Abstract
Transport of the co-substrate UDPGA (UDP-glucuronic acid) into the lumen of the endoplasmic reticulum is an essential step in glucuronidation reactions due to the intraluminal location of the catalytic site of the enzyme UGT (UDP-glucuronosyltransferase). In the present study, we have characterized the function of several NSTs (nucleotide sugar transporters) and UGTs as potential carriers of UDPGA for glucuronidation reactions. UDPGlcNAc (UDP-N-acetylglucosamine)-dependent UDPGA uptake was found both in rat liver microsomes and in microsomes prepared from the rat hepatoma cell line H4IIE. The latency of UGT activity in microsomes derived from rat liver and V79 cells expressing UGT1A6 correlated well with mannose-6-phosphatase latency, confirming the UGT in the recombinant cells retained a physiology similar to rat liver microsomes. In the present study, four cDNAs coding for NSTs were obtained; two were previously reported (UGTrel1 and UGTrel7) and two newly identified (huYEA4 and huYEA4S). Localization of NSTs within the human genome sequence revealed that huYEA4S is an alternatively spliced form of huYEA4. All the cloned NSTs were stably expressed in V79 (Chinese hamster fibroblast) cells, and were able to transport UDPGA after preloading of isolated microsomal vesicles with UDPGlcNAc. The highest uptake was seen with UGTrel7, which displayed a Vmax approx. 1% of rat liver microsomes. Treatment of H4IIE cells with β-naphthoflavone induced UGT protein expression but did not affect the rate of UDPGA uptake. Furthermore, microsomes from UGT1-deficient Gunn rat liver showed UDPGA uptake similar to those from control rats. These data show that NSTs can act as UDPGA transporters for glucuronidation reactions, and indicate that UGTs of the 1A family do not function as UDPGA carriers in microsomes. The cell line H4IIE is a useful model for the study of UDPGA transporters for glucuronidation reactions.
Keywords: microsome, nucleotide sugar transporter (NST), UDP-glucuronic acid, UDP-glucuronosyltransferase, UDP-galactose transporter-related protein 7 (UGTrel7), YEA4
Abbreviations: βNF, β-naphthoflavone; DAPI, 4′,6-diamino-2-phenylindole; DMEM, Dulbecco's modified Eagle's medium; ER, endoplasmic reticulum; EST, expressed sequence tag; H4IIE, rat hepatoma cell line; HA, haemagglutinin; NST, nucleotide sugar transporter; ORF, open reading frame; RACE, rapid amplification of cDNA ends; RT, reverse transcriptase; sORF, short ORF; UDPGA, UDP-glucuronic acid; UDPGlcNAc, UDP-N-acetylglucosamine; UGT, UDP-glucuronosyltransferase; UGTrel, UDP-galactose transporter-related protein; UTR, untranslated region; V79, Chinese hamster lung fibroblast cell line
INTRODUCTION
Glucuronidation, catalysed by UGTs (UDP-glucuronosyltransferases), is an important detoxification reaction where the resulting soluble glucuronides are generally more readily excreted from the body via urine and bile [1]. UGTs catalyse the transfer of glucuronic acid from the co-substrate UDPGA (UDP-glucuronic acid) to numerous xeno- and endobiotic aglycone substrates. The catalytic domain of UGT is located on the luminal side of the ER (endoplasmic reticulum) membrane [2,3], and therefore the water-soluble UDPGA synthesized in the cytosol needs to be transported into the ER lumen [4]. The mechanism for UDPGA supply into the ER lumen has been a source of controversy from the earliest stages of research on UGTs, and several models have been proposed (see [2] for discussion). The generally favoured compartment model assumes there is an independent ER UDPGA transporter(s) that supplies UDPGA to the active site of the UGTs within the lumen of the ER for glucuronidation [5–8]. Another hypothesis suggests UGTs are dimers [9] and proposes that dimerized UGT can act as a carrier for UDPGA [10]. Some evidence for dimerization and possible oligomerization of UGT has been reported [11,12]. A lack of significant direct experimental evidence hampers our ability to confirm or refute the various hypotheses, and the intraluminal catalytic site of the enzymes makes it difficult to fully understand the molecular mechanism of glucuronidation.
A family of NSTs (nucleotide sugar transporters) important for sugar chain and proteoglycan synthesis was identified by complementation of a deficiency in murine cells [13,14] and in the yeast Kluyveromyces lactis [15]. A human NST isoform called UGTrel7 (UDP-galactose tranporter-related protein), which transports UDPGA and UDPGalNAc (UDP-N-acetylgalactosamine) and exists in the ER, has also been found [16] and, due to its cellular localization and substrate specificity, it has been suggested that this transporter functions as a supplier of UDPGA for glucuronidation.
In the present study, we examined the possibility that NSTs contribute to the supply of UDPGA for glucuronidation in the ER lumen. We obtained two novel and two known NST cDNAs encoding putative ER UDPGA transporters and found that these cloned transporters transport UDPGA when stably expressed in a mammalian cell line. The potential role of UGTs for glucuronidation was assessed with a cell line expressing UGT1A6, a major phenol UGT in human liver, as well as Gunn rat liver microsomes in which UGT1A enzymes are absent. We also explored various cell lines retaining physiological UDPGA transport activity and found that microsomes from H4IIE cells had UDPGlcNAc (UDP-N-acetylglucosamine)-dependent UDPGA transport activity similar to that of rat liver microsomes.
EXPERIMENTAL
Materials
1-[1-14C]Naphthol and [U-14C]UDPGA were from Amersham Pharmacia Biotech and PerkinElmer respectively. PVDF and nitrocellulose membranes were obtained from Millipore. Nylon membranes were obtained from Roche Diagnostics, medium and antibiotics for cell culture were from Invitrogen, and tissue culture serum replacement (NuSerum) was purchased from BD Biosciences. Enzymes, vectors, kits and other molecular biology reagents were obtained from the following suppliers: Invitrogen, Roche Diagnostics, Promega, New England Biolabs, Stratagene and Pierce. Gunn rat livers were obtained from Harlan Sera-Lab. Most chemicals were obtained from Sigma–Aldrich. All other materials used were analytical grade or of the highest grade available.
Cells
V79 (Chinese hamster lung fibroblast), HeLa (human cervical carcinoma) and H4IIE (rat hepatoma) cell lines were grown under an atmosphere of 95% O2/5% CO2 with 100% relative humidity. DMEM (Dulbecco's modified Eagle's medium) was used as a medium for all of the cell lines, and was supplemented with 10% (v/v) NuSerum, 100 μg/ml streptomycin and 100 units/ml penicillin. Cells were passaged at approx. 70% confluence, with 2–3 day intervals for V79 cells and 5–7 day intervals for H4IIE cells. Established transfectants were maintained in the presence of 200 μg/ml G418 (Geneticin).
Buffer solutions
SH buffer comprised of 5 mM Hepes/KOH (pH 7.4) and 250 mM sucrose. Transport assay buffer comprised of 20 mM Tris/maleic acid (pH 7.4), 250 mM sucrose, 5 mM magnesium chloride and 2 mM NADH. PBS comprised of 150 mM sodium chloride and 10 mM sodium phosphate (pH 7.2). PHEM buffer comprised of 60 mM Hepes/KOH, 10 mM EGTA and 2 mM magnesium chloride (pH 6.9).
Preparation of microsomes from rat liver and cell lines
All procedures for preparing microsomes were as described previously [17]. For the preparation of microsomes from cultured cells, cells were grown to 80% confluence and scraped off by the method of Marsh et al. [18], resuspended in SH buffer and homogenized with a tightly fitting Dounce homogenizer (15 strokes for V79 cells, 5 strokes for H4IIE cells), and P1 (3000 g), P2 (13000 g) and P3 (100000 g) pellets were then obtained by differential centrifugation. The final microsomal pellets were resuspended in SH buffer and aliquots stored at −80 °C.
UDPGA uptake assay
Uptake of UDPGA into microsomal vesicles was analysed essentially as described in [19]. Microsomes (100 μg/assay) were pre-incubated for 90 min at 37 °C, with or without 1 mM of UDPGlcNAc, in transport assay buffer. Uptake assays were initiated by the addition of [14C]UDPGA (10 μM, 6.6 dpm/pmol) unless stated. At each time point, a portion of the reaction product (20 μl) was diluted into 1 ml of the ice-cold assay buffer and immediately poured onto a nitrocellulose filter (MF-membrane filter, pore size 0.45 μm; Millipore) attached to a vacuum manifold (Type 1225; Millipore), followed by washing of filters with 5 ml of ice-cold transport assay buffer. The amount of [14C]UDPGA taken into the microsomes and collected on the filter was measured with a liquid scintillation counter (LS6500; Beckman) after being dried and dissolved in liquid scintillation fluid (Filter-count). Kinetic parameters for UDPGA transport were analysed by direct curve fitting (Prism, GraphPad Software).
Cloning of NST cDNAs
All of the EST (expressed sequence tag) clones used in this study were obtained from the IMAGE consortium (UK HGMP Resource Centre). For UGTrel1 (UDP-galactose transporter-related protein 1), an EST clone (IMAGE 1589817) was identified by an homology search with a human Golgi UDP-galactose transporter [13]. The sequence of IMAGE 1589817 was identical with UGTrel1 (GenBank accession No. D87989), the function of which is not yet known [20], but was 73 bp shorter than reported and no upstream in-frame stop codon was observed. Therefore a full-length clone was obtained by screening a human liver cDNA library (Stratagene) with this EST clone as a probe.
For huYEA4, three EST clones (IMAGE 1032400, IMAGE 1373956 and IMAGE 1658129) were identified based on homology with the Kluyveromyces lactis nmm2-2 gene [15], which is involved in UDPGA uptake [21] and the Saccharomyces cerevisiae YEA4 gene, an ER UDPGlcNAc transporter [22]. IMAGE 1032400 had a complete coding sequence, and the other two clones were identical. Approx. 400 bp of these two clones corresponded to a middle part of the IMAGE 1032400. However, there was no in-frame stop codon upstream of the first ATG in both clones. Therefore 5′-RACE (5′-rapid amplification of cDNA ends) was applied using human liver cDNA (Marathon-Ready cDNA; BD Biosciences). Sequence analysis of the product obtained confirmed that the missing region corresponds to the 5′-portion of IMAGE 1032400. The full-length cDNAs obtained from the 5′-RACE and IMAGE 1032400 were named huYEA4 and huYEA4S respectively. To exclude the possibility of cloning artefacts, we performed RT (reverse transcriptase)-PCR for the full-length huYEA4 and huYEA4S cDNAs using human liver mRNA.
During the early part of the present study, the first NST which is able to transport UDPGA and UDPGalNAc was reported by another group [16]. Based on this NST cDNA, an identical EST clone (IMAGE 1989970) was obtained and the sequence confirmed. Phylogenetic analysis was carried out by the neighbour-joining method [23]. All DNA sequencing was performed at the Molecular Genetics Analysis Facility, University of Dundee, Dundee, Scotland, U.K.
Production of antibodies against NSTs
A partial cDNA fragment coding for the unique amino acid sequence residues 170–250 of UGTrel7 was subcloned into the Escherichia coli expression vector, pMALc2x (New England Biolabs). The BL21 E. coli strain transformed with the above construct was cultured in Luria-Broth medium and protein expression induced with 0.3 mM of isopropyl thio-β-D-galactoside. The expressed fusion protein was affinity purified using amylose resin (New England Biolabs) and the purity confirmed by SDS/PAGE. Peptides corresponding to residues 305–322 of UGTrel1 (C-terminus) and residues 175–194 of huYEA4 (between putative transmembrane domains 4 and 5) were synthesized at the University of Aberdeen (Department of Molecular and Cell Biology, Aberdeen, Scotland, U.K.) and were coupled to keyhole limpet haemocyanin (Pierce). The antibodies were raised in sheep at Alba Bioscience. The antisera obtained were affinity purified using protein- or peptide-affinity chromatography as appropriate.
Functional expression of NSTs in V79 cells
Up to nucleotide-6 of the 5′-UTRs (untranslated regions) and all of the 3′-UTRs of the cloned NST cDNAs were removed by PCR amplification of the coding sequences. The products were cloned into pGemT (Promega). A UGTrel7 cDNA containing three HA (haemagglutinin) tag sequences at the C-terminus was also generated by PCR. After confirming sequence identity, these fragments were subcloned into pCIneo (Promega). Expression constructs were transfected into V79 cells using Lipofectin® (Invitrogen), and after 2 weeks of selection with 800 μg/ml of G418, resistant colonies were screened by Northern blotting (clones without HA tags) or by Western blotting (for tagged constructs) using an anti-HA antibody.
Immunocytochemistry
HeLa cells were grown on glass cover slips. A UGTrel7 expression construct with three C-terminal HA-tags was transfected into cells using Effectene (Qiagen) following the manufacturer's instructions. Cells were processed at room temperature. Cells were fixed for 10 min in 3.7% (v/v) paraformaldehyde dissolved in PHEM buffer. Following a 10 min permeabilization with 1% (v/v) Triton X-100 in PBS and blocking with 1% (v/v) goat serum in PBS for 20 min, cells were treated with primary antibodies, either an anti-HA monoclonal antibody (Roche Diagnostics) or a rabbit anti-calreticulin polyclonal antibody (SPA-600; Stressgen) for 30 min. Cells were subjected to three 10 min washing steps in PBS, and stained with secondary antibodies (FITC-conjugated anti-mouse IgG or TRITC-conjugated anti-rabbit IgG; Jackson ImmunoResearch) for 30 min. Following a further three 10 min washes with PBS, cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole; 0.3 μg/ml) and mounted in Vectashield medium (Vector Laboratories). Immunostained samples were examined using a Zeiss 63×objective and images recorded using a Hamamatsu Orca camera. Optical sections separated by 200 nm were recorded using a binning of 2×2. Images were restored using an iterative constrained deconvolution algorithm (Volocity, Improvision) using a calculated point-spread function.
Induction of UGT expression in H4IIE cells
H4IIE cells were cultured using the same conditions as V79 cells. βNF (β-naphthoflavone; 100 μM) dissolved in DMSO (0.1% v/v) was added to the culture medium and incubated for a further 48 h. As a control, the same volume of DMSO alone was added. Preparation of microsomes was as described for V79 cells except that 5 strokes of homogenization were used. Western blot analysis employed a broad-spectrum sheep anti-rat UGT antiserum [24].
Measurement of latency
The latency of microsomal UGT and glucose 6-phosphatase activities toward glucose 6-phosphate and mannose 6-phosphate were measured using 1-naphthol, mannose 6-phosphate and glucose 6-phosphate respectively. Microsomes prepared from rat liver microsomes and recombinant V79 cells were treated with various concentrations of the non-ionic detergent Lubrol PX or the pore-forming peptide antibiotic alamethicin for 30 min on ice. 1-Naphthol glucuronidation activity was measured using 1-[1-14C]-naphthol as a substrate [25]. As known ER marker enzymes, glucose 6-phosphatase activity toward glucose 6-phosphate or mannose 6-phosphate was measured as described [26]. Both assays were performed at 37 °C for 10–30 min, which is within the range of linearity. The degree of latency was expressed as a percentage, which was calculated by the following equation: Latency (%)=100 [1−(activity obtained in microsomes without activation)/(maximal activity obtained in activated microsomes)].
RESULTS
Microsomes prepared from rat liver and H4IIE cells transport UDPGA with UDPGlcNAc dependence
Uptake of [14C]UDPGA was observed with rat liver microsomes. In the present study, we used three cell lines: V79, H4IIE and HeLa cells. For functional studies, V79 and H4IIE cells were used in addition to rat liver microsomes. HeLa cells were used for investigating intracellular localization by immunofluorescence microscopy.
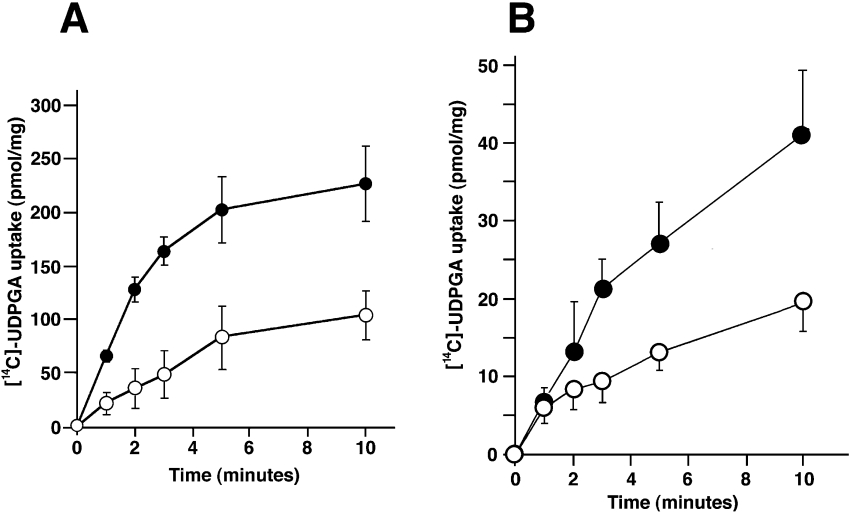
UDPGA uptake was partially UDPGlcNAc-dependent [27], as the pre-incubation step with UDPGlcNAc enhanced microsomal UDPGA transport approximately 2-fold (Figure 1A). In both cases, initial velocity of uptake was linear up to 2 min. Uptake of UDPGA could also be observed in microsomes prepared from H4IIE cells (Figure 1B). The pattern of uptake and the magnitude of increase in uptake in UDPGlcNAc-preloaded microsomes were qualitatively similar to that of rat liver microsomes (approx. 2-fold at 10 min). In contrast, in microsomes prepared from V79 cells, there was no UDPGA uptake, either in UDPGlcNAc-preloaded or water-preloaded microsomes (see Figure 4). These results suggest that H4IIE cells retain a UDPGA uptake system similar to rat liver microsomes but that V79 cells do not possess such a transport system.
Figure 1. Transport of UDPGA into rat liver and hepatoma cell line microsomes.
The time- and UDPGlcNAc-dependent uptake of [14C]UDPGA was investigated both in rat liver microsomes (A) and in microsomes prepared from H4IIEcells (B). Rapid filtration assays were started after 90 min of pre-incubation with 1mM UDPGlcNAc (●) or water (○) at 37 °C. Error bars represent the means±S.D. (n=4–6).
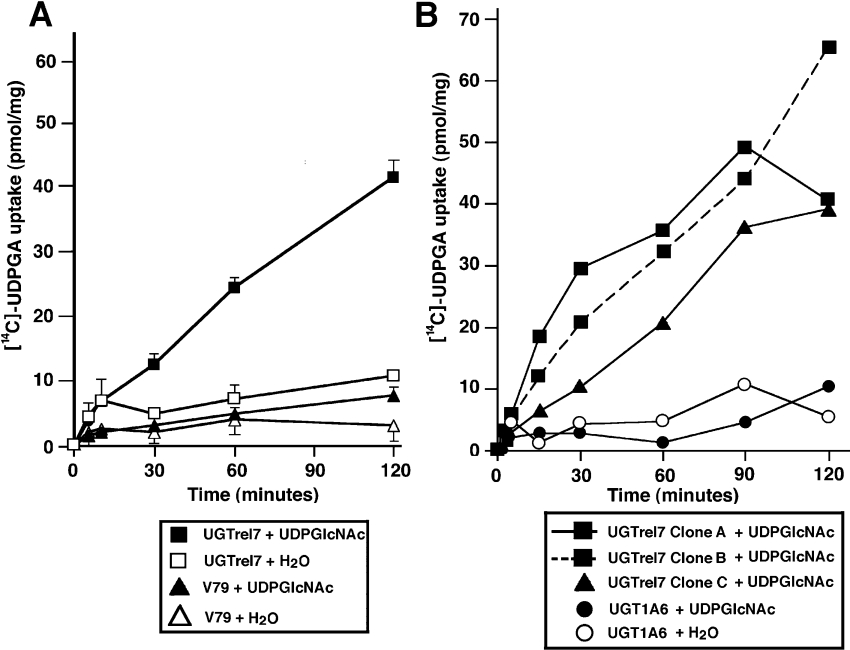
Figure 4. Functional expression of UGTrel7 in V79 cells.
Three V79 cell lines stably expressing UGTrel7 were established. Clones A and B have three HA tag sequences at the C-terminal end whereas Clone C does not have a tag sequence. (A) Uptake of [14C]UDPGA was analysed in Clone B and V79 (parental cell line) cells. Assay conditions were as for Figure 1. Microsomes were from UGTrel7-expressing V79 cells (squares) and parental V79 cells (triangles). Assays were started after pre-incubation with 1 mM UDPGlcNAc (filled points) or water (empty points) as for Figure 1. (B) Uptake of [14C]UDPGA in various established cell lines. Clone A (■), Clone B (■ with a dotted line), Clone C (▲). Assays were also carried out with microsomes prepared from UGT1A6-expressing V79 cells preloaded with 1 mM UDPGlcNAc (●) or water (○).
Retention of latency both in rat liver microsomes and in microsomes from a UGT1A6-expressing cell line
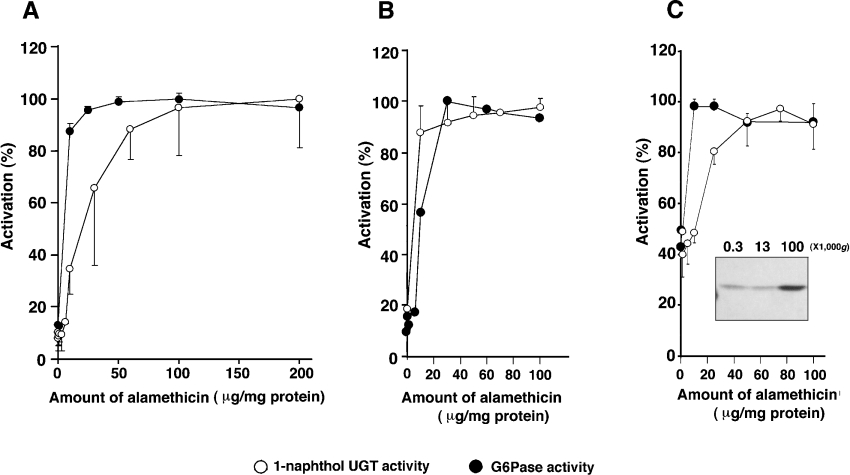
It is well established that to obtain full microsomal UGT activity, an activation step which disrupts the microsomal membrane is required, e.g. by using detergent, sonication or by use of pore-forming peptides such as alamethicin. Without these procedures, the UGT maintains a ‘latent’ state. The detergent treatment of microsomes prepared from V79 cells expressing UGT1A6 [28] inhibited UGT activity whereas addition of alamethicin resulted in a substantial increase in enzyme activity (results not shown). In rat liver microsomes and microsomes prepared from H4IIE cells, greater than 90% latency was observed (Figures 2A and 2B) and at least 55% latency could be observed with the microsomes prepared from the V79/UGT1A6 cell line (Figure 2C). In all sets of microsomes, latency of 1-naphthol UGT activity and of the activity of the ER marker enzyme, glucose 6-phosphatase, was almost identical (Figures 2A, 2B and 2C). Distribution of expressed UGT1A6 protein in the UGT1A6-expressing V79 cells was examined by Western blotting (using an anti-rat liver UGT antibody [24]) following differential centrifugation, and we found the UGT1A6 protein was almost exclusively localized within the 100000 g pellet fraction, i.e. in the microsomes (Figure 2C inset).
Figure 2. Activation of glucose 6-phosphatase (G6Pase) and 1-naphthol UGT activities by alamethicin.
Microsomes prepared from rat liver (A), H4IIE cells (B) and V79 cells expressing human UGT1A6 (C) were pre-treated with various concentrations of alamethicin and assayed for enzyme activity. ○ denote 1-naphthol UGT activity and ● show G6Pase activity. Mannose 6-phosphate was used as a substrate in (A) and (C) whereas glucose 6-phosphate was used in (B). Data are shown as a percentage of maximum activity (n=3±S.D.). The degree of latency was expressed as a percentage, calculated using the formula described in the Experimental section. The inset (C) shows a representative immunoblot probed with sheep anti-rat UGT antibody. Each lane contains 10 μg of protein. The numbers at the top of each lane show the gravitation forces (g) used for precipitation of each fraction.
cDNA cloning of human NSTs
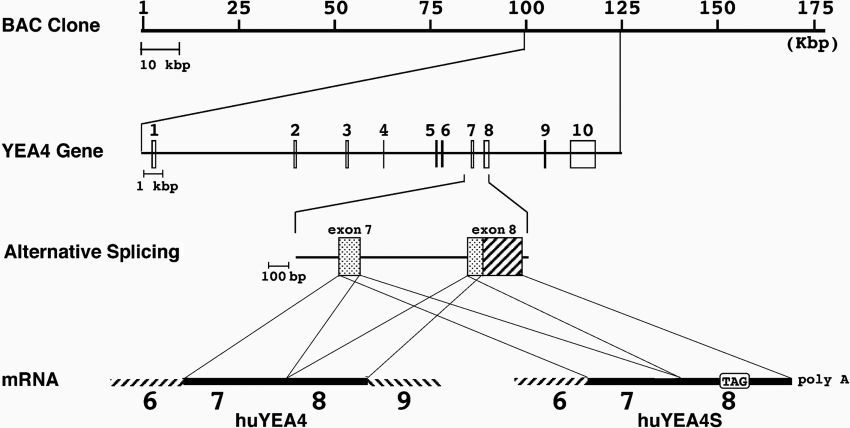
NSTs were originally identified as suppliers (transporters) of substrates required for sugar chain/proteoglycan synthesis pathways. Four NSTs were obtained in the present study (Table 1). For UGTrel1 the cDNA obtained had an entire ORF (open reading frame) coding sequence as reported [20] (GenBank accession number D87979). In addition, there was a second ATG within the 5′-UTR, which creates a sORF (short ORF) in the 5′-UTR (see Table 1). Two cDNAs coding for an apparent homologue of S. cerevisiae YEA4 were obtained from human liver (huYEA4 and huYEA4S; GenBank accession numbers AJ315497 and AJ315498 respectively). Comparison of deduced amino acid sequences of these two cDNAs revealed that huYEA4S lacks the C-terminal 103 residues of huYEA4, which are replaced with seven unique residues. Searching for the location of huYEA4(S) in the human genome, it appeared that huYEA4 and huYEA4S are splice variants (Figure 3). The huYEA4 and the huYEA4S cDNAs comprise the products of 10 exons and 8 exons respectively, with usage of a shorter portion of exon 8 in huYEA4 (Figure 3, third row). One in-frame stop codon and one sORF were identified in the 5′-UTR of each cDNA. The UGTrel7 cDNA obtained as an EST clone had exactly the same sequence as that reported [13]. There were no in-frame stop codons or sORFs in the 5′-UTR. Cloned NSTs in the present study, except for huYEA4S, had a di-lysine motif or a similar lysine-rich ER retention sequence [29–31] at the C-terminal end (Table 1).
Table 1. NST cDNAs obtained in the present study.
| Name | Chromosome | Number of amino acids in sORF at 5′-UTR | Total number of amino acids | Last 10 amino acids | Putative transmembrane domains |
|---|---|---|---|---|---|
| UGTrel7 | 1p31 | 0 | 355 | KLDIKGKGAV | 8 |
| UGTrel1 | 17q21 | 17 | 322 | FGKGAKKTSH | 8 |
| huYEA4 | 7q31 | 14 | 321 | SEPQKDSKKN | 8 |
| huYEA4S | 7q31 | 14 | 231 | NKSGEFGVRQ | 5 |
Figure 3. Two human YEA4 isoforms appeared as alternatively spliced transcripts.
Comparison between two huYEA4 cDNA sequences and the human genome sequence revealed the positions of each exon and the alternative splicing mechanism. From the top row: BAC clone No. RPC11–98D12 (GenBank Accession number AC008154); expanded view for huYEA4 locus; expanded view for exons 7–8 showing alternative splicing site at exon 8; mRNAs for huYEA4 and huYEA4S.
Uptake of UDPGA by NSTs depends on UDPGlcNAc
We first established V79 cell lines stably expressing UGTrel7 carrying three HA tags. Transcribed mRNA and the translated protein of the expected sizes could be detected in these cells (results not shown), and UDPGA uptake could be detected only after preloading with UDPGlcNAc in microsomes (Figures 4A, 4B and 5B). The transport of UDPGA in microsomes from UGTrel7-expressing V79 cells was completely dependent on UDPGlcNAc (Figures 4A, 4B and 5B), whereas rat liver microsomes transported approx. 50% of UDPGA in the absence of this compound (Figure 1A).
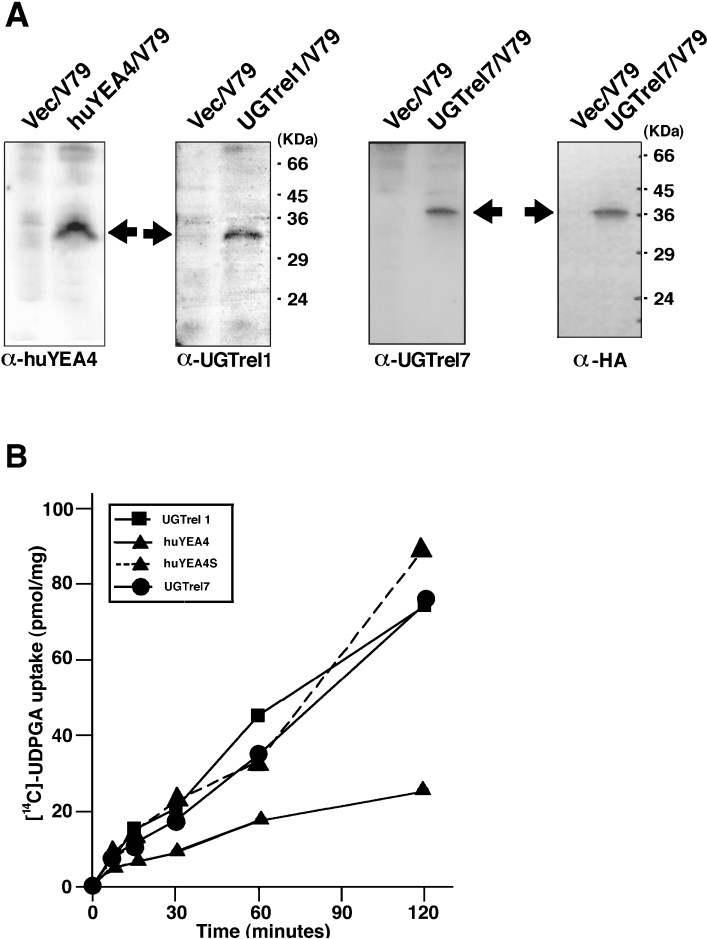
Figure 5. Detection of expressed NST proteins and UDPGlcNAc-dependent uptake of UDPGA by microsomes prepared from huYEA4, huYEA4S, UGTrel7 and UGTrel1.
(A) Assessment of antibodies raised against each NST and the existence of protein in established cell lines. Each lane contained 150 μg (UGTrel7–HA panel) or 200 μg (other panels) of microsomal protein. To induce protein expression, 10 mM of butyric acid was added 24 h before harvesting of cells. The positions of the expressed NST proteins are marked with arrows. Microsomes prepared from untagged UGTrel7 also gave similar result (results not shown). Note that the exposure time required for chemiluminescence detection with the α-HA (monoclonal antibody against an HA tag) blot was a few seconds whereas the other three were 15–30 min. (B) UDPGA uptake assay: huUGTrel7 (●), huYEA4 (▲), huYEA4S (▲ with a dotted line), UGTrel1 (■). Assays were performed after preloading of microsomes with 1 mM UDPGlcNAc for 90 min. There was no detectable uptake in water-preloaded microsomes (results not shown). The other assay conditions were the same as for Figure 1. Data shown are means (n=3) and S.D. was within 10%.
Other cell lines expressing untagged UGTrel7, UGTrel1, huYEA4 and huYEA4S were also established, and polyclonal antibodies were developed (Figure 5). Established cell lines showed corresponding mRNA with the predicted size, and no NST mRNAs or proteins were detected in untransfected V79 cells (results not shown). Western blot analysis of established cell lines with the antibodies prepared showed faint bands at the appropriate molecular mass (Figure 5A). All of these cell lines showed qualitatively similar UDPGA transport characteristics in that UDPGA uptake could be observed only after the preloading of microsomes with UDPGlcNAc (Figure 5B). Although the rate of uptake varied among the cloned cell lines, transport activities are clearly attributable to expressed NSTs, since parental V79 cells and the UGT1A6 (a major xenobiotic metabolizing UGT isoform)-expressing V79 cell line [28] did not show any detectable UDPGA uptake, even in UDPGlcNAc-preloaded microsomes (Figure 4A) and the uptake was time dependent (at least up to 1 h). The highest uptake was seen in the V79 cell line expressing UGTrel7 (80 pmol/mg protein in 2 h).
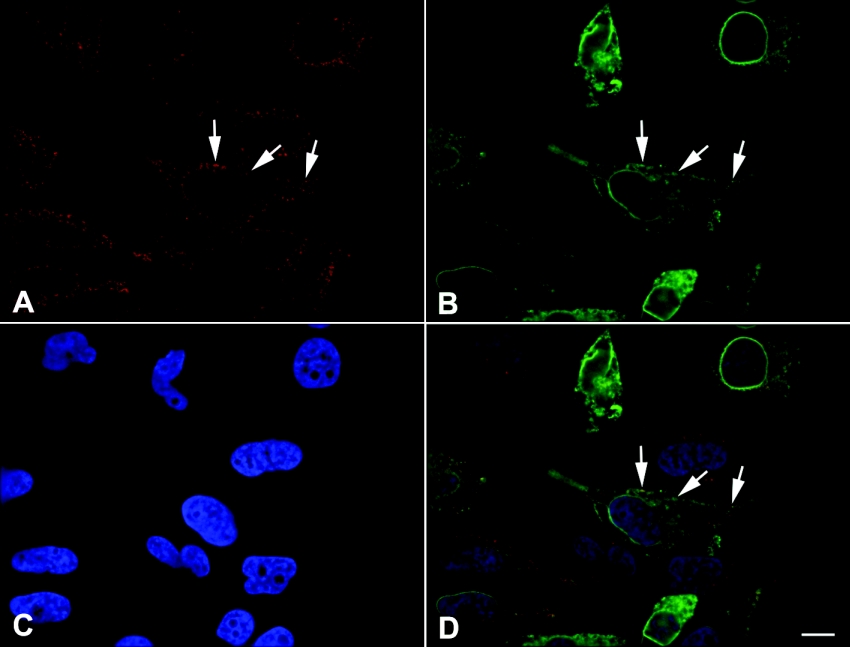
To determine whether UGTrel7 localized to the ER, which is where UGT proteins are principally located, we performed immunofluorescence microscopy on cells transiently transfected with the UGTrel7 expression construct containing three HA tag sequences. UGTrel7 protein was localized both in the nuclear envelope and in the ER (Figure 6B). The antibody against calreticulin demonstrated that UGTrel7 co-localized with this ER marker protein [32] (Figure 6A and 6D), confirming that this transporter is expressed at least partially in the ER.
Figure 6. Cellular localization of UGTrel7 protein.
Digitally deconvolved section of paraformaldehyde-fixed HeLa cells expressing HA-tagged UGTrel7. (A) Localization of calreticulin. (B) Localization of HA-tagged UGTrel7. (C) DAPI staining of nuclei. (D) Overlay of calreticulin (red), HA-tagged UGTrel7 (green) and DAPI (blue). Arrows highlight structures containing both calreticulin and HA-tagged UGTrel7. Bar=10 μm.
Comparison of kinetic parameters for UDPGA uptake between rat liver microsomes and expressed cell line
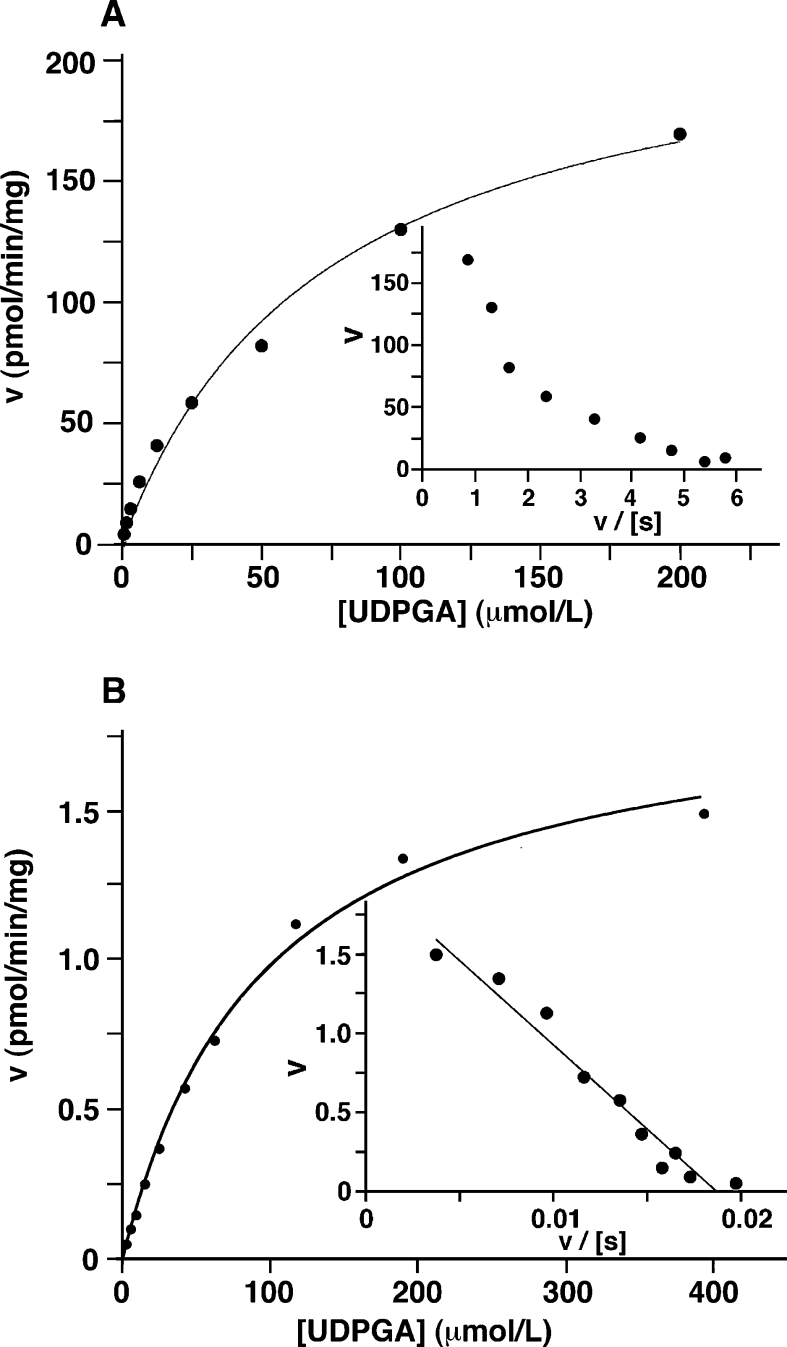
Kinetic parameters (Km and Vmax) for UDPGA uptake into rat liver microsomes and microsomes from UGTrel7-expressing V79 cells were determined (Figure 7). In rat liver microsomes, biphasic kinetics were observed (Figure 7A inset), suggesting the presence of more than one transporter capable of accepting UDPGA as a substrate. The Km of the high affinity component in rat liver microsomes and microsomes prepared from the UGTrel7 cell line were similar, whereas over a 100-fold difference in the Vmax was observed between rat liver microsomes and UGTrel7 microsomes (see legend to Figure 7).
Figure 7. Kinetic analysis of UDPGA uptake in rat liver (A) and UGTrel7/V79 (B) microsomes.
Maximum velocities (Vmax) and kinetic constants (Km) for UDPGA were determined from non-linear regression analysis of Michaelis–Menten plots using various concentrations of UDPGA. (A) Rat liver microsomes: Vmax=259 and 119 pmol/mg/min; Km=104 and 23 μM. (B) Microsomes prepared from UGTrel7 expressing V79 cells (Clone B of Figure 4): Vmax=2.3 pmol/mg/min; Km=66 μM. Data were also analysed using Eadie–Hofstee transformations (insets; n=3).
Effect of UGTs on UDPGA uptake
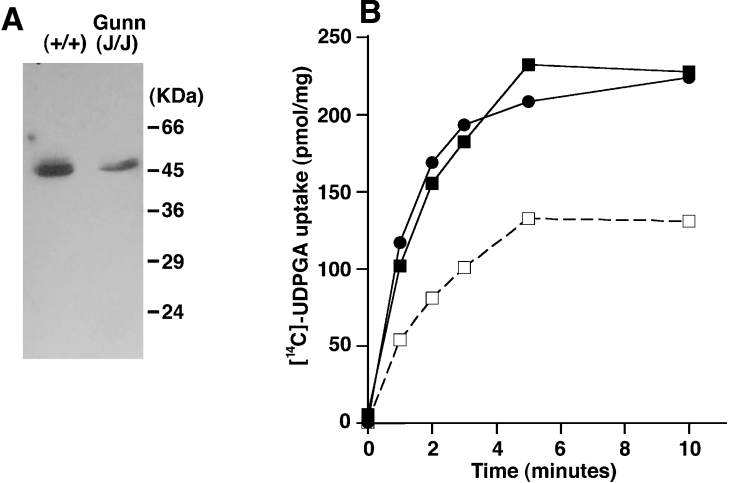
To investigate the possibility that UGT proteins themselves can function as carriers for UDPGA uptake into the ER lumen, Gunn rat liver microsomes, in which all UGT1 isoforms are deficient due to a one nucleotide deletion in exon 4 of the UGT1 gene complex [33], were used for uptake assays. The homozygous frame-shift mutation in these samples was confirmed by direct sequencing of a PCR product of the affected region (results not shown) and by the decreased total UGT protein expression observed (Figure 8A). In Gunn rat liver microsomes, UGT activities toward bilirubin and 1-naphthol were reduced by approx. 90% and 40% respectively (results not shown), consistent with previous observations. Microsomes from both the homozygous deficient strain (j/j) and the wild-type strain (+/+) showed almost identical UDPGA uptake (Figure 8B), suggesting that at least the UGT1 family members do not significantly contribute to UDPGA uptake into the ER.
Figure 8. UDPGA uptake in Gunn rat liver microsomes.
Effects of UGT proteins on UDPGA uptake were examined with UGT1-deficient rat liver microsomes prepared from Gunn rats. (A) UGT protein in microsomes detected by immunoblotting. (B) Uptake of [14C]UDPGA in wild-type and UGT1-deficient rat liver microsomes. Gunn rat (j/j) homozygote (squares); wild-type (+/+) normal (circles). Solid lines and dotted lines show the time course of uptake after 90 min of preload with UDPGlcNAc and with water respectively. The uptake of water-preloaded wild-type rat liver microsomes was the same as Figure 1 and Gunn rat (j/j) liver microsomes (results not shown; n=2).
We found that H4IIE cells show similar UDPGlcNAc-dependent UDPGA uptake properties to rat liver microsomes (Figure 1B). Thus this cell line was used to see whether induction of UGT enzyme protein expression could affect UDPGA uptake into the ER. Cells were treated with the inducer βNF and increased UGT protein expression was observed in the βNF-treated cells compared with DMSO-treated cells (results not shown). On quantifying UDPGA uptake using the rapid filtration assay we found no obvious difference between these two treatments (results not shown), again indicating that the UGT proteins do not significantly influence UDPGA transport.
DISCUSSION
More than 50 years after the discovery of UDPGA [34,35], the mechanism(s) whereby this co-substrate for glucuronidation gains access to the UGT active site remain unclear. In the present study, we set out to test the hypotheses that members of the NST family may supply UDPGA into the ER lumen for small molecule glucuronidation, and that the UGT proteins themselves are involved in UDPGA uptake. We obtained cDNA clones coding for four NSTs and assessed their ability to transport UDPGA by establishing stable (V79) cell lines expressing these proteins. We also explored the endogenous UDPGA uptake activity in cell lines and found that H4IIE cells could be a useful model for the study of UDPA uptake.
In cells over-expressing the NSTs, UGTrel7 showed the highest level of UDPGA transport, however certain UDPGA uptake characteristics of this transport were different from those initially reported [16]. For example, we detected UDPGA uptake only after preloading with UDPGlcNAc (Figures 4C, 4D and 5B), whereas Muraoka et al. demonstrated 8 pmol of UDPGA uptake in 30 min without preloading [16]. It is possible that this discrepancy results from the different cell type used to express the protein in the present study (mammalian cells compared with S. cerevisiae), and therefore reflects the impact of e.g. different post-translational modifications and lipid composition of the ER membrane, on transport characteristics. In the present study, the cellular localization of UGTrel7 revealed by immunocytochemistry was broadly the same as that described in previous reports [16,36], with staining in the ER and nuclear envelope that overlapped with the ER marker calreticulin. Additionally, the effect of the di-lysine motif in the C-terminus on the intracellular localization of various NSTs has been studied [37]. These authors found that the UDP-galactose transporter 2 localized to the ER and Golgi and that this was associated with the di-lysine-like motif KVKGS (single letter amino acid code). UGTrel7 has a similar motif (KGKGAV; single letter amino acid code) at the C-terminus.
Kinetic analysis (Figure 7) suggested that, in rat liver microsomes, there are probably multiple transport system(s) for UDPGA, as proposed in an earlier report [38]. There is also a report showing differences in UDPGA uptake between ER and Golgi membranes [39]. Therefore our result obtained with V79/UGTrel7 cells, demonstrating that transport of UDPGA is UDPGlcNAc-dependent, may be important in the future to assist in revealing the UDPGA transporters essential for glucuronidation. In microsomes prepared from rat liver and H4IIE cells, there was a high degree of latency associated with UGT and glucose 6-phosphatase activities, therefore the orientation of vesicles is likely to be ‘inside-in’, i.e. in the correct orientation. In V79 cells expressing UGT1A6, latency was not as extensive, and so it is possible that some vesicles could be ‘inside-out’, or the membranes are more susceptible to damage during the preparation of the microsomes. However, we at least can say that expressed UGT1A6 in V79 cells demonstrates latency which is qualitatively similar to that of rat liver microsomes, although to a different degree.
It was hypothesized that dimerized and/or oligomerized UGT proteins are able to function as UDPGA carriers in the ER membrane [9,10]. We tested this hypothesis using three different approaches. First, in microsomes prepared from V79 cells there was no difference in the uptake of UDPGA between the parental (i.e. untransfected) V79 cells and the cell line expressing UGT1A6, (Figure 4A). Secondly, we used liver microsomes prepared from Gunn rats, which are deficient in the expression of all members of the UGT1 family due to a mutation in the shared common exon 4, to show that the absence of these UGT1 enzymes did not influence the transport of UDPGA. Thirdly, microsomes prepared from H4IIE cells pre-treated with the UGT inducer βNF showed no increase in transport of UDPGA despite a significant increase in the expression of the UGT1 family. It is of course possible that UGT2 protein(s) could be involved in ER transport of UDPGA, although βNF can induce certain members of the UGT2 family as well as several UGT1 isoforms, and levels of UGT2 proteins are essentially unaffected in the Gunn rat.
In conclusion, we have characterized various NSTs for their ability to transport the glucuronidation reaction co-substrate UDPGA in model in vitro systems, and have provided a body of evidence which suggests the UGTs themselves are not significantly involved in the transport of UDPGA. Our results confirm that the uptake of this compound into the ER lumen is a complex process, which most likely involves more than one transport molecule. Future studies aimed at further identification of the key features of this process will be able to use these model systems and the substantial data in the present study.
Acknowledgments
This work was funded by the Wellcome Trust. J.E.S. is a Royal Society University Research Fellow. We are grateful to Dr Alan Prescott, School of Life Sciences, University of Dundee, Dundee, Scotland, U.K. for helpful advice.
References
- 1.Burchell B., Coughtrie M. W. H. UDP-glucuronosyltransferases. Pharmacol. Ther. 1989;43:261–289. doi: 10.1016/0163-7258(89)90122-8. [DOI] [PubMed] [Google Scholar]
- 2.Dutton G. J., Burchell B. Newer aspects of glucuronidation. Prog. Drug Metab. 1977;2:1–70. [Google Scholar]
- 3.Dutton G. J. Boca Raton: CRC Press; 1980. Glucuronidation of drugs and other compounds. [Google Scholar]
- 4.Burchell B., Weatherill P. J., Berry C. Evidence indicating that UDP-N-acetylglucosamine does not appear to stimulate hepatic microsomal UDP-glucuronosyltransferase by interaction with the catalytic unit of the enzyme. Biochim. Biophys. Acta. 1983;735:309–313. doi: 10.1016/0005-2736(83)90143-8. [DOI] [PubMed] [Google Scholar]
- 5.Winsnes A. Kinetic properties of different forms of hepatic UDP-glucuronyltransferase. Biochim. Biophys. Acta. 1972;284:394–405. doi: 10.1016/0005-2744(72)90135-0. [DOI] [PubMed] [Google Scholar]
- 6.Berry C., Stellon A., Hallinan T. Guinea pig liver microsomal UDP-glucuronyltransferase. Biochim. Biophys. Acta. 1975;403:335–344. doi: 10.1016/0005-2744(75)90063-7. [DOI] [PubMed] [Google Scholar]
- 7.Berry C., Hallinan T. Summary of a novel three-component regulatory model for uridine diphosphate glucuronosyltransferase. Biochem. Soc. Trans. 1976;4:650–652. doi: 10.1042/bst0040650. [DOI] [PubMed] [Google Scholar]
- 8.Tephly T. R., Burchell B. UDP-glucuronosyltransferases: a family of detoxifying enzymes. Trends Pharmacol. Sci. 1990;11:276–279. doi: 10.1016/0165-6147(90)90008-v. [DOI] [PubMed] [Google Scholar]
- 9.Ikushiro S., Emi Y., Iyanagi T. Protein–protein interactions between UDP-glucuronosyltransferase isozymes in rat hepatic microsomes. Biochemistry. 1997;36:7154–7161. doi: 10.1021/bi9702344. [DOI] [PubMed] [Google Scholar]
- 10.Ikushiro S., Emi Y., Kimura S., Iyanagi T. Chemical modification of rat hepatic microsomes with N-ethylmaleimide results in inactivation of both UDP-N-acetylglucosamine-dependent stimulation of glucuronidation and UDP-glucuronic acid uptake. Biochim. Biophys. Acta. 1999;1428:388–396. doi: 10.1016/s0304-4165(99)00066-5. [DOI] [PubMed] [Google Scholar]
- 11.Meech R., Mackenzie P. I. UDP-glucuronosyltransferase, the role of the amino terminus in dimerization. J. Biol. Chem. 1997;272:26913–26917. doi: 10.1074/jbc.272.43.26913. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh S. S., Sappal B. S., Kalpana G. V., Lee S. W., Chowdhury J. R., Chowdhury N. R. Homodimerization of human bilirubin-uridine-diphosphoglucuronate glucuronosyltransferase-1 (UGT1A1) and its functional implications. J. Biol. Chem. 2001;276:42108–42115. doi: 10.1074/jbc.M106742200. [DOI] [PubMed] [Google Scholar]
- 13.Miura N., Ishida N., Hoshino M., Yamauchi M., Hara T., Ayusawa D., Kawakita M. Human UDP-galactose translocator: molecular cloning of a complementary DNA that complements the genetic defect of a mutant cell line deficient in UDP-galactose translocator. J. Biochem. (Tokyo) 1996;120:236–241. doi: 10.1093/oxfordjournals.jbchem.a021404. [DOI] [PubMed] [Google Scholar]
- 14.Eckhardt M., Gerardy-Schahn R. Molecular cloning of the hamster CMP-sialic acid transporter. Eur. J. Biochem. 1997;248:187–192. doi: 10.1111/j.1432-1033.1997.00187.x. [DOI] [PubMed] [Google Scholar]
- 15.Guillen E., Abeijon C., Hirschberg C. B. Mammalian golgi apparatus UDP-N-acetylglucosamine transporter: molecular cloning by phenotypic correction of a yeast mutant. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7888–7892. doi: 10.1073/pnas.95.14.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muraoka M., Kawakita M., Ishida N. Molecular characterization of human UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, a novel nucleotide sugar transporter with dual substrate specificity. FEBS Lett. 2001;495:87–93. doi: 10.1016/s0014-5793(01)02358-4. [DOI] [PubMed] [Google Scholar]
- 17.Csala M., Staines A. G., Banhegyi G., Mandl J., Coughtrie M. W. H., Burchell B. Evidence for multiple glucuronide transporters in rat liver microsomes. Biochem. Pharmacol. 2004;68:1353–1362. doi: 10.1016/j.bcp.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 18.Marsh M., Schmid S., Kern H., Harms E., Male P., Mellman I., Helenius A. Rapid analytical and preparative isolation of functional endosomes by free flow electrophoresis. J. Cell Biol. 1987;104:875–886. doi: 10.1083/jcb.104.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser S. C., Zuirys J. C., Gollan J. L. A membrane transporter mediates access of uridine 5′-diphosphoglucuronic acid from the cytosol into the endoplasmic reticulum of rat hepatocytes: implications for glucuronidation reactions. Biochim. Biophys. Acta. 1988;967:149–157. doi: 10.1016/0304-4165(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 20.Ishida N., Miura N., Yoshioka S., Kawakita M. Molecular cloning and characterization of a novel isoform of the human UDP-galactose transporter, and of related complementary DNAs belonging to the nucleotide-sugar transporter gene family. J. Biochem. (Tokyo) 1996;120:1074–1078. doi: 10.1093/oxfordjournals.jbchem.a021523. [DOI] [PubMed] [Google Scholar]
- 21.Bossuyt X., Blanckaert N. A mutant yeast deficient in transport of uridine diphosphate glucuronic acid. J. Hepatol. 2000;33:1023–1024. doi: 10.1016/s0168-8278(00)80141-5. [DOI] [PubMed] [Google Scholar]
- 22.Roy S. K., Chiba Y., Takeuchi M., Jigami Y. Characterization of yeast Yea4p, a uridine diphosphate-N-acetylglucosamine transporter localized in the endoplasmic reticulum and required for chitin synthesis. J. Biol. Chem. 2000;275:13580–13587. doi: 10.1074/jbc.275.18.13580. [DOI] [PubMed] [Google Scholar]
- 23.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Burchell B., Kennedy S. M. E., Jackson M. R., McCarthy L. The biosynthesis and induction of microsomal UDP-glucuronyltransferase in avian liver. Biochem. Soc. Trans. 1984;12:50–53. doi: 10.1042/bst0120050. [DOI] [PubMed] [Google Scholar]
- 25.Otani G., Abou-El-Makarem M. M., Bock K. W. UDP glucuronyltranferase in perfused rat liver and in microsomes iii effects of galactosamine and carbon. Biochem. Pharmacol. 1975;25:1293–1297. doi: 10.1016/0006-2952(76)90092-7. [DOI] [PubMed] [Google Scholar]
- 26.Burchell A., Hume R., Burchell B. A new microtechnique for the analysis of the human hepatic microsomal glucose-6-phosphatase system. Clin. Chim. Acta. 1988;173:183–191. doi: 10.1016/0009-8981(88)90256-2. [DOI] [PubMed] [Google Scholar]
- 27.Bossuyt X., Blanckaert N. Mechanism of stimulation of microsomal UDP-glucuronosyltransferase by UDP-N-acetylglucosamine. Biochem. J. 1995;305:321–328. doi: 10.1042/bj3050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fournel-Gigleux S., Sutherland L., Sabolovic N., Burchell B., Siest G. Stable expression of two human UDP-glucuronosyltransferase cDNAs in V79 cell cultures. Mol. Pharmacol. 1991;39:177–183. [PubMed] [Google Scholar]
- 29.Munro S., Pelham H. R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 30.Jackson M. R., Nilsson T., Peterson P. A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothman J. E., Wieland F. T. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 32.Johnson S., Michalak M., Opas M., Eggleton P. The ins and outs of calreticulin: from the ER lumen to the extracellular space. Trends Cell Biol. 2001;11:122–129. doi: 10.1016/s0962-8924(01)01926-2. [DOI] [PubMed] [Google Scholar]
- 33.Iyanagi T. Molecular basis of multiple UDP-glucuronosyltransferase isoenzyme deficiencies in the hyperbilirubinemic rat (Gunn rat) J. Biol. Chem. 1991;266:24048–24052. [PubMed] [Google Scholar]
- 34.Dutton G. J., Storey I. D. E. The isolation of a compound of uridine diphosphate and glucuronic acid from liver. Biochem. J. 1953;53:xxxvii–xxxviii. [PubMed] [Google Scholar]
- 35.Storey I. D. E., Dutton G. J. Uridine compounds in glucuronic acid metabolism. 2. The isolation and structure of ‘uridine-diphosphate-glucuronic acid’. Biochem. J. 1955;59:279–288. doi: 10.1042/bj0590279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suda T., Kamiyama S., Suzuki M., Kikuchi N., Nakayama K., Narimatsu H., Jigami Y., Aoki T., Nishihara S. Molecular cloning and characterization of a human multisubstrate specific nucleotide-sugar transporter homologous to Drosophila fringe connection. J. Biol. Chem. 2004;279:26469–26474. doi: 10.1074/jbc.M311353200. [DOI] [PubMed] [Google Scholar]
- 37.Kabuss R., Ashikov A., Oelmann S., Gerardy-Schahn R., Bakker H. Endoplasmic reticulum retention of the large splice variant of the UDP-galactose transporter is caused by a dilysine motif. Glycobiology. 2005;15:905–911. doi: 10.1093/glycob/cwi085. [DOI] [PubMed] [Google Scholar]
- 38.Battaglia E., Nowell S., Drake R. R., Mizeracka M., Berg C. L., Magdalou J., Fournel-Gigleux S., Gollan J. L., Lester R., Radominska A. Two kinetically-distinct components of UDP-glucuronic acid transport in rat liver endoplasmic reticulum. Biochim. Biophys. Acta. 1996;1283:223–231. doi: 10.1016/0005-2736(96)00098-3. [DOI] [PubMed] [Google Scholar]
- 39.Bossuyt X., Blanckaert N. Differential regulation of UDP-GlcUA transport in endoplasmic reticulum and in Golgi membranes. J. Hepatol. 2001;34:210–214. doi: 10.1016/s0168-8278(00)00083-0. [DOI] [PubMed] [Google Scholar]