Abstract
Recently, we have described the first human case of AdoHcyase (S-adenosylhomocysteine hydrolase) deficiency. Two point mutations in the AdoHcyase gene, the missense mutation p.Y143C (AdoHcyase in which Tyr143 is replaced by cysteine) and the truncation mutation p.W112stop (AdoHcyase in which Trp112 is replaced by opal stop codon) were identified [Barić, Fumić, Glenn, Ćuk, Schulze, Finkelstein, James, Mejaški-Bošnjak, Pažanin, Pogribny et al. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4234–4239]. To elucidate the molecular and catalytic properties of AdoHcyase, we have made recombinant wild-type and mutant p.Y143C (AdoHcyase in which Tyr143 is replaced by cysteine) enzymes for a comparative analysis. The catalytic rates of p.Y143C protein in the directions of S-adenosylhomocysteine synthesis or hydrolysis are decreased from 65% to 75%. Further, the oxidation states of coenzyme NAD differ between mutant and wild-type protein, with an increased NADH accumulation in the mutant p.Y143C enzyme of 88% NADH (wild-type contains 18% NADH). Quantitative binding of NAD is not affected. Native polyacrylamide gel electrophoresis showed, that mutant p.Y143C subunits are able to form the tetrameric complex as is the wild-type enzyme. CD analysis showed that the p.Y143C mutation renders the recombinant protein thermosensitive, with an unfolding temperature significantly reduced by 7 °C compared with wild-type protein. Change of Glu115 to lysine in wild-type protein causes a change in thermosensitivity almost identical with that found in the p.Y143C enzyme, indicating that the thermosensitivity is due to a missing hydrogen bond between Tyr143 and Glu115. We emphasize involvement of this particular hydrogen bond for subunit folding and/or holoenyzme stability. In summary, a single mutation in the AdoHcyase affecting both the oxidation state of bound co-factor NAD and enzyme stability is present in a human with AdoHcyase deficiency.
Keywords: CD; 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB); hydrogen bond; in vitro mutagenesis; S-adenosylhomocysteine hydrolase (AdoHcyase) deficiency; thermosensitivity
Abbreviations: AdoHcy, S-adenosyl-L-homocysteine; AdoHcyase, S-adenosylhomocysteine hydrolase; AdoMet, S-adenosylmethionine; DLS, dynamic light scattering; DTNB, 5′,5-dithiobis-(2-nitrobenzoic acid); DTT, dithiothreitol; Hcy, homocysteine; IPTG, isopropyl β-D-thiogalactoside; NAD, NAD+ or NADH; Ni-NTA, Ni2+-nitrilotriacetate; p.E115L, AdoHcyase in which Glu115 is replaced by leucine; p.W112stop, AdoHcyase in which Trp112 is replaced by opal stop codon; p.Y143C, AdoHcyase in which Tyr143 is replaced by cysteine
INTRODUCTION
AdoHcyase (S-adenosylhomocysteine hydrolase; EC 3.3.1.1) catalyses the hydrolysis of AdoHcy (S-adenosyl-L-homocysteine) to adenosine and Hcy (homocysteine) [1]. AdoHcy is formed as a product of AdoMet (S-adenosylmethionine) through transmethylation reactions [2]. AdoMet is the major methyl donor for delivery of methyl groups to DNA, RNA, proteins and cellular metabolites in eukaryotes [3], and AdoHcy hydrolysis is the only source of Hcy (homocysteine) in mammals. The reaction is reversible, with an equilibrium favouring AdoHcy formation, but proceeds under physiological conditions in the hydrolytic direction due to the rapid removal of both adenosine and Hcy. Adenosine is removed by adenosine deaminase and adenosine kinase, and Hcy is used for the synthesis of cystathionine and the regeneration of methionine [4]. AdoMet/AdoHcy turnover is believed to play a critical role in methionine metabolism and the regulation of biological methylation processes. The importance of AdoHcyase for mammalian survival is suggested by the fact that a chromosomal deletion that includes the gene encoding AdoHcyase causes embryo lethality in mice [5]. Elevated Hcy levels have been reported as a risk factor for dementia and Alzheimer's disease [6], and are discussed as a possible risk marker for vascular disease [7,8]. Human AdoHcyase deficiency results in severe biochemical abnormalities, including plasma elevations of as much as 150-fold in AdoHcy, 30-fold in AdoMet, and 12-fold in methionine, with significant decreases in the AdoMet/AdoHcy ratio [9,10]. As shown previously, AdoHcy is a strong competitive inhibitor of many AdoMet-dependent methyltransferases [4]. Therefore aberrant methylation processes resulting from the significant imbalance in the AdoMet/AdoHcy ratio might be the reason for other abnormalities, such as myopathy and retarded psychomotor development [9].
Human AdoHcyase is a tetramer consisting of chemically identical and functionally equivalent subunits [11]. Each subunit is built of 432 amino acid residues and contains one molecule of tightly bound NAD (NAD+ or NADH) [12]. The proposed mechanism for reversible hydrolysis of AdoHcy catalysed by AdoHcyase from bovine liver [12,13] involves initial oxidation by the cofactor NAD+ of AdoHcy to the 3′-keto derivative, followed by β-elimination of L-Hcy resulting in 3′-keto-4′,5′-didehydro-adenosine. Michael addition of water to the 5′-position of the tightly bound intermediate and reduction of the 3′-keto by NADH then forms the final product, adenosine.
The three-dimensional structures of rat [14] and human [15,16] AdoHcyases have been resolved and mutational studies have allowed detailed insights into the processes important for catalytic activity [17–22]. Amino acids His54, Asp130, Glu155, Lys185, Asp189 and Asn190 have been pinpointed as crucial for the catalytic reaction of rat AdoHcyase, and a catalytic mechanism at single amino acid resolution has been proposed [22]. The geometry of the active site is also important for the catalytic mechanism of AdoHcyase and can be described as a substrate-free open structure [14] and as a substrate-bound closed structure [15,16,20,23]. During the catalytic reaction a 17° rigid body movement of the catalytic domain occurs upon substrate binding [20]. Thus the conformational change is considered as an important catalytic capability of the enzyme [21].
In the present study, we elucidate the molecular and catalytic properties of recombinant mutant AdoHcyase. We investigated the effects of mutations of Trp112 and Tyr143 identified in our previous study [23a] on enzymatic activity of AdoHcyase, measured the oxidation state of bound cofactor NAD, and used CD to determine structural aspects such as protein unfolding and stability during thermal induction. Further, we conducted site-directed mutagenesis studies to investigate the importance of the hydrogen-bonding of Tyr143 with Glu155 in wild-type AdoHcyase. For this purpose we mutated Glu155 to leucine and investigated the effects on enyzme activity, subunit folding and protein stability of AdoHcyase.
EXPERIMENTAL
Cloning of recombinant wild-type and mutant forms of AdoHcyase for expression in Escherichia coli
Mutational analysis of the human AdoHcyase gene was performed as described previously [9].
A combination of PCR-based cloning and oligonucleotide-directed mutagenesis was used to prepare cDNAs encoding wild-type and mutated forms of AdoHcyase. Oligonucleotides (listed in Table 1) were purchased from Invitrogen (San Diego, CA, U.S.A.).
Table 1. Oligonucleotides used for PCR-based cloning and site-directed mutagenesis.
The codons changed are underlined, and the nucleotides changed are shown in boldface. The cDNA sizes of amplification products are for wild-type 1337 bp, for p.Y143C 949 bp and for p.W112stop 367 bp.
| Oligonucleotide (5′–3′) | |||
|---|---|---|---|
| Construct | Forward | Reverse | Codon change |
| Wild-type | CGCCGGTACCTCTGACAAACTGCCCT | GCATCTAGAGGGTGAAACGCAGA | − |
| p.Y143C | CGCCGGTACCTCTGACAAACTGCCCT | ATGTTCACCTTCTCCACGGC | [8] |
| p.E115L | − | AGGGTCTGCAGAATGCACCACAGGT | GAG→CTG |
| p.W112stop | CGCCGGTACCTCTGACAAACTGCCCT | GCTGGATCCATCACAGGTACT | TGG→TGA |
The complete coding region of the wild-type human AdoHcyase gene was amplified from a full-length cDNA clone (IRALp962A0727Q2) obtained from RZPD (Berlin, Germany) and subcloned into pBluescriptSK+ (Stratagene, Heidelberg, Germany).
Mutant p.Y143C cDNA was partially amplified as described previously [9] and was used to replace the corresponding region in the subcloned wild-type cDNA. The truncation mutation p.W112stop was generated by PCR using wild-type cDNA as template and subcloned as above. Expression vectors for each of the cDNAs were prepared using vector pET32b (Novagen–Merck Biosciences, Nottingham, U.K.).
Wild-type expression vector was used for site-directed mutagenesis to generate the vector harbouring the p.E115L (AdoHcyase in which Glu115 is replaced by leucine) mutation. Following the PCR-based approach of Shenoy and Visweswariah [24], a single oligonucleotide (Table 1) was used to convert a glutamic acid residue into a leucine residue. Sequence analysis was performed for all expression vector constructs.
Overexpression and purification of wild-type and mutant forms of AdoHcyase
E. coli BL21 (DE3) RIL carrying the expression vectors containing the respective wild-type and mutated cDNAs was grown in 1 litre of 2YT medium [1.6% (w/v) tryptone, 1% (w/v) yeast extract and 0.5% (w/v) NaCl] containing 100 μg/ml of ampicillin and 34 μg/ml chloramphenicol at 37 °C. When the cell turbidity measured at 600 nm reached an attenuance of 0.5, IPTG (isopropyl β-D-thiogalactoside) was added to a final concentration of 0.5 mM. Culture was continued for an appropriate time at various temperatures depending on the expression plasmid (16 h at 30 °C for wild-type and p.W112stop; 40 h at 18 °C for p.Y143C; and 18 h at 29 °C for p.E115L).
Cells were harvested by centrifugation at 4500 g for 30 min, suspended in 20 ml of buffer A [50 mM potassium phosphate buffer (pH 7.2) and 1 mM EDTA] and lysed by treatment with hen's-egg white lysozyme (1 mg/ml) for 30 min at 4 °C, followed by freezing and thawing. The mixture was made up to 40 ml with the same buffer and subjected to a brief sonication. Cell-free extract was prepared by centrifuging the mixture at 20000 g for 1 h. To remove nucleic acids, each extract was passed through a column (1 cm×10 cm) of DEAE-cellulose (Bio-Rad Laboratories, Munich, Germany) equilibrated and eluted with buffer A. The effluent was fractionated with ammonium sulfate, and the precipitate obtained between 35 and 60% saturation was dissolved in 20 ml of potassium phosphate buffer (pH 7.2) 300 mM NaCl and 10 mM imidazole. After removal of insoluble material by centrifugation, the enzyme solution was subjected to Ni-NTA (Ni2+-nitrilotriacetate) affinity chromatography. Namely, 2–3 ml of a saturated Ni-NTA agarose slurry (Bio-Rad Laboratories) was added to the enzyme solution and incubated for 30 min at 4 °C under constant stirring. Each mixture was then poured into a chromatography column, allowed to settle and washed with 50 ml of a buffer containing 50 mM potassium phosphate buffer (pH 7.2), 300 mM NaCl and 20 mM imidazole. Bound enzyme was eluted with 50 mM potassium phosphate buffer (pH 7.2), 300 mM NaCl and 300 mM imidazole. Eluates from separate protein extractions were analysed for AdoHcyase activity and desalted with buffer A using Nap-10 columns (GE Healthcare, Vienna, Austria) containing Sephadex G-25.
Aditionally, purified AdoHcyase protein was digested with thrombin (1 unit/mg of protein; Sigma–Aldrich, St. Louis, MO, U.S.A.) to remove most of the fusion peptide containing specific tags used for protein purification. Cleaved or uncleaved AdoHcyase protein was purified further using gel-filtration chromatography in buffer B [50 mM potassium phosphate buffer (pH 7.2) and 150 mM NaCl] and on a BIO-SIL SEC 250-5 column (Bio-Rad Laboratories).
PAGE
SDS/PAGE was carried out according to the method of Laemmli [25] using a 10% (w/v) polyacrylamide gel. Samples were treated with 1% SDS and 5% (v/v) 2-mercaptoethanol at 100 °C for 5 min before electrophoresis in a vertical Mini Gel system (Bio-Rad Laboratories). Proteins were stained with Coomassie Brilliant Blue R250 (GE Healthcare).
Additionally, purity and electrophoretic behaviour of the recombinant AdoHcyase protein was analysed using native PAGE, consisting of a 7% (w/v) polyacrylamide separating gel with a 5% (w/v) polyacrylamide stacking gel. Electrophoresis was performed in a buffer containing 25 mM Tris/HCl and 192 mM glycine (pH 8.3). Staining was performed as described above for SDS/PAGE.
The molecular mass of native AdoHcyase protein was estimated by 5–10% native PAGE using a non-denatured protein molecular-mass-marker kit (Sigma–Aldrich). The retardation coefficient for each protein was determined and plotted against the logarithm of the molecular mass of each protein.
CD analysis and DLS (dynamic light scattering)
CD measurements were performed on a Jasco J-715 spectro-polarimeter using a 0.02 cm (wild-type and p.Y143C) and 0.1 cm (p.E115L) water-jacketted cylindrical cell, thermostatically controlled by an external computer-controlled water bath. The far-UV spectra were recorded at 20 °C from 185–260 nm as an average of three scans with the following parameters: step resolution, 0.2 nm; speed, 50 nm/min; response, 2 s; and bandwidth, 1 nm. Thermal denaturation data were recorded in the temperature range 20–90 °C at 208 nm by a step scan procedure: heating rate, 1 °C/min; response, 1 s; recorded interval, 0.2 °C. The protein concentrations used were 0.73 mg/ml for the wild-type protein, and 0.55 and 0.1 mg/ml for the p.Y143C and p.E115L mutants respectively. The thermal denaturation curves were fitted with a sigmoidal function, and Tm (‘melting’ temperature) was determined from the point of inflection using the Microcal™ Origin™ verision 5.0 program (Microcal Software, Northampton, MA, U.S.A.). Results were expressed as the mean residue elliplicity at a given wavelength. Analysis of protein secondary structure from CD spectra was performed using the CDSSTR program included at the DICHROWEB site (http://public-1.cryst.bbk.ac.uk/cdweb/html) [27,28].
DLS measurements were performed on DynaPro (Wyatt Technology, Santa Barbara, CA, U.S.A.) using 45 μl cell, at 4, 25, 35 and 45 °C. Ten acquisitions were included in each measurement. The protein concentrations used for measurements were 3.20 mg/ml for wild-type, and 0.55 and 0.77 mg/ml for the p.Y143C and p.E115L proteins respectively.
Besides the CD analysis we performed heat inactivation experiments with wild-type and mutant proteins p.E115L and p.Y143C. Therefore we incubated 5 μg of recombinant protein for 15 min at various temperatures ranging from 37–57 °C in buffer A. Following heat inactivation, enzymatic activity was determined as described below.
Enzymatic assays
AdoHcyase activity in purified enzyme preparations was assayed as described by Takata et al. [21].
The reaction mixture (0.5 ml of buffer A) contained 1–2 μg of recombinant protein and an excess amount (>2.0 units) of calf intestine adenosine deaminase (Roche, Basel, Switzerland). The reaction for specific determination of AdoHcyase activity was started by the addition of various amounts of a 1 mM stock solution of AdoHcy (Sigma–Aldrich). The formation of adenosine and subsequently inosine was measured at 265 nm over a period of 30 min. Data were the average of two determinations, and the Km and enzyme activity values were obtained by directly fitting the data into the Michaelis–Menten equation using a Lineweaver–Burk linearization.
The synthetic activity of the purified AdoHcyase was determined in duplicate by the rate of disappearance of adenosine under the following conditions. Recombinant protein was incubated at 25 °C with 100 μM adenosine and 50 mM D/L-Hcy in 0.1 ml of buffer A. At an appropriate time, the reaction was terminated by heat denaturation at 75 °C for 15 min. The remaining adenosine was determined by the coupled assay as described above for the hydrolytic activity measurement with the exclusion of recombinant AdoHcyase. In addition, the amount of synthesized AdoHcy could be determined by using identical conditions as described above for AdoHcyase hydrolytic activity.
Effects of thiol-reducing or thiol-modifying compounds [DTT (dithiothreitol) or DTNB (5′,5-dithiobis-(2-nitrobenzoic acid); Sigma–Aldrich] on enzymatic activity were monitored. Recombinant AdoHcyases were incubated with various concentrations of either DTT (10–100 mM) or DTNB (10–50 μM), or a combination of both over different incubation periods (30–60 min).
Determination of AdoHcyase-bound NAD+ and NADH
Quantification of enzyme-bound NAD+ and NADH was performed in duplicate using a fluorescence technique described by Hohman et al. [26]. Nucleosides were released from 200 μg of recombinant enzyme by the addition of 2.5 vol. of 97% (v/v) ethanol and centrifugation for 5 min at 4 °C. The precipitate was washed with 200 μl of 97% (v/v) ethanol and centrifuged again. Supernatants were pooled and freeze-dried following resuspension in 200 μl of water and 300 μl of 0.1 M sodium pyrophosphate buffer (pH 8.8) containing 0.5% semicarbazide. NADH was measured directly, whereas NAD+ was first converted into NADH by adding 10 μl of a 1% solution of bakers' yeast alcohol dehydrogenase and 20 μl of 97% (v/v) ethanol. Detection of NADH was accomplished by excitation at 340 nm and measurement of emission at 460 nm. The calibration curve was constructed by measuring the fluorescence of known quantities of NADH under the same conditions.
Bioinformatics
We used software applications nnPredict (Dr Donald Kneller, Department of Cellular and Molecular Pharmacology, University of California, San Francisco, CA, U.S.A.) for secondary structure prediction. Model building was performed with the SWISS-MODEL server [29] by using the structure of human AdoHcyase as template (PDB 1A7A) [14]. Deepview/Swiss-PdbViewer [30] was used for in silico mutation and three-dimensional structural analysis of AdoHcyase protein (http://www.expasy.org/spdbv/). PeptideMass (http://www.expasy.ch/tools) was used for molecular mass prediction of recombinant AdoHcyase protein.
RESULTS
Purification and characterization of recombinant wild-type and mutant AdoHcyase
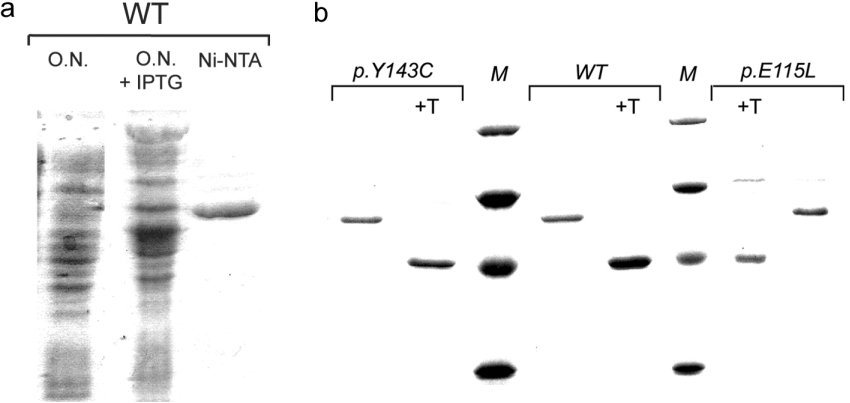
The cell-free extract prepared from E. coli transformed with the vector p32AHHwt and grown in the presence of IPTG exhibited a high level of AdoHcyase activity (results not shown). SDS/PAGE showed induction of a protein with the electrophoretic behaviour expected for recombinant AdoHcyase (Figure 1a). Compared with the human serum protein subunit, recombinant wild-type subunit has an increased molecular mass (63.9 kDa) due to the additional 152 amino acid residues contributed by the plasmid expression vector. The recombinant protein is reactive with anti-AdoHcyase antibody (a gift from Dr Doris Kloor, Department of Pharmacology and Toxicology, Faculty of Medicine, University of Tübingen, Tübingen, Germany), and with a monoclonal anti-polyhistidine antibody (Sigma–Aldrich) after electrotransfer to a nitrocellulose membrane (results not shown). Cleavage with thrombin decreased the size of the recombinant protein by 130 residues (∼14 kDa), leaving only a small peptide of 22 amino acids fused to the AdoHcyase protein. The fusion part is identical for all recombinant AdoHcyase proteins investigated as proved by sequencing. Cleaved protein migrates slightly above the 43 kDa ovalbumin standard (Figure 1b). Purification of recombinant wild-type enzyme was achieved by a four-step procedure as described in the Experimental section. Approximately 10 mg of homogeneous enzyme was obtained from a 1 litre culture.
Figure 1. SDS/PAGE of recombinant wild-type and mutant AdoHcyases.
(a) Crude extracts from overnight (O.N.) cultures of non-induced and IPTG-induced E. coli harbouring the wild-type expression plasmid, and Ni-NTA affinity-purified wild-type AdoHcyase, (b) Ni-NTA affinity-purified thrombin-cleaved or uncleaved recombinant AdoHcyases (1 μg). M, molecular-mass standards (Roti-Mark Standard; Carl Roth, Karlsruhe, Germany), from top to bottom: β-galactosidase (119 kDa), serum albunium (66 kDa), ovalbumin (43 kDa) and carbonic anhydrase (29 kDa). +T, thrombin-cleaved protein.
We constructed vectors that expressed recombinant protein with single mutations in the AdoHcyase gene at amino acid positions 112 (p.W122X), 115 (p.E115L) and 143 (p.Y143C). Mutations in codons 112 and 143 have been detected in AdoHcyase-deficient patients, whereas the codon 115 mutation was deliberately introduced into the AdoHcyase cDNA. The same purification steps used to purify the wild-type enzyme were used in the purification of the mutant enzymes.
Significant differences were observed in the amounts of mutant enzymes recovered during purification compared with the wild-type enzyme. Interestingly, expression of significant amounts of mutant p.Y143C protein could be achieved only at a bacterial growth temperature below 20 °C. Temperatures above 22 °C resulted in low yield and decreased enzymatic activity of recombinant protein. Approximately 5–7 mg of homogeneous enzyme was obtained from a 1 litre culture.
Mutant protein p.E115L could efficiently be expressed at 29 °C with yields of approx. 6 mg of homogeneous enzyme from a 1 litre culture. Growth at 37 °C resulted in similar observations as described above for p.Y143C protein, i.e. low yield and low enzymatic activity.
Figure 1(b) shows SDS/PAGE analysis of purified wild-type and mutant proteins. Both wild-type and mutant forms of the enzyme were purified to virtual homogeneity as judged by a single band on SDS/PAGE. Mutant p.Y143C protein is indistinguishable from wild-type protein but shows a tendency to precipitate at low protein concentration.
Quaternary structure and thermal stability of wild-type and mutant AdoHcyase protein
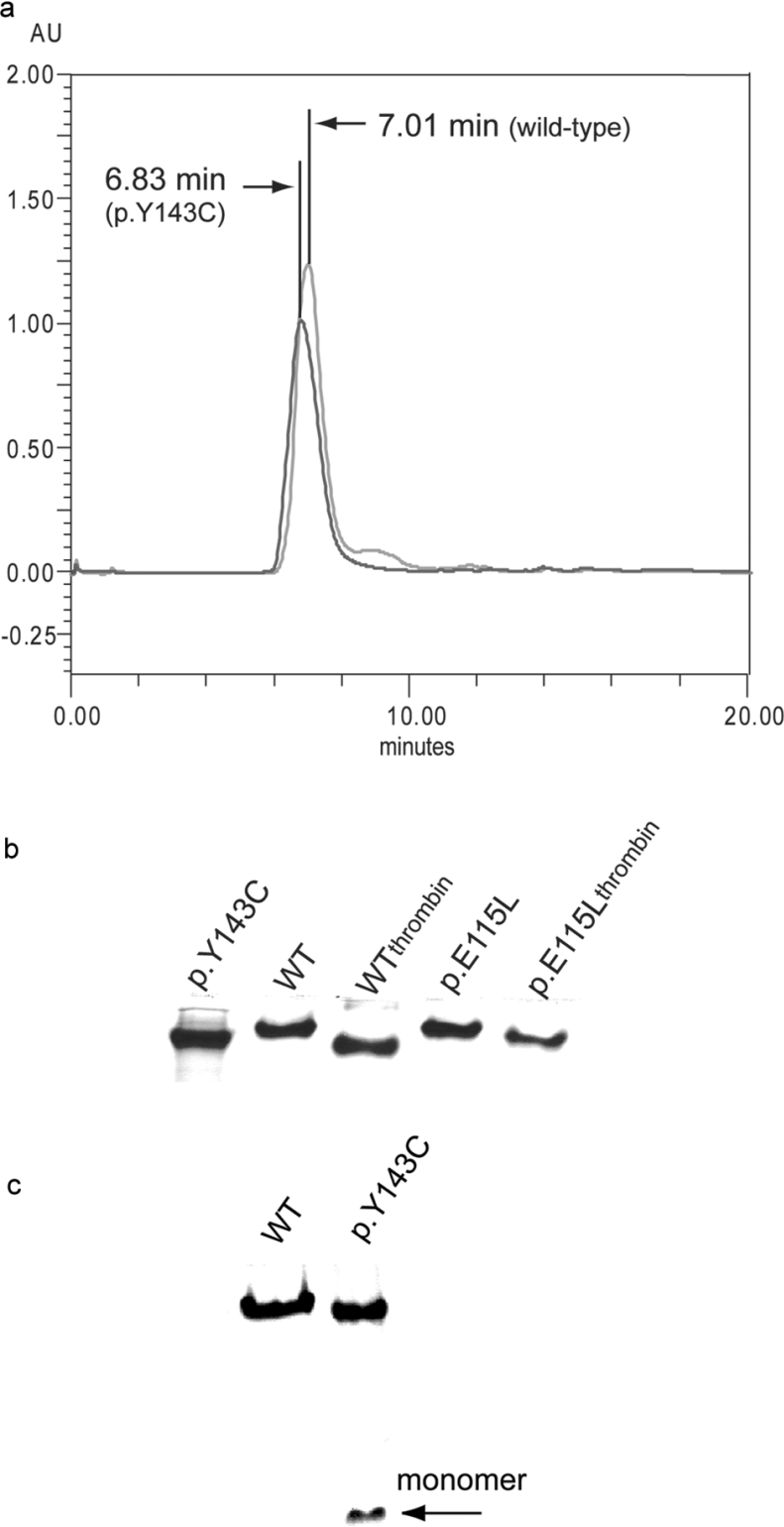
The molecular masses of cleaved and uncleaved mutant and wild-type forms of AdoHcyase were analysed by gel filtration chromatography. Uncleaved wild-type enzyme was eluted as a single symmetrical peak at the position corresponding to Mr 260000 (Figure 2a), indicating that the recombinant enzyme is a tetramer as is the native enzyme found in human tissues. Uncleaved mutant p.Y143C elutes also with a single symmetrical peak, but with a slightly different retention time compared with wild-type. Additionally, PAGE using native conditions gave a single band having a similar mobility for uncleaved wild-type and mutant p.Y143C or p.E115L (Figure 2b). The same behaviour was observed for thrombin-digested AdoHcyase proteins (Figures 2b and 6), indicating that the fusion part of the recombinant protein does not interfere with holoenzyme formation. However, unlike wild-type and p.E115L protein, p.Y143C exhibits a significantly higher instability, i.e. irreversible disassembly into monomers (Figure 2c). Higher protein concentrations (10 mg/ml) counteract the disassembly process to some extent.
Figure 2. Tetrameric forms of recombinant AdoHcyases as observed by gel-filtration chromatography and native PAGE.
(a) 1 mg of Ni-NTA affinity purified uncleaved wild-type and p.Y143C, suspended in 1 ml of buffer B, was applied to a BIO-SIL SEC 250-5 column (Bio-Rad), equilibrated with buffer B. Elution was performed with equilibration buffer at a flow rate of 1 ml/min. The protein was monitored at 280 nm. (b) 5 μg of thrombin-cleaved or uncleaved recombinant proteins. (c) 5 μg of recombinant wild-type and p.Y143C proteins; p.Y143C mutant is partially disassembled. Proteins were resolved on 7.5% native PAGE.
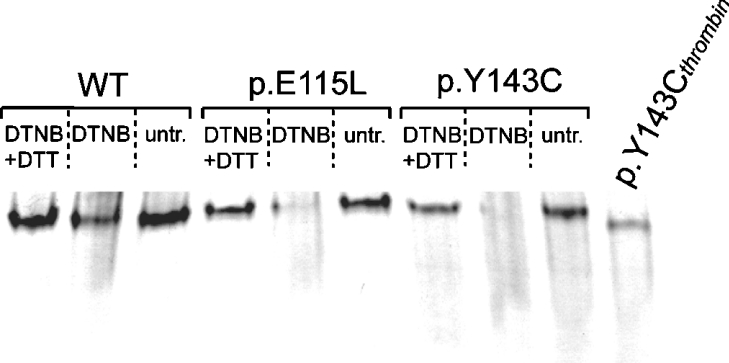
Figure 6. Tetramer disassembly as a result of DTNB treatment.
Recombinant AdoHcyases (5 μg) were treated with 50 μM DTNB for 60 min. Subsequently, 100 mM DTT was added to aliquots of the DNTB-treated enzymes and compared with untreated (untr.) enzymes. Proteins were resolved on 7.5% native PAGE.
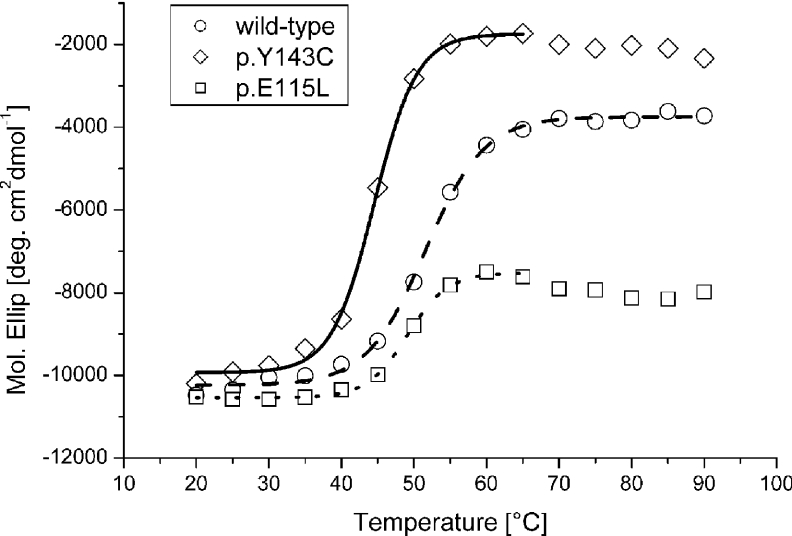
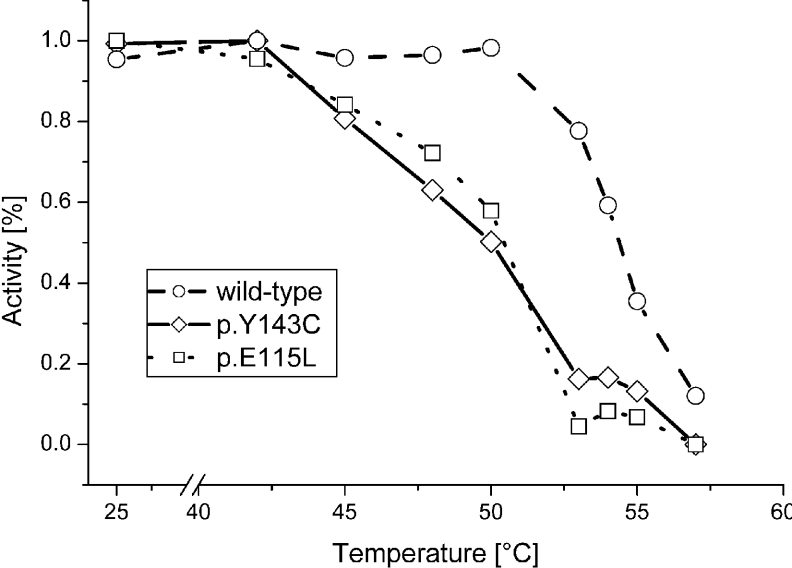
The far-UV CD spectra of wild-type, p.Y143C and p.E115L recombinant proteins in water at room temperature (20 °C) were almost identical (results not shown). The CD analysis showed, in agreement with the known X-ray structure (PDB 1A7A and 1LI4) [14,15], that the observed secondary structure is dominated by α-helices, but includes some β-strands. A striking difference was observed in the thermal stability of the recombinant proteins (Figure 3). The wild-type protein undergoes an unfolding process between 46.0 and 57.2 °C, with a Tm of 51.5 °C, while the p.Y143C mutant unfolds at lower temperatures (40.5–48.6 °C) with Tm of 44.5 °C. Unfolding of p.E115L lies between 45.2 and 52.9 °C with a Tm of 49.0 °C. Refolding ability is not observed for either wild-type or mutant proteins.
Figure 3. CD analysis.
Heat-induced unfolding of recombinant wild-type and mutant proteins monitored at 208 nm. The data were fitted to a Boltzman function shown as straight, dotted and dashed lines.
DLS measurements showed no presence of aggregates for the wild-type or for the mutants at 4, 25 and 35 °C. Aggregates were first observed at 45 °C. All three proteins tend to be tetrameric with a slight increase in the radius of hydration for the p.Y143C mutant (6.6–7.5 nm).
Results for heat inactivation of wild-type and mutant proteins p.E115L and p.Y143C are shown in Figure 4. A 15 min incubation in buffer A at 50 °C results in a 43% loss of enzymatic activity for p.E115L, and a 50% loss for p.Y143C. Enzymatic activity of wild-type protein is not affected at 50 °C, but is reduced to 50% at an incubation temperature of 55 °C. Mutant proteins are fully inactivated above 55 °C, whereas wild-type retains 12% activity even after incubation at 57 °C.
Figure 4. Kinetics of thermal inactivation of wild-type and mutant AdoHcyases.
Proteins were incubated for 15 min in buffer A at various temperatures. Inactivation is shown as a function of remaining AdoHcy hydrolysis.
Effects of mutations on enzyme activities and oxidation states of bound coenzyme NAD
The overall catalytic activity was determined in the directions of AdoHcy synthesis or hydrolysis. Truncation mutation p.W112stop (AdoHcyase in which Trp112 is replaced by opal stop codon) was catalytically completely inactive. Table 2 summarizes the catalytic properties of wild-type and mutant proteins.
Table 2. Kinetic parameters of wild-type and mutated enzymes in the directions of hydrolysis and synthesis.
Values in parentheses are percentage of wild-type. Truncation mutant p.W112stop is completely inactive. N/A, not assessed.
| Enzymatic activity (μmol·min−1·mg−1) | ||||
|---|---|---|---|---|
| Km (μM) Hydrolysis | Hydrolysis | Synthesis | NAD/tetramer | |
| Wild-type | 15.09 | 0.748 (100%) | 1.23 (100%) | 3.5 |
| p.Y143C | 11.0 | 0.185 (25%) | 0.42 (34%) | 3.1 |
| p.E115L | 11.47 | 0.500 (68%) | 0.75 (61%) | 4.6 |
| p.W112stop | N/A | N/A | N/A | N/A |
The p.E115L mutant retained a substantial amount of the overall catalytic activity in both directions (68% of the wild-type). In contrast, mutant p.Y143C exhibits only 25–30% catalytic activity in either direction.
In the AdoHcy hydrolysis mechanism, the bound NAD+ oxidizes C3′-AdoHcy, becoming NADH, which then reduces the 3′-keto-adenosine in the last step of the hydrolysis and is reconverted into NAD+ [22]. NADH has a characteristic absorption band around 340 nm. Therefore we measured the absorbance at 340 nm to monitor the oxidation state of the bound NAD.
The p.Y143C mutated enzyme exhibits significant NADH accumulation during AdoHcy hydrolysis. Approximately 88% of the coenzyme bound to p.Y143C was found to be NADH, whereas mutant protein p.E115L showed a value of 54% and wild-type protein a value of only 18% NADH respectively.
We determined the coenzyme content of recombinant AdoHcyase to 3.5 molecules NAD per tetramer for wild-type, 3.1 molecules for p.Y143C, and 4.6 molecules for p.E115L, in agreement with previously published results (3.2–3.6 molecules per tetramer in bovine liver) [31].
Reducing agents and disulfide bonds
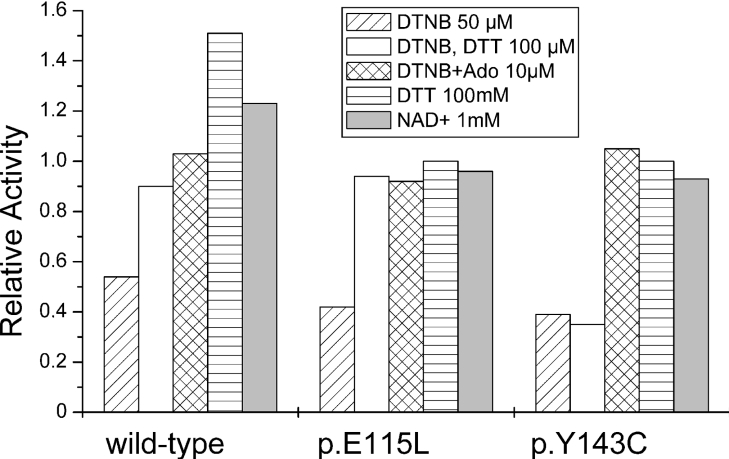
The presence of an additional cysteine residue in p.Y143C mutant protein raises the possibility that this residue might be involved in formation of a novel disulfide bridge. To test this hypothesis, the catalytic properties of p.Y143C mutant protein were investigated in the presence of the reducing agent DTT, using wild-type protein as reference. As shown in Figure 5, after 15 min of pre-incubation at 25 °C with various concentrations of DTT, no significant change in enzymatic activity was observed for p.Y143C and p.E115L mutant protein. On the other hand the catalytic capability of wild-type protein is enhanced 50% by addition of 100 mM DTT.
Figure 5. Kinetic parameters of wild-type and mutant AdoHcyases upon treatment with DTNB.
A summary of effects of thiol-reducing and modifying compounds on enzymatic activity is given for wild-type and mutant AdoHcyases. Values are calculated to a relative activity of 1 (100% enzymatic activity) as determined before treatment with reagents. Ado, adenosine.
Both wild-type and mutant proteins are partially inactivated by the thiol-modifying compound DTNB. Enzymatic activity of wild-type protein is decreased to 54% after 60 min DTNB (50 μM) treatment, whereas p.E115L protein retains 42% activity and p.Y143C 39% activity respectively. Inactivation of wild-type and p.E115L can be reversed by DTT (100 mM), whereas p.Y143C is unable to recover to the original values. Figure 6 shows that mutant proteins have undergone significant tetramer disassembly during DNTB treatment, whereas wild-type is more resistant and only partially disassembled. Addition of 100 mM DTT after DTNB treatment results in full reassembly of wild-type and p.E115L tetramer. Densitometric analysis shows reassembly of only 45% of p.Y143C tetramer. Inactivation of wild-type and both mutant proteins is prevented by addition of 10 μM adenosine prior to DTNB treatment (Figure 5).
Bioinformatics
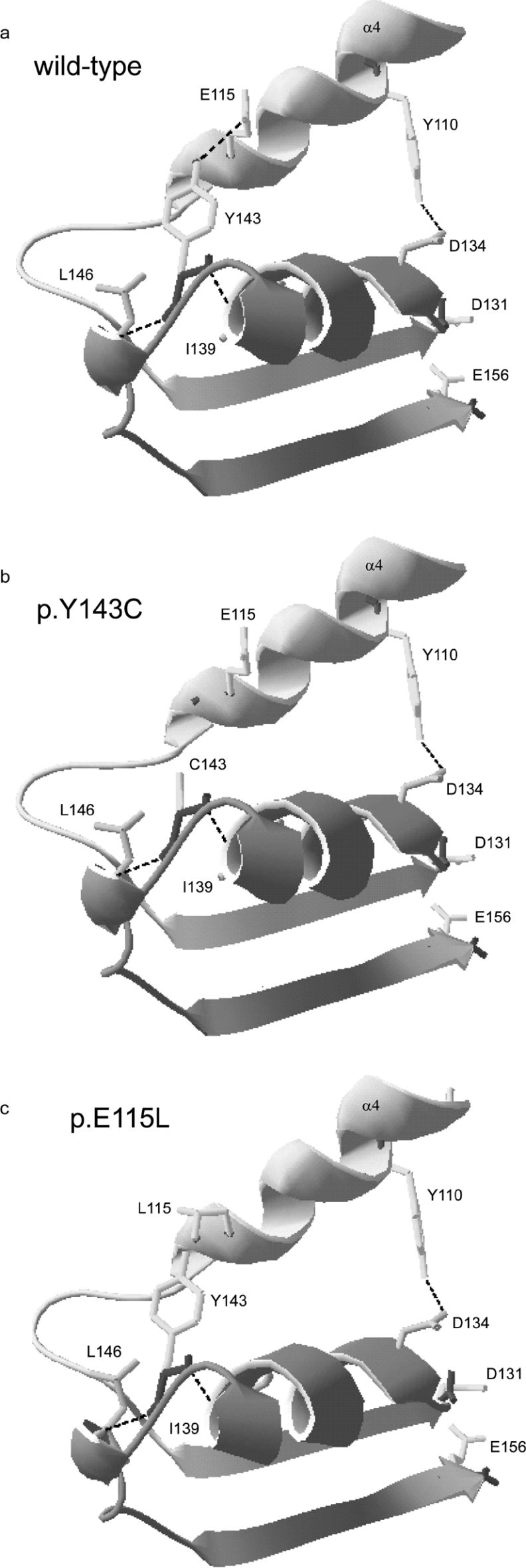
Analysis for possible changes in the number of hydrogen bonds between mutated and surrounding amino acid residues suggests that the Y143C mutation has lost a single hydrogen bond with Glu115 (Figure 7b). The same situation occurs in the p.E115L mutant, where Tyr143 cannot establish a hydrogen bond with Leu115 (Figure 7c). On the other hand, secondary structure prediction does not indicate changes in the region containing Tyr143 after conversion into cysteine. Nor does conversion of Glu115 to Leu115 bring about changes to the α-helical conformation.
Figure 7. Ribbon diagrams of the region containing Tyr143 in human AdoHcyase.
The enlargements show the region containing residues Tyr106 to Glu156 of human AdoHcyase. Mutated amino acids or residues involved in enzymatic activity and hydrogen-bonding are represented with side chains. Existing hydrogen bonds are shown as broken lines. (a) Wild-type; (b) p.Y143C is lacking hydrogen bond between Tyr143 and Glu115; (c) p.E115L is lacking hydrogen bond between Tyr143 and Leu115.
We have summarized some of the differences between the amino acids relevant to the present studies. Cysteine exhibits a much smaller surface area and residue volume than tyrosine, but is highly hydrophobic. As a replacement for Glu115 we chose leucine because of its similar size residue volume and surface area. High hydrophobicity of leucine was deliberately chosen to imitate the properties of Cys143 in the region containing α-helices 4 and 5. Values were retrieved from http://prowl.rockefeller.edu/.
DISCUSSION
Heterologous expression of wild-type human AdoHcyase cDNA yields satisfactory amounts of recombinant AdoHcyase protein. The resultant enzyme is a fusion protein with an additional 152 amino acids containing specific tags useful for protein solubility and purification. Regardless of the presence or absence of the tag region, recombinant protein appears to fold into a three-dimensional structure very similar to that of the human serum enzyme. Furthermore, the kinetic parameters of the recombinant wild-type AdoHcyase are very similar to those of the AdoHcyases of rat [17], bovine [32] or human [19] origin. Therefore our recombinant human AdoHcyase protein can be used as a reference for comparative mutational analyses.
The single amino acid exchange Y143C in the coding sequence of AdoHcyase brought about a significant change in expression efficiency and catalytic activity. Our results show that tetramer formation of p.Y143C protein is unlikely to be impaired. However, irreversible disassembly of mutant protein into monomers followed by a complete loss of enzymatic activity occurs spontaneously. So far, we have been unable to prevent this process. Freeze-drying is the only alternative to preserve mutant p.Y143C protein. Wild-type protein is extremely stable and loss of enzymatic activity of frozen stock was not observed over at least 1 year.
To seek clues to the reason for protein instability we studied protein unfolding behaviour during thermal induction. CD analysis of the p.Y143C mutant protein showed a significantly lower unfolding temperature of 7 °C compared with wild-type protein. DLS experiments showed accelerated aggregation of mutant protein at rising temperature. Also, mutant holoenzyme diameter was shown to be enlarged, indicating a less compact quaternary structure of mutant AdoHcyase. Taken together, our results show that the Y143C mutation renders the enzyme temperature-sensitive and implicates Tyr143 in protein subunit folding.
The AdoHcyase subunit is composed of three domains with the peptide chain organized into 17 α-helices and 15 β-strands [20]. The Y143C mutation is located in the catalytic domain (residues 1–183 and 357–390), which has the following topology: α0-α1-β1-α2-β2-α3-α4-β3-α5-α6-β4-α7-β5 (and α14–β15). Tyr143 is located at the end of α-helix 5, which is a neighbour of α-helix 4. In the three-dimensional model of human AdoHcyase (PDB 1A7A), a hydrogen bond between Tyr143 and Glu115 is connecting both helices. The hydrogen bond in question is absent from the calculated model of mutant p.Y143C protein. Therefore it seems likely that the loss of this particular hydrogen bond results, upon increase in temperature, in conformational changes in the proximity of the active site of the AdoHcyase subunit.
To probe this hypothesis, we imitated the situation in the p.Y143C mutant by changing the wild-type protein from Glu115 to Leu115 to prevent hydrogen-bonding with Tyr143. Our results show that the resulting mutant p.E115L possesses thermosensitivity almost identical with that of the p.Y143C protein. Enzymatic activity of mutant protein p.E115L is 30% less at temperatures below 37 °C. This indicates that the hydrogen bond between Tyr143 and Glu115 is not essential for enzymatic activity, but plays an important role in subunit folding and protein integrity. We conclude that in vivo at physiological temperatures of 37 °C, protein folding and tetramer assembly of p.Y143C and probably p.E115L are impaired. Additionally, the CD measurements showed that neither recombinant wild-type nor mutant proteins are able to completely refold, indicating that irregularly folded protein is unable to reconstitute into its active form. This obstacle can be overcome when the temperature is lowered to permissive conditions, as shown by heterologous expression at 18 °C for p.Y143C or at 29 °C for p.E115L. Once the subunits have assembled into a tetramer, the protein complex provides stability and reasonable enzymatic activity (depending on the type of mutation). These observations are in agreement with the properties of mutant protein p.Y143C produced under permissive conditions (25–30% enzymatic activity of wild-type), and with the AdoHcyase activities from patients' tissues (where restrictive conditions prevail) in which enzymatic activities of only 3% in liver or 5–10% in whole blood or fibroblasts were found [9].
Interestingly, sequence comparison of rat AdoHcyase with its human or mouse homologue shows that His142 is equivalent to human Tyr143 (mouse Tyr142). The crystal structure of rat AdoHcyase (PDB: 1b3rA) [14] shows that His142 does not form a hydrogen bond with Glu114 (human Glu115). Nevertheless, there may be interaction via a C–H hydrogen bond between His142 and Glu114. This special type of hydrogen bond is much weaker than the usual one and exhibits a bond length of 2.3–2.5 Å (1 Å=0.1 nm) [33]. Such a bond occurs chiefly when the carbon hydrogen-bond donor is part of an aromatic ring, which might be the case for His142 in the rat AdoHcyase. Thus it would be worthwhile to compare temperature dependent unfolding of rat and mouse AdoHcyases with those of our mutant proteins.
Although covalent bonds are mostly associated with the primary structure of proteins, sometimes such bonds may influence tertiary structures. An example is when a disulfide bridge is formed between two cysteine residues due to a redox reaction. Because the cytoplasm possesses a reducing environment, to our knowledge there is no intra-cytoplasmic protein with disulfide bonds. However, in view of the additional cysteine residue in p.Y143C protein, we investigated the effect of thiol-reducing reagent DTT on catalytic activities of recombinant wild-type and mutant proteins. We found that enzymatic activity of wild-type enzyme is enhanced by the addition of DTT. This is in agreement with reports that one cysteine residue is involved in the catalytic mechanism of rat AdoHcyase, where it modulates the oxidation state of bound NAD [34]. However, activity of mutant p.Y143C and p.E115L is unchanged in the presence of DTT.
Both wild-type and mutant proteins are partially inactivated by thiol-modifying compound DTNB. Inactivation is due to disassembly of the tetramer. Wild-type enzyme exhibits higher resistance to DTNB than mutant proteins, which indicates lower accessibility of cysteine residues because of a more compact structure. This is in agreement with the DLS studies, where enlarged diameter of mutant protein was detected. Nevertheless, kinetics of deactivation for both wild-type and mutant proteins follow the same principles, indicating that the same cysteine residues with free thiol groups are located near the substrate binding site. Inactivation of both wild-type and mutant proteins can be prevented by addition of adenosine. Interestingly, inactivation is reversed by addition of DTT only for wild-type and p.E115L, whereas enzymatic activity of p.Y143C does not recover and remains in a ‘status quo’ situation. This is because tetramer reassembly is significantly impaired only for p.Y143C, but not for wild-type and p.E115L. Thus we assume that the additional cysteine residue in p.Y143C is modified as a result of the reaction with DTNB leading to irreversible conformational changes and loss of enzymatic activity. Consequently, irregular disulfide bonding in p.Y143C protein is not considered to be likely due to the accessibility of Cys143 to DTNB.
The activity of wild-type enzyme can be increased by 21% upon incubation with exogenous NAD. As reported by Hohman et al. [26], native AdoHcyase from Dictyostelium can bind one additional NAD per tetramer, which serves a regulatory function and binds to a site distinct from the catalytic site. Activity of p.Y143C and p.E115L is unaffected by addition of exogenous NAD, indicating that conditions used for purification and storage support association of NAD with mutant proteins or that additional NAD is not bound by mutant proteins.
However, there is a striking difference in the oxidation states of NAD between mutant and wild-type AdoHcyase, with significant NADH accumulation occurring in mutant p.Y143C protein. NADH accumulation is less prominent for p.E115L protein, in which Tyr143 is unchanged.
A similar situation regarding NADH accumulation has been described for both human placental K426R [19] and rat D189N mutant AdoHcyases [21]. This feature is accompanied by loss of overall catalytic activity due to a shift in favour of the oxidation rates.
The mechanism of oxidation of AdoHcy and adenosine involves several steps. Upon substrate binding the enzyme undergoes conformational changes and the cleft between the catalytic (AdoHcy and adenosine-binding domain) and the NAD-binding domain is closed. Deduced from the three-dimensional structure [15], H-bond-forming residues Tyr143 and Glu115 are part of a helix–sheet–helix–helix–sheet (α4-b3-α5-α6-b4; Figure 7a) topology in the catalytic domain. Located at the ends of β-sheets 3 and 4 are amino acids Asp131 and Glu156 (rat Asp130 and Glu155), with their side chains facing the catalytic cleft. Both residues are crucial for substrate binding and catalytic activity, and specifically proximity of Asp131 to the C4′-H of adenosine is essential for the reaction [22]. Also, His55 (rat His54), which donates a proton necessary for cleavage of the C5′-SD bond of AdoHcy, is located close to D131 in the catalytic domain.
Thus it might be possible that loss of the hydrogen bond connecting helices 4 and 5 might lead to conformational changes which could slightly change the geometry of the active site and move Asp131 away from His55 into an unfavourable position. As a consequence, the C5′-SD bond cleavage of 3′-keto-4′-dehydro-AdoHcy to produce 3′-keto-4′5′-dehydro-adenosine and Hcy may be inefficient and NADH accumulation occurs. It has been shown that the catalytic domain of the substrate-free enzyme vibrates with a frequency of 40 MHz [35]. During the catalytic reaction slow open–closed conformational changes occur, which allow release of Hcy and access of a water molecule to the catalytic domain. Addition of water to 3′-keto-4′5′-dehydro-adenosine and subsequent reduction of this intermediate by NADH produce adenosine. Opening the cleft and releasing adenosine from the active site completes the catalytic cycle. However, in view of the missing hydrogen bond in mutant proteins the slow open–closed conformational changes may be impaired and product release slowed down. This is indicated by our Km measurements for mutant proteins, suggesting higher affinity for AdoHcy. Subsequently, decreased catalytic activity for both mutant proteins p.Y143C and p.E115L is observed.
Interestingly, despite intense research on AdoHcyase structure and its catalytic mechanism, amino acid Tyr143 has not been considered relevant for enzyme activity. However, we show that Tyr143 might be crucial not only for subunit folding and geometry of the active site but also important for the oxidation states of NAD in the AdoHcyase protein. Thus a combination of structural and biochemical abnormalities in the mutant p.Y143C protein represent one basis for severe human AdoHcyase deficiency and its pathological effects. The extent of AdoHcyase activity necessary to guarantee proper AdoHcy metabolism and AdoMet/AdoHcy turnover in humans remains unknown. The clinical well-being and the metabolic normality on normal diets of the known heterozygous parents of AdoHcyase-deficient patients [9,10] suggest that activities probably close to 50% of normal are likely to be adequate. If it becomes possible to increase the in vivo AdoHcyase activity of deficient patients to 25–30% (as the present work shows is feasible in the laboratory), the adverse clinical effects of AdoHcyase deficieny might be mitigated. A deeper understanding of the catalytic mechanism of AdoHcyase and the amino acids involved in substrate conversion might be of considerable importance to improve therapy.
Acknowledgments
This work was supported by grants 0098086 (O.V.) and 0108016 (I.B.) of the Ministry of Science, Education and Sports of the Republic of Croatia, and by the Austrian Science Fund (FWF) project P17885 (T.P.). Thrombin was a gift from Antonija J. Begonja (Institute of Clinical Biochemistry and Pathobiochemistry, University of Würzburg, Würzburg, Germany).
References
- 1.Dela Haba G., Cantoni G. L. The enzymatic synthesis of S-adenosyl-L-homocysteine from adenosine and homocysteine. J. Biol. Chem. 1959;234:603–608. [PubMed] [Google Scholar]
- 2.Cantoni G. L. Biological methylation: selected aspects. Annu. Rev. Biochem. 1975;44:435–451. doi: 10.1146/annurev.bi.44.070175.002251. [DOI] [PubMed] [Google Scholar]
- 3.Fontecave M., Atta M., Mulliez E. S-adenosyl methionine: nothing goes to waste. Trends Biochem. Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Prigge S. T., Chiang P. K. S-Adenosylhomocysteine hydrolase. In: Carmel R., Jacobsen D. W., editors. Homocysteine in Health and Disease. Cambridge: Cambridge University Press; 2001. pp. 79–90. [Google Scholar]
- 5.Miller M. W., Duhl D. M., Winkes B. M., Arredondo-Vega F., Saxon P. J., Wolff G. L., Epstein C. J., Hershfield M. S., Barsh G. S. The mouse lethal nonagouti (ax) mutation deletes the S-adenosylhomocysteine hydrolase (Ahcy) gene. EMBO J. 1994;13:1806–1816. doi: 10.1002/j.1460-2075.1994.tb06449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seshadri S., Beiser A., Selhub J., Jacques P. F., Rosenberg I. H., D'Agostino R. B., Wilson P. W., Wolf P. A. Plasma homocysteine as a risk factor for dementia and Alzheimer's disease. N. Engl. J. Med. 2002;346:476–483. doi: 10.1056/NEJMoa011613. [DOI] [PubMed] [Google Scholar]
- 7.Nygard O., Nordrehaug J. E., Refsum H., Ueland P. M., Farstad M., Vollset S. E. Plasma homocysteine levels and mortality in patients with coronary artery disease. N. Engl. J. Med. 1997;337:230–236. doi: 10.1056/NEJM199707243370403. [DOI] [PubMed] [Google Scholar]
- 8.Loscalzo J. Homocysteine trials: clear outcomes for complex reasons. N. Engl. J. Med. 2006;354:1629–1632. doi: 10.1056/NEJMe068060. [DOI] [PubMed] [Google Scholar]
- 9.Barić I., Ćuk M., Fumić K., Vugrek O., Allen R. H., Glenn B., Maradin M., Pažanin L., Pogribny I., Radoš M., et al. S-adenosylhomocysteine hydrolase deficiency: a second patient, the younger brother of the index patient, and outcomes during therapy. J. Inherit. Metab. Dis. 2005;28:885–902. doi: 10.1007/s10545-005-0192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buist N. R. M., Glenn B., Vugrek O., Wagner C., Stabler S., Allen R. H., Pogribny I., Schulze A., Zeisel S. H., Barić I., Mudd S. H. S-Adenosylhomocysteine hydrolase deficiency in a 26-year-old man. J. Inherit. Metab. Dis. 2006;29:538–545. doi: 10.1007/s10545-006-0240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulter-Karis D. E., Hershfield M. S. Sequence of full length cDNA for human S-adenosylhomocysteine hydrolase. Ann. Hum. Genet. 1989;53:169–175. doi: 10.1111/j.1469-1809.1989.tb01781.x. [DOI] [PubMed] [Google Scholar]
- 12.Palmer J. L., Abeles R. H. Mechanism for enzymatic thioether formation: mechanism of action of S-adenosylhomocysteinase. J. Biol. Chem. 1976;251:5817–5819. [PubMed] [Google Scholar]
- 13.Palmer J. L., Abeles R. H. The mechanism of action of S-adenosylhomocysteinase. J. Biol. Chem. 1979;254:1217–1226. [PubMed] [Google Scholar]
- 14.Hu Y., Komoto J., Huang Y., Gomi T., Ogawa H., Takata Y., Fujioka M., Takusagawa F. Crystal structure of S-adenosylhomocysteine hydrolase from rat liver. Biochemistry. 1999;38:8323–8333. doi: 10.1021/bi990332k. [DOI] [PubMed] [Google Scholar]
- 15.Turner M. A., Yuan C. S., Borchardt R. T., Hershfield M. S., Smith G. D., Howell P. L. Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength. Nat. Struct. Biol. 1998;5:369–376. doi: 10.1038/nsb0598-369. [DOI] [PubMed] [Google Scholar]
- 16.Yang X., Hu Y., Yin D. H., Turner M. A., Wang M., Borchardt R. T., Howell P. L., Kuczera K., Schowen R. L. Catalytic strategy of S-adenosyl-L-homocysteine hydrolase: transition-state stabilization and the avoidance of abortive reactions. Biochemistry. 2003;42:1900–1909. doi: 10.1021/bi0262350. [DOI] [PubMed] [Google Scholar]
- 17.Gomi T., Date T., Ogawa H., Fujioka M., Aksamit R. R., Backlund P. S., Jr, Cantoni G. L. Expression of rat liver S-adenosylhomocysteinase cDNA in Escherichia coli and mutagenesis at the putative NAD binding site. J. Biol. Chem. 1989;264:16138–16142. [PubMed] [Google Scholar]
- 18.Gomi T., Takata Y., Date T., Fujioka M., Aksamit R. R., Backlund P. S., Jr, Cantoni G. L. Site-directed mutagenesis of rat liver S-adenosylhomocysteinase: effect of conversion of aspartic acid 244 to glutamic acid on coenzyme binding. J. Biol. Chem. 1990;265:16102–16107. [PubMed] [Google Scholar]
- 19.Ault-Riche D. B., Yuan C. S., Borchardt R. T. A single mutation at lysine 426 of human placental S-adenosylhomocysteine hydrolase inactivates the enzyme. J. Biol. Chem. 1994;269:31472–31478. [PubMed] [Google Scholar]
- 20.Komoto J., Huang Y., Gomi T., Ogawa H., Takata Y., Fujioka M., Takusagawa F. Effects of site-directed mutagenesis on the structure of S-adenosylhomocysteine hydrolase: Crystal structure of D244E mutant enzyme. J. Biol. Chem. 2000;275:32147–32156. doi: 10.1074/jbc.M003725200. [DOI] [PubMed] [Google Scholar]
- 21.Takata Y., Yamada T., Huang Y., Komoto J., Gomi T., Ogawa H., Fujioka M., Takusagawa F. Catalytic mechanism of S-adenosylhomocysteine hydrolase: site-directed mutagenesis of Asp-130, Lys-185, Asp-189, and Asn-190. J. Biol. Chem. 2002;277:22670–22676. doi: 10.1074/jbc.M201116200. [DOI] [PubMed] [Google Scholar]
- 22.Yamada T., Takata Y., Komoto J., Huang Y., Gomi T., Ogawa H., Fujioka M., Takusagawa F. Catalytic mechanism of S-adenosylhomocysteine hydrolase: roles of His 54, Asp130, Glu155, Lys185, and Aspl89. Int. J. Biochem. Cell Biol. 2005;37:2417–2435. doi: 10.1016/j.biocel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y., Komoto J., Takata Y., Powell D. R., Gomi T., Ogawa H., Fujioka M., Takusagawa F. Inhibition of S-adenosylhomocysteine hydrolase by ‘acyclic sugar’ adenosine analogue d-eritadenine: crystal structure of S-adenosylhomocysteine hydrolase complexed with d-eritadenine. J. Biol. Chem. 2002;277:7477–7482. doi: 10.1074/jbc.M109187200. [DOI] [PubMed] [Google Scholar]
- 23a.Barić I., Fumić K., Glenn B., Ćuk M., Schulze A., Finkelstein J. D., James S. J., Mejaški-Bošnjak V., Pažania L., Pagribny I. P., et al. S-adenosylhomocysteine hydrolase deficiency in a human: a genetic disorder of methionine metabolism. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4234–4239. doi: 10.1073/pnas.0400658101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shenoy A. R., Visweswariah S. S. Site-directed mutagenesis using a single mutagenic oligonucleotide and DpnI digestion of template DNA. Anal. Biochem. 2003;319:335–336. doi: 10.1016/s0003-2697(03)00286-0. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Hohman R. J., Guitton M. C., Veron M. Inactivation of S-adenosyl-L-homocysteine hydrolase by cAMP results from dissociation of enzyme-bound NAD+ Proc. Natl. Acad. Sci. U.S.A. 1985;82:4578–4581. doi: 10.1073/pnas.82.14.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobley A., Whitmore L., Wallace B. A. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 28.Whitmore L., Wallace B. A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwede T., Kopp J., Guex N., Peitsch M. C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 31.Matuszewska B., Borchardt R. T. The role of nicotinamide adenine dinucleotide in the inhibition of bovine liver S-adenosylhomocysteine hydrolase by neplanocin A. J. Biol. Chem. 1987;262:265–268. [PubMed] [Google Scholar]
- 32.Abeles R. H., Fish S., Lipinskas B. S-adenosylhomocysteinase: mechanism of inactivation by 2′-deoxyadenosine and interaction with other nucleosides. Biochemistry. 1982;21:5557–5562. doi: 10.1021/bi00265a027. [DOI] [PubMed] [Google Scholar]
- 33.Derewenda Z. S., Lee L., Derewenda U. The occurrence of C–HO hydrogen bonds in proteins. J. Mol. Biol. 1995;252:248–262. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]
- 34.Aksamit R. R., Backlund P. S., Jr, Moos M., Jr, Caryk T., Gomi T., Ogawa H., Fujioka M., Cantoni G. L. The role of cysteine 78 in fluorosulfonylbenzoyladenosine inactivation of rat liver S-adenosylhomocysteine hydrolase. J. Biol. Chem. 1994;269:4084–4091. [PubMed] [Google Scholar]
- 35.Yin D., Yang X., Hu Y., Kuczera K., Schowen R. L., Borchardt R. T., Squier T. C. Substrate binding stabilizes S-adenosylhomocysteine hydrolase in a closed conformation. Biochemistry. 2000;39:9811–9818. doi: 10.1021/bi000595a. [DOI] [PubMed] [Google Scholar]