VBSs, representing the range of matching tolerance settings evaluated, are shown in Table 3. For each VBS, total isolates and non-VSG isolates within the dendrogram group decreased until the VBS of 0.925% and then increased with the VBS of 1.25%. Sensitivity was 100% by definition across all VBSs (all VSG isolates were included in the dendrogram group). The specificity of the dendrogram group increased until the 0.925% VBS and then decreased at 1.25%. Of all VBSs tested, the dendrogram group at 0.925% yielded the highest match similarity (74%), the highest specificity, while maintaining 100% sensitivity, and the lowest number of dissimilar isolates. Therefore, the optimal VBS for the E. faecium PFGE library was 0.925%. Figure 3 shows the dendrogram at a VBS of 0.925%.
TABLE 3.
Range of VBSs tested to determine optimum matching tolerance for E. faecium isolates
| Parameter | Result at VBS of:
|
||||
|---|---|---|---|---|---|
| 0.125 | 0.500 | 0.750 | 0.925a | 1.250 | |
| Match similarity percentage defining dendrogram group | 7 | 44 | 66 | 74 | 73 |
| Within dendrogram group | |||||
| Total no. of isolatesb | 59 | 55 | 37 | 31 | 43 |
| No. of VSG isolatesc | 26 | 26 | 26 | 26 | 26 |
| No. of dissimilar isolates (non-VSG) | 33 | 29 | 11 | 5 | 17 |
| % Sensitivityc | 100 | 100 | 100 | 100 | 100 |
| % Specificityd | 6 | 17 | 31 | 86 | 49 |
Optimum VBS setting for E. faecium PFGE library.
Excludes reference E. faecium isolate.
By definition, the dendrogram group included all VSG isolates.
Specificity = [true negatives/(false positives + true negatives) × 100], where true negatives are non-VSG isolates located outside of the dendrogram group.
FIG. 3.
UPGMA dendrogram generated from Dice coefficients that contains E. faecium PFGE library isolates compared at a VBS of 0.925%. Isolates marked with an asterisk indicate those in the VSG. The single isolate indicated by a “^” symbol (designated 1435 in the middle of the dendrogram group) is the reference (index) isolate. Of 31 isolates in the dendrogram group (marked with a bracket), 26 were visually similar, and 5 were dissimilar. The match similarity for the dendrogram group was 74%.
Matching tolerance optimization for the S. aureus PFGE library.
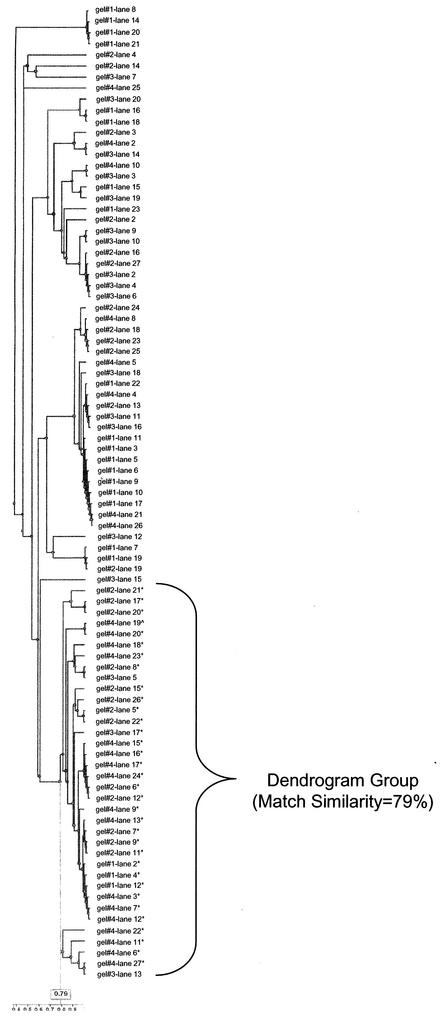
Four gels containing SmaI-digested DNA restriction patterns from 89 S. aureus isolates obtained from the organism collection at the Centers for Disease Control and Prevention (the S. aureus PFGE library) were analyzed by using Phoretix software. All fragment positions within the S. aureus PFGE library were normalized by using the Rf method. A group of 33 S. aureus isolates were designated the VSG. UPGMA dendrograms were generated from Dice coefficients by using 10 different VBSs that included the manufacturer's recommended setting (0.500%). Six of the ten VBSs, representing the range of the results, are shown in Table 4. Within the dendrogram group, total and non-VSG (n = 55) isolates decreased while the specificity of the dendrogram group increased until 0.300% VBS and then remained unchanged thereafter. Although the specificity reached its peak at 0.300% VBS, the match similarity continued to increase throughout the remaining VBSs tested. Given that the 0.300% VBS had only a 57% match similarity, 0.500% was considered to be the optimal VBS for the S. aureus PFGE library based upon its higher 79% match similarity and the desire to keep the vector box as small as possible. The 79, 83, and 84% match similarities for the dendrogram group at VBSs of 0.500, 0.925, and 1.25%, respectively, were higher than the match similarity for the optimal VBS setting of the E. faecium PFGE library (74%). The dendrogram group shown in Fig. 4 was similar to the dendrogram group found for all VBSs of ≥0.300%.
TABLE 4.
Range of VBSs tested to determine optimum matching tolerance for S. aureus isolates
| Parameter | Result at VBS of:
|
|||||
|---|---|---|---|---|---|---|
| 0.125 | 0.250 | 0.300 | 0.500a | 0.925 | 1.25 | |
| Match similarity percentage defining dendrogram group | 11 | 40 | 57 | 79 | 83 | 84 |
| Within dendrogram group | ||||||
| Total no. of isolatesb | 74 | 37 | 35 | 35 | 35 | 35 |
| No. of VSG isolatesc | 33 | 33 | 33 | 33 | 33 | 33 |
| No. of dissimilar isolates (non-VSG) | 41 | 4 | 2 | 2 | 2 | 2 |
| % Sensitivityc | 100 | 100 | 100 | 100 | 100 | 100 |
| % Specificityd | 25 | 93 | 96 | 96 | 96 | 96 |
Optimum VBS setting for S. aureus PFGE library.
Excludes reference S. aureus isolate.
By definition, the dendrogram group included all VSG isolates.
Specificity = [true negatives/(false positives + true negatives) × 100], where true negatives are non-VSG isolates located outside of the dendrogram group.
FIG. 4.
UPGMA dendrogram generated from Dice coefficients that contains S. aureus PFGE library isolates compared at a VBS of 0.500%. Isolates marked with an asterisk indicate those in the visually similar group (VSG). The single isolate indicated by a “^” symbol (designated lane 19 toward the top of the dendrogram group) is the reference (index) isolate. Of 35 isolates in the dendrogram group (marked with a bracket), 33 were visually similar, and 2 were dissimilar. The match similarity for the dendrogram group was 79%.
Comparison of VBS and vector percentage.
Both the Rf and MW methods were used to normalize the fragment positions in the E. faecium PFGE library. Thus, the relative impact was assessed of the two normalizing methods on the optimization of matching tolerance settings. In Phoretix, the MW matching tolerance algorithm was defined as the vector percentage, rather than the VBS (the matching tolerance algorithm for Rf). The approximate relationship between the vector percentage (MW) and VBS (Rf) for this particular analysis and the database was as follows: 1% of the vector percentage = 0.025% of a vector box. In addition to optimizing the Rf matching tolerance (VBS) of the four-gel PFGE library of 62 E. faecium isolates as discussed previously, the library was also optimized for the MW matching tolerance of the vector percentage. Eleven vector percentage settings were investigated. The optimized vector percentage was determined from the dendrogram group that had the highest specificity. The comparison between VBS and vector percentage was based on the total number of isolates, the number of dissimilar isolates, and the specificity (%) within the optimized dendrogram group.
For MW, the optimal vector percentage of the E. faecium library was 18% (data not shown). Upon comparison of the optimal VBS (0.925%) and the vector percentage (18%), the dendrogram group normalized by Rf (VBS) as opposed to MW (vector percentage) contained fewer total (31 versus 43) and dissimilar (5 versus 17) isolates while it maintained a higher match similarity (74% versus 58%) and specificity (86% versus 51%).
DISCUSSION
Commercial computer software packages can aid in strain identification, but it is imperative that investigators optimize the software parameters before any gels are analyzed. In the present study, the position of the standards on the gel and the age of the agarose gel plugs affected the software setting (VBS) required to identify 32 identical E. faecalis OG1RF isolates as the same PFGE type. This analysis was performed specifically to determine the matching tolerance levels that would be used in subsequent analyses. A previous study using a different gel analysis software identified the first, middle, and last lanes of gels 1 and 2 as optimal for the standards (E. C. Felicione, C. D. Steward, J. E. McGowan, Jr., and F. C. Tenover, Abstr. Prog. Int. Conf. Emerg. Infect. Dis., abstr. 4.7, 1998). In the present study, standards placed in the third, middle, and last lanes of the two gels appeared to be the preferred standard lane placement, although it was not possible to judge whether differences noted in VBSs were statistically significant. For subsequent analysis, we placed the standards in the first, middle, and last lane positions because our data indicated that clinical isolates placed outside of the standard lanes led to unreliable fragment position normalization (data not shown).
No pattern of gel plug lot-to-lot variation was detected in gels 1 and 2; thus, plug lot did not contribute to the intragel variation found within either gel. However, for intergel variation, lot 2 lanes yielded a much lower minimum VBS than did lot 1 lanes. This phenomenon could be related to the fact that lot 1 lanes contained more lane distortions. Baseline VBSs required to match gels 1 and 2 to their duplicate image at the highest match similarity were higher than some of the VBSs produced in lot-specific, intergel analyses. The inability of software-based algorithms to match identical isolates at 100% is an indication that algorithms of mathematic rigidity cannot yet replace visual interpretation of PFGE fragment patterns. Although software programs aid in the analysis of PFGE patterns across multiple gels, user intervention is still required during the steps of computerized analysis to produce accurate results (7, 16). Typing techniques, such as amplified fragment length polymorphism, that use automated sequence equipment for gel reading and analysis potentially have less variability in band finding and greater run-to-run reproducibility than PFGE.
The generation of mean Rf values provided a single set of “gold standard” values that could be entered manually into all E. faecalis OG1RF standard Rf values to normalize fragment position and minimize intra- and intergel variation within a PFGE library. This was how the E. faecium and S. aureus PFGE libraries were analyzed in the present study. Mean Rf values generated from a PFGE library capture dynamic variations in fragment position, whereas predetermined MW standards (with known fragment MWs) are not representative of the unique fragment pattern variations that reside within a particular PFGE library. However, generating the mean Rf values required the additional effort of running the same strain 38 times on two gels to determine minimum matching tolerance settings. An alternate method of accomplishing the same task would be to place an additional standard isolate (e.g., E. faecalis OG1RF in the present study) in a nonstandard lane on each gel of a PFGE library (11).
CV was a better indicator of variability between the same fragment on different lanes for MW and Rf because it measured the variability relative to the mean (for each fragment, the mean was higher for MW than for Rf due to the units of measure involved). Comparisons of CVMW and 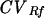 for each fragment indicated that the same variability existed no matter what normalization method was used. However, P values for fragments 7 to 9 were smaller and closer to the significance level (0.05) than P values for the other fragments. Interestingly, fragments 7 to 9 were closer to the actual MW log curve used to estimate the MWs of all of the other fragments on the gel. This suggests that the statistical variation between the estimated MWs and the actual MWs decreases as the estimated MWs approach the real MW values used to generate the log curve.
for each fragment indicated that the same variability existed no matter what normalization method was used. However, P values for fragments 7 to 9 were smaller and closer to the significance level (0.05) than P values for the other fragments. Interestingly, fragments 7 to 9 were closer to the actual MW log curve used to estimate the MWs of all of the other fragments on the gel. This suggests that the statistical variation between the estimated MWs and the actual MWs decreases as the estimated MWs approach the real MW values used to generate the log curve.
Even though the between-lane variability was basically the same for both Rf and MW normalized values, MW normalization showed less specificity than did Rf normalization for the E. faecium PFGE library analyzed in the present study. Thus, software users should be aware of the differences between Rf and MW normalization before performing matching operations on any given PFGE library. In our study at optimized matching tolerance settings, the results favored the use of Rf over MW normalization methods for our E. faecium PFGE library analyzed with Phoretix software. Comparison of MW and Rf was possible by using one software package in the present study because Phoretix software allows the user a choice of normalization method (MW or Rf), although different algorithms are used to calculate matching tolerances for each normalization method. Other software packages typically use either the MW or the Rf normalization method. Phoretix is similar to other software packages in most other respects, offering similar choices of regression method, similarity coefficient (e.g., Dice, Jaccard, and Pearson), and dendrogram calculation method (e.g., UPGMA and neighbor joining).
VBS matching tolerance algorithms remain constant for all fragments at any given VBS. This can result in unnecessary fragment overlap when the VBS is not optimized prior to analyzing a PFGE library. In the current study, suboptimal matching tolerance settings decreased the specificity with which visually similar isolates were grouped together. The manufacturer's recommended setting of 0.500% decreased the specificity of the organism clustering for the E. faecium PFGE library but yielded acceptable results for the S. aureus PFGE library. The degree of alteration of the matching tolerance from its recommended or default setting is dependent on the magnitude of intra- and intergel variation due to organism-specific fragment or lane distortions. Thus, optimizing the VBS setting should be performed before subsequent matching analysis takes place. The E. faecium PFGE library contained a high degree of fragment pattern variation and made computer-assisted analysis more difficult compared to S. aureus. Heterogeneity of restriction fragment patterns in E. faecium has been discussed by other researchers (10, 12) and may have played a role in the apparent lower match similarity of the dendrogram group in the E. faecium PFGE library compared to the S. aureus library. However, the influence of other factors on the match similarities, such as the variety of patterns in each library and the choice of reference pattern for the VSG, cannot be determined in the present study.
Our data showed that, when optimized, a gel analysis software package could compensate for the various sources of intra- and intergel variation in PFGE libraries. However, in agreement with the findings of Cardinali et al. (3), there is a clear need for further development of software packages and algorithms used for gel analysis. Improvements, combined with optimization of the software algorithms prior to use, will further facilitate fragment pattern comparisons on multiple gels.
Acknowledgments
We thank Elize Felicione Lundeen for producing the E. faecalis and E. faecium gels and Molly Kellum for producing the S. aureus gels. We thank the laboratories that submitted enterococci to Project ICARE. We thank Andy Borthwick, Stuart Cullen, and Ashley Frieze for Phoretix technical support.
Phase IV of Project ICARE is supported in part by unrestricted grants to the Rollins School of Public Health of Emory University by Astra-Zeneca Pharmaceuticals, Wilmington, Del.; Bayer Corporation, Pharmaceuticals Division, West Haven, Conn.; Cubist Pharmaceuticals, Inc., Lexington, Mass.; Elan Pharmaceuticals, San Diego, Calif.; Pharmacia Corporation, Peapeck, N.J.; and Roche Laboratories, Nutley, N.J.
The use of trade names is for identification purposes only and does not constitute endorsement by the Public Health Service or the U.S. Department of Health and Human Services.
REFERENCES
- 1.Archibald, L., L. Phillips, D. Monnet, J. E. McGowan, Jr., F. Tenover, and R. Gaynes. 1997. Antimicrobial resistance in isolates from inpatients and outpatients in the United States: increasing importance of the intensive care unit. Clin. Infect. Dis. 24:211-215. [DOI] [PubMed] [Google Scholar]
- 2.Birren, B. W., and E. Lai. 1993. Pulsed field gel electrophoresis: a practical guide. Academic Press, Inc., San Diego, Calif.
- 3.Cardinali, G., A. Martini, R. Preziosi, F. Bistoni, and F. Baldelli. 2002. Multicenter comparison of three different analytical systems for evaluation of DNA banding patterns from Cryptococcus neoformans. J. Clin. Microbiol. 40:2095-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aries de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sá-Leão, I. Santos Sanches, J. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Facklam, R. R., D. F. Sahm, and L. M. Teixeira. 1999. Enterococcus, p. 297-305. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology. American Society for Microbiology, Washington, D.C.
- 6.Fridkin, S. K., C. D. Steward, J. R. Edwards, E. R. Pryor, J. E. McGowan, Jr., L. K. Archibald, R. P. Gaynes, and F. C. Tenover. 1999. Surveillance of antimicrobial use and antimicrobial resistance in United States hospitals: project ICARE phase 2. Clin. Infect. Dis. 29:245-252. [DOI] [PubMed] [Google Scholar]
- 7.Gerner-Smidt, P., L. M. Graves, S. Hunter, and B. Swaminathan. 1998. Computerized analysis of restriction fragment length polymorphism patterns: comparative evaluation of two commercial software packages. J. Clin. Microbiol. 36:1318-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordillo, M. E., K. V. Singh, and B. E. Murray. 1993. Comparison of ribotyping and pulsed-field gel electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. J. Clin. Microbiol. 31:1570-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloos, W. E., and T. L. Bannerman. 1999. Staphylococcus and Micrococcus, p. 264-282. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology. American Society for Microbiology, Washington, D.C.
- 10.Mato, R., H. de Lencastre, R. B. Roberts, and A. Tomasz. 1996. Multiplicity of genetic backgrounds among vancomycin-resistant Enterococcus faecium isolates recovered from an outbreak in a New York City hospital. Microb. Drug Resist. 2:309-317. [DOI] [PubMed] [Google Scholar]
- 11.Michaud, S., S. Menard, C. Gaudreau, and R. D. Arbeit. 2001. Comparison of SmaI-defined genotypes of Campylobacter jejuni examined by KpnI: a population-based study. J. Med. Microbiol. 50:1075-1081. [DOI] [PubMed] [Google Scholar]
- 12.Morrison, D., N. Woodford, S. P. Barrett, P. Sisson, and B. D. Cookson. 1999. DNA banding pattern polymorphism in vancomycin-resistant Enterococcus faecium and criteria for defining strains. J. Clin. Microbiol. 37:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray, B. F., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.NCCLS. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 5th ed. NCCLS document M7-A5. NCCLS, Wayne, Pa.
- 15.NCCLS. 2002. Performance standards for antimicrobial susceptibility testing; 12th informational supplement. NCCLS document M100-S12. NCCLS, Wayne, Pa.
- 16.Rementeria, A., L. Gallego, G. Quindós, and J. Garaizar. 2001. Comparative evaluation of three commercial software packages for analysis of DNA polymorphism patterns. Clin. Microbiol. Infect. 7:331-336. [DOI] [PubMed] [Google Scholar]
- 17.Snedecor, G. H., and W. A. Cochran. 1989. Statistical methods. Iowa State University Press, Ames.
- 18.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover, F. C., R. D. Arbeit, and R. V. Goering. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect. Control Hosp. Epidemiol. 18:426-439. [DOI] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]