Abstract
The liver is exposed to a wide variety of toxic agents, many of which damage DNA and result in increased levels of the tumour suppressor protein p53. We have previously shown that p53 inhibits the transactivation function of HNF (hepatocyte nuclear factor) 4α1, a nuclear receptor known to be critical for early development and liver differentiation. In the present study we demonstrate that p53 also down-regulates expression of the human HNF4α gene via the proximal P1 promoter. Overexpression of wild-type p53 down-regulated endogenous levels of both HNF4α protein and mRNA in Hep3B cells. This decrease was also observed when HepG2 cells were exposed to UV irradiation or doxorubicin, both of which increased endogenous p53 protein levels. Ectopically expressed p53, but not a mutant p53 defective in DNA binding (R249S), down-regulated HNF4α P1 promoter activity. Chromatin immunoprecipitation also showed that endogenous p53 bound the HNF4α P1 promoter in vivo after doxorubicin treatment. The mechanism by which p53 down-regulates the P1 promoter appears to be multifaceted. The down-regulation was partially recovered by inhibition of HDAC activity and appears to involve the positive regulator HNF6α. p53 bound HNF6α in vivo and in vitro and prevented HNF6α from binding DNA in vitro. p53 also repressed stimulation of the P1 promoter by HNF6α in vivo. However, since the R249S p53 mutant also bound HNF6α, binding HNF6α is apparently not sufficient for the repression. Implications of the p53-mediated repression of HNF4α expression in response to cellular stress are discussed.
Keywords: doxorubicin, hepatocyte nuclear factor 4α (HNF4α), hepatocyte nuclear factor 6 (HNF6), nuclear receptor, p53
Abbreviations: Ad, adenovirus; β-gal, β-galactosidase; ChIP, chromatin immunoprecipitation; Co-IP, co-immunoprecipitation; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; GST, glutathione S-transferase; HA, haemagglutinin; HDAC, histone deacetylase; HNF, hepatocyte nuclear factor; MOI, multiplicity of infection; RLU, relative light units; Sp1, specificity protein 1; TSA, trichostatin acid
INTRODUCTION
The liver, as the major detoxification organ in the body, is exposed to a wide variety of toxic agents, many of which damage DNA and result in increased levels of the tumour suppressor protein p53. p53 is one of the most commonly mutated genes in human cancer and has a key role in repressing cell growth and inducing apoptosis in response to DNA damage, hypoxia and other stress signals [1]. Much of the effect of p53 is due to its ability to activate or repress transcription of target genes. Whereas the mechanism of activation is fairly well characterized, the mechanism by which p53 represses transcription remains somewhat elusive [2–5]. In some cases, p53 binds a specific DNA response element and prevents binding of a transcriptional activator [e.g. HNF (hepatocyte nuclear factor) 3, Sp1 (specificity protein-1) and E2F]. In other cases, p53 recruits HDAC (histone deacetylase) or DNMT1 (DNA methyltransferase) to the target promoter, thereby inducing a closed, transcriptionally inactive chromatin conformation [e.g. Map4 (microtubule-associated protein 4) and survivin] [2,6].
p53 can also repress gene expression through mechanisms that do not involve binding to cis-acting promoter elements (FOS, JUN, RB1, IL6, BCL2 [3]). In the case of nuclear receptors, p53 binds directly to the transcriptional activator and blocks DNA binding [7–9]. We have also previously observed that p53 can repress the transactivation function of HNF4α1 by binding the HNF4α activation domain and recruiting HDAC activity to HNF4α target genes [10]. This represents a novel combination of the above mechanisms and suggests a role for p53 as an important regulator of HNF4α activity.
HNF4α, a member of the nuclear receptor superfamily of ligand-dependent transcription factors (NR2A1), is found primarily in the liver, kidney, intestine and, to a lesser extent, in the pancreas and stomach in the adult [11]. Repression of HNF4α activity by p53 is potentially important in the response to liver injury, since HNF4α has been shown to be critical for the adult liver phenotype and to act as a linchpin in a transcription factor network that drives hepatocyte differentiation [12–14]. HNF4α stimulates the expression of transcriptional activators such as the cut-homeodomain protein HNF6α and the pou-homeodomain protein HNF1α, which in turn activate HNF4α gene expression in a positive feedback loop [14]. Overall, HNF4α plays an essential role in the regulation of over 60 genes critical to metabolism and nutrient transport and has been linked to several human diseases including diabetes, haemophilia and hepatitis [11,15]. HNF4α is also known to be critical during development where it is first found in the primary endoderm at embryonic day 4.5 in mouse and is essential for survival beyond embryonic day 10.5 [14].
There are two promoters in the HNF4α gene which yield a total of nine potential splice variants [11]. The proximal P1 promoter drives the expression of the major isoforms in the adult hepatocyte, intestine and kidney (primarily HNF4α1 and HNF4α2); HNF4α2 differs from HNF4α1 by a 10 amino acid insertion in the C-terminal region (see Figure 1C). The distal P2 promoter drives the expression of isoforms present primarily in the embryonic liver and adult stomach and pancreas (primarily HNF4α7/8); HNF4α7/8 are 13 amino acids shorter than HNF4α1/2 due to a different N-terminus [16–19].
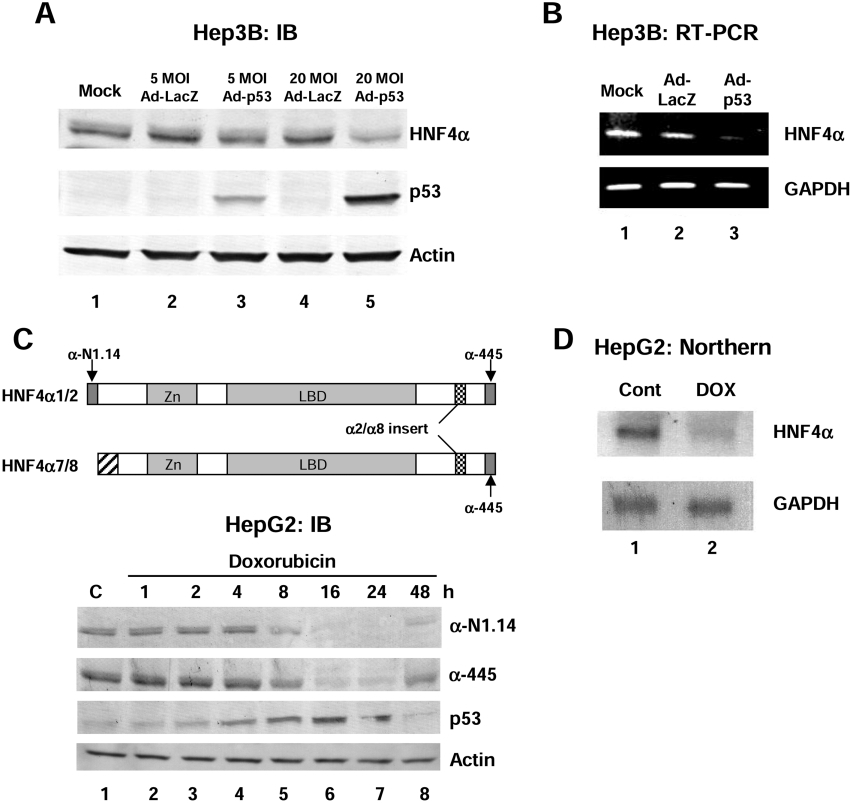
Figure 1. Over expression of wild-type p53 in human liver cancer cell lines results in a decrease in HNF4α1/2 protein and mRNA levels.
(A) IB (immunoblot) analysis using the colour reaction of whole cell extracts from Hep3B cells (60 μg of protein per lane) harvested 48 h after infection of recombinant adenovirus expressing wild-type p53 (Ad-p53) using antibodies against HNF4α (α-445), p53 (DO-1) and β-actin as indicated. Recombinant adenovirus expressing LacZ was used as a control (Ad-LacZ). Similar results were observed using the αN1.14 antibody [results not shown, see (C) below]. Mock, media only added. (B) RT-PCR analysis using HNF4α or GAPDH primers. The template was 0.5 μg of total RNA from Hep3B cells 36 h after infection with Ad-p53 or Ad-LacZ. (C) Upper panel: schematic diagram of HNF4α1/2 and HNF4α7/8 isoforms and the antibodies that recognize them. α-445 recognizes all four isoforms. α-N1.14 recognizes only the isoforms from the P1 promoter (HNF4α1 and HNF4α2). Lower panel: IB (immunoblot) analysis using the colour reaction of whole cell extracts from HepG2 cells (50 μg of total protein per lane) harvested after exposure to doxorubicin (0.5 μg/ml) for the times indicated using antibodies against HNF4α (α-445 and α-N1.14), p53 (DO-1) and β-actin as indicated. C, control (not treated). (D) Northern blot analysis with HNF4α or GAPDH cDNA probes and 8 μg of total RNA from HepG2 cells treated with 0.5 μg/ml doxorubicin (DOX) for 24 h or untreated (Cont).
Previous studies have shown that the proximal P1 promoter region of the human HNF4α gene contains binding sites for HNF1α, Sp1, HNF6α and GATA-6, and is sufficient to drive high levels of expression in cultured human hepatocellular carcinoma/hepatoblastoma HepG2 cells [20]. The P1 promoter region has also been shown to be involved in a dynamic process with the upstream enhancer regions during the differentiation of human Caco-2 (colon carcinoma cells) cells [21]. In the present study we show that the P1 promoter of the human HNF4α gene is down-regulated by elevated p53 protein levels induced by recombinant adenovirus infection, DOX (doxorubicin) treatment and UV irradiation. We also present evidence indicating that the mechanism by which p53 represses the HNF4α promoter is complex and may include both HDAC recruitment and interaction of p53 with HNF6α, an activator of HNF4α transcription.
EXPERIMENTAL
Plasmids
Human wild-type p53 in pcDNA3.1 (p53wt) has been previously described [10]. The R249S mutant in human p53 was subcloned from the appropriate BamHI fragment of CMV.p53mt249 (a gift from Dr Bert Vogelstein, The John Hopkins University, Baltimore, MD, U.S.A.) into pcDNA3.1 (p53mt249). The luciferase reporter construct containing the human HNF4α P1 promoter from −560 to +67 bp (pGL3.hHNF4) [20] and the expression vector containing rat HNF6α (pECE.HNF6α and pGEX.HNF6α) [22,23] were generous gifts from Dr Iannis Talianidis (Foundation for Research and Technology Hellas, Crete, Greece) and Dr Frédéric Lemaigre (Université Catholique de Louvain, Brussels, Belgium) respectively. A potential p53 binding site (−329 to −304) was deleted using XhoI digestion which removed 26 nucleotides spanning −330 to −305 from the pGL3.hHNF4α reporter (pGL3.hHNF4α.Δp53). The pcDNA3.HNF6α construct was made by inserting a PCR product from pECE.ratHNF6α using appropriate primers. All constructs were verified by dideoxy sequencing (University of California Riverside Core Instrumentation Facility).
Cell culture, cell extracts and RNA preparation
Human hepatocellular carcinoma/hepatoblastoma HepG2 and Hep3B cell lines were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FBS (foetal bovine serum), 0.1 mM non-essential amino acids, 1 mM sodium pyruvate (Hep3B only) and penicillin/streptomycin. All cells were grown at 37 °C in 5% CO2. Cells were treated with doxorubicin (Sigma) at a final concentration of 0.5 μg/ml for 24 h unless otherwise indicated. After removing the media, HepG2 cells in 100 mm dishes were UV irradiated by exposure to a germicidal UV lamp (254 nm) and the cells were harvested after 12 or 24 h as indicated (Spectrolinker, XL-1000UV Crosslinker, Spectronics). Whole cell extracts from six-well plates were prepared by gentle scraping in 0.3 ml per well of Nonidet P40 lysis buffer [50 mM Tris/HCl (pH.7.4), 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P40, 0.5 mM PMSF and 1 mM DTT (dithiothreitol)] followed by centrifugation at 13000 g for 15 min at 4 °C to pellet the debris. RNA for Northern blotting and RT (reverse transcriptase)-PCR analysis was harvested from the cells using TRIzol® reagent (Life Technologies) as specified by the manufacturer.
Immunoblot analysis
Immunoblot analysis was performed as previously described [10] with a 1:5000 dilution of antisera raised in rabbits against peptides corresponding to the C-terminus of HNF4α (α-445) or the N-terminus of HNF4α1/2 (α-N1.14) [24,25], a 1:1000 dilution of an antibody against β-actin (Sigma), 0.5 μg/ml final concentration of antibodies for p53 (DO-1 or FL-393), HNF6α (H-100), HNF1α (C-19) and Sp1 (PEP 2) (Santa Cruz Biotechnology), a 1:5000 dilution of HNF6α N-terminus antibody (a gift from Dr R. Costa, University of Illinois at Chicago) and a 1:5000 dilution of AP (alkaline phosphatase) or HRP (horseradish peroxidase)-conjugated goat anti-rabbit secondary antibodies (Jackson ImmunoResearch). Signals were detected using the colour reaction or ECL® Western blotting detection reagent kit (Amersham Bioscience).
Northern blot analyses
Total RNA (8 μg) was fractionated on a 1.2% agarose–formaldehyde gel, transferred to GeneScreen® membrane (PerkinElmer) and hybridized with random primed 32P-labelled cDNA probes for HNF4α (rat HNF4α1, amino acids 127–455) or human GAPDH (approx. 1 kb) in ULTRAhyb hybridization buffer (Ambion) at 42 °C overnight. Blots were washed twice for 5 min at room temperature (25 °C) in 2× SSC buffer (0.3 M NaCl and 30 mM sodium citrate) and once at 42 °C in 0.1× SSC and 0.1% SDS, dried and subjected to autoradiography.
Transient transfection assays
One day before transfection, HepG2 cells were plated in six- or twelve-well plates (0.5×106 and 2.4×105 cells respectively) (Becton Dickinson). DNA mixtures containing pGL3.hHNF4α, CMV.β-gal and expression vectors were added using Lipofectamine™ 2000 (Invitrogen). After 24–30 h, transfected cells were harvested and luciferase and β-gal (β-galactosidase) activities were determined as previously described [10]. The data are presented as the mean±S.D. of the luciferase activity [RLU (relative light unit) normalized to β-gal activity (normalized RLU) from triplicate wells from one of at least three representative experiments.
Ad (adenovirus) infection of Hep3B cells
Ad-CMV/LacZ (Ad-LacZ control) and Ad-CMV/p53wt (human) (Ad-p53) [26] were kindly provided by Dr Jack A. Roth (Department of Thoracic and Cardiovascular Surgery, University of Texas M.D. Anderson Cancer Center, Houston, TX, U.S.A.). Hep3B cells were plated in a six-well plate with 1.0×106/well and infected 24 h later with 5 or 20 MOI (multiplicity of infection; determined in HEK-293 cells) of recombinant Ad (Ad-LacZ or Ad-p53) in 0.4 ml medium for 2 h, after which time 1.5 ml of medium were added and the incubation was continued for 36 h or 48 h until the time of harvest.
RT-PCR
RT-PCR was performed using the Access RT-PCR System (Promega). The RT reaction was performed at 48 °C for 45 min, followed by incubation at 94 °C for 2 min and then amplification by 18 cycles of PCR (94 °C for 30 sec, 60 °C for 1 min and 68 °C for 2 min) followed by a 7 min extension at 68 °C in a DNA Thermal Cycler 480 (PerkinElmer). Aliquots of each PCR reaction mixture were run on 1.5% agarose gels and visualized by ethidium bromide staining. Primers for RT-PCR were 5′-CGACCACTTTGTCAAGCTCA-3′ (forward, +964 nt) and 5′-AGGGGAGATTCAGTGTGGTG-3′ (reverse, +1166 nt) for GAPDH and 5′-CGAGCAGATCCAGTTCATCA-3′ (forward, +1145 nt) and 5′-TCACACATCTGTCCGTTGCT-3′ (reverse, +1345 nt) for HNF4α.
EMSA (electrophoretic mobility-shift analysis) and GST (glutathione-S-transferase) pulldown assay
Shift reactions and gel analysis with crude nuclear extracts of HepG2 cells were carried out essentially as previously described [27]. Purified wild-type human p53 protein was prepared from HeLa cells infected with a recombinant vaccinia virus containing HA (haemagglutinin) epitope-tagged wild-type human p53 by immunoaffinity purification with the 12CA5 antibody (recognizing the HA tag at the N-terminus) [28,29]. In vitro protein–protein interaction assays were performed using GST, GST.p53wt or GST.HNF6α fusion protein [23,30] as previously described [31]. GST protein (5 μg) was incubated at 4 °C with 4 μl of in vitro-translated 35S-labelled HNF6α, p53wt or p53.mt249 protein in rabbit reticulocyte lysate (TNT, Promega) for 4–8 h with gentle agitation before extensive washing and elution with SDS buffer and analysis by SDS/PAGE (10% gels) followed by autoradiography.
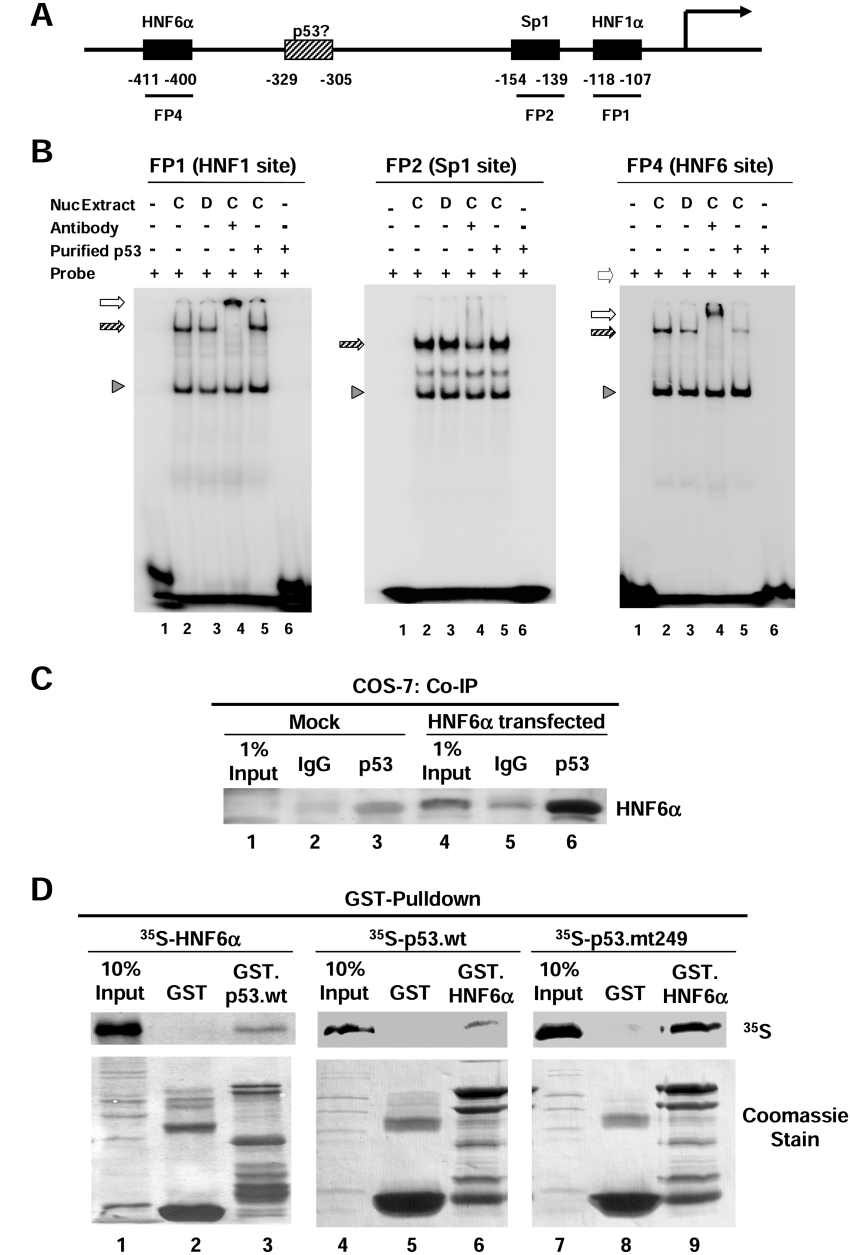
Co-IP (co-immunoprecipitation) assay
For Co-IP assays, 3.75×106 COS-7 cells were seeded in 150 mm plates 24 h before transfection with 25 μg of pcDNA3.HNF6α. Whole cell lysates were collected at 30 h after treatment using an Nonidet P40 lysis buffer [50 mM Tris/HCl (pH 8), 120 mM NaCl, 0.5% Nonidet P40, 1 mM DTT, 2 μg/ml aprotinin and 2 μg/ml leupeptin]. Crude whole cell extract (1 ml) was incubated with 5 μg of anti-p53 antibody (DO-1) or control mouse IgG at 4 °C for 2 h. Then 40 μl of pre-washed Protein A beads (Pierce) were added to the mixture and incubated at 4 °C overnight with gentle agitation. After extensive washing with a 0.1% Nonidet P40 lysis buffer, p53-interacting proteins were eluted with SDS buffer, and analysed by immunoblot analysis using a HNF6α N-terminus antibody.
ChIP (chromatin immunoprecipitation) assay
HepG2 cells were treated with 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by the addition of 0.125 M glycine (final concentration). Cells were harvested in cold PBS and lysed in ChIP sonication buffer [1% Triton X-100, 0.1% deoxycholate, 50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 2 μg/ml aprotinin, 2 μg/ml leupeptin and 0.2 mM PMSF]. The DNA fragments were sonicated to an average size of 500–1000 bp using a Sonifier Model 450 (Branson). Immunoprecipitations were performed with α-Sp1 (PEP2), α-HNF6α (H-100) and α-p53 (FL-393) antibodies, and DNA–protein complexes were eluted in 1% SDS elution buffer (1% SDS, 0.1 M NaHCO3 and 0.01 mg/ml herring sperm DNA). The crosslinks were reversed by heating at 65 °C overnight, proteins were digested by proteinase K (0.17 μg/μl), and the DNA was extracted with phenol/chloroform, precipitated with ethanol and dissolved in 100 μl TE buffer [10 mM Tris/HCl (pH 8.0) and 1 mM EDTA]. PCR amplification (35 cycles) was performed with 5 μl template DNA and primers spanning the P1 promoter region of the human HNF4α gene (−472 nt 5′- ATTTAGGTTTCTAAATCGTGGGCC-3′ and −25 nt 5′-AACCGTCCTCTGGGAAGATCTGCT-3′). The products were analysed by agarose gel electrophoresis and visualized by ethidium bromide staining.
Data Analysis
Statistical analyses were performed by the Student's t test and all differences reported as significant have P<0.05.
RESULTS
Ectopically expressed p53 decreases HNF4α protein and mRNA levels
We have previously shown that p53 inhibited the transactivation function of HNF4α1 protein [10]. To determine whether p53 also affected HNF4α gene expression, we overexpressed wild-type human p53 in a human hepatocellular carcinoma/hepatoblastoma cell line (Hep3B) using recombinant adenovirus. Hep3B cells express endogenous HNF4α but not p53. When p53 was ectopically expressed (Figure 1A, lanes 3 and 5, middle panel), the level of endogenous HNF4α protein was significantly decreased (Figure 1A, lanes 3 and 5, top panel). p53 overexpression also decreased HNF4α mRNA (Figure 1B), suggesting that the effect of p53 was on the level of transcription of the HNF4α gene.
Elevated levels of endogenous p53 protein are associated with a decrease in endogenous HNF4α1/2 protein and mRNA levels
To determine whether elevated levels of endogenous p53 were also associated with a decrease in HNF4α protein expression, we treated HepG2 cells with doxorubicin, an anti-cancer agent known to increase p53 protein levels [32]. HepG2 is a human hepatocellular carcinoma/hepatoblastoma cell line known to express both HNF4α and wild-type p53. Doxorubicin treatment increased p53 protein levels in a time-dependent fashion starting at 4 h (Figure 1C, panel labelled p53) and decreased the level of the endogenous HNF4α protein starting at approx. 8 h. Since a decrease in signal was observed using antibodies that recognized both the C-terminus (Figure 1C, panel labelled α-445) and the N-terminus of HNF4α1/2 (Figure 1C, panel labelled α-N1.14), we concluded that the protein being identified in these blots was HNF4α1/2, and not HNF4α7/8 (Figure 1C). Northern blot analysis confirmed that doxorubicin treatment decreased HNF4α mRNA levels (Figure 1D), suggesting that, just as in Hep3B cells, the effect of elevated p53 is on the level of HNF4α gene transcription. Since similar results with the α-N1.14 antibody were obtained in the Hep3B Ad experiment (results not shown), we concluded that elevated levels of exogenously or endogenously expressed p53 protein affected the HNF4α P1 promoter that drives expression of the primary adult liver forms of HNF4α, HNF4α1/2. It is difficult to distinguish between HNF4α1 and HNF4α2 because they differ by only 10 amino acids, hence the HNF4α1/2 designation. The multiple bands occasionally seen in the blots can also be due to differentially phosphorylated forms of HNF4α [33].
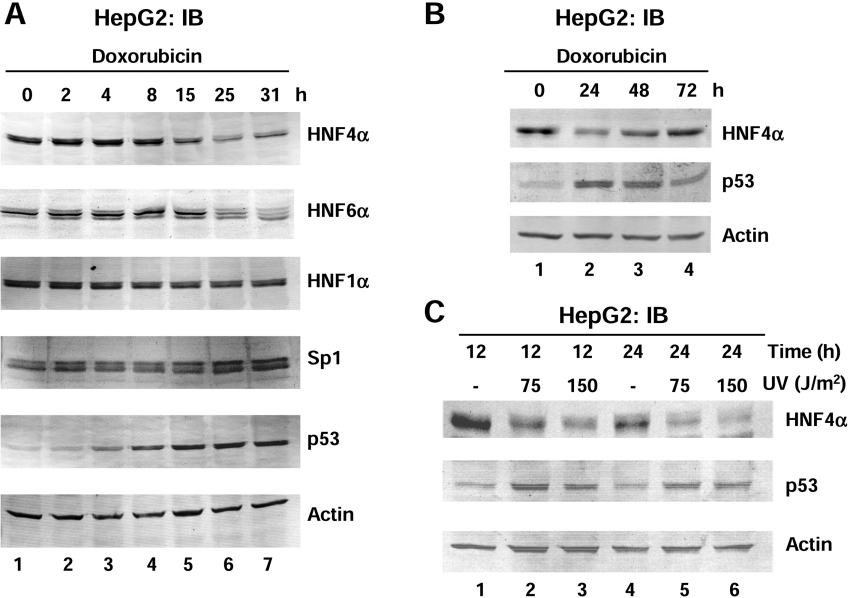
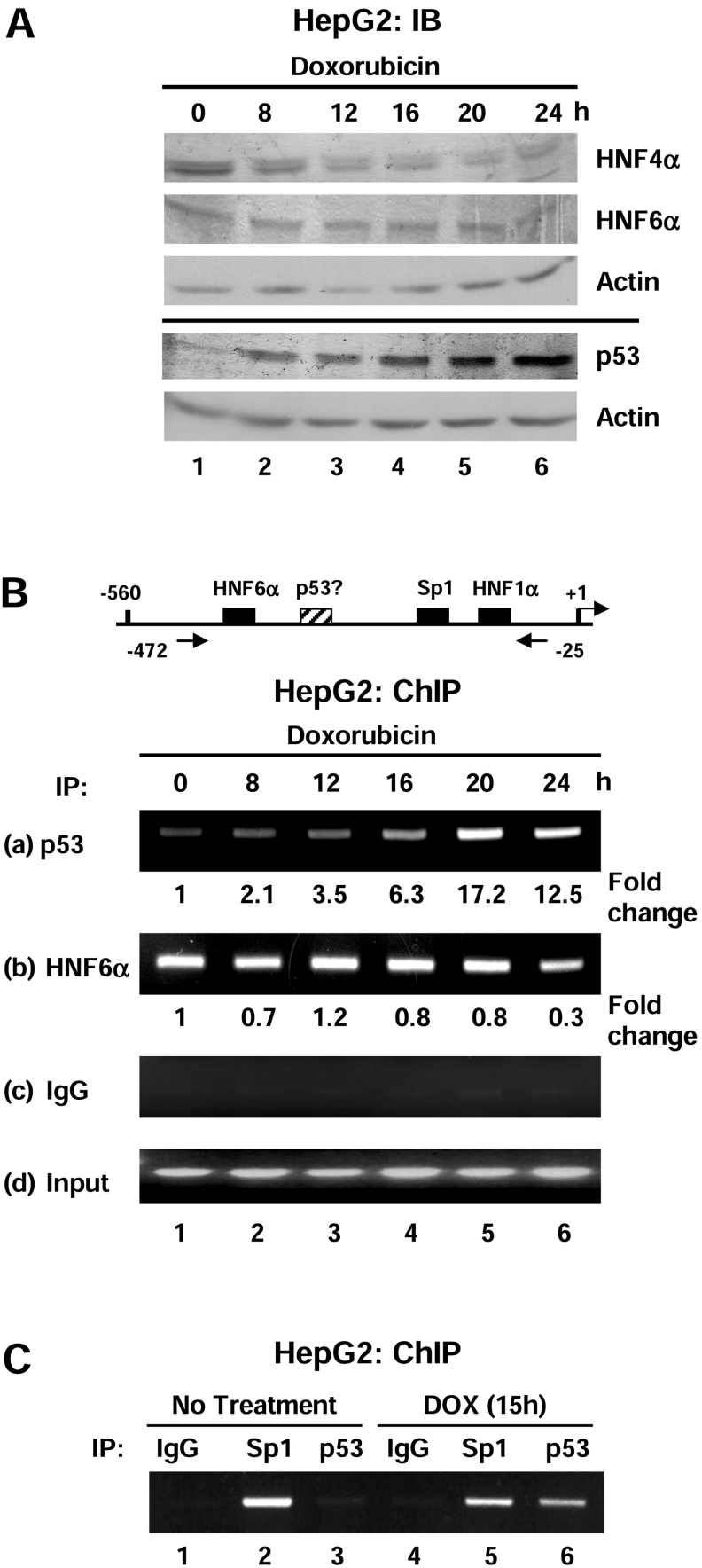
To determine whether the effect on HNF4α1/2 protein and mRNA levels might be due to a decrease in transcription factors known to activate the proximal P1 promoter of the HNF4α gene, the levels of liver-enriched transcription factors HNF6α and HNF1α and the ubiquitous transcription factor Sp1 were monitored after doxorubicin treatment. Again, p53 protein levels increased and HNF4α1/2 decreased in a similar manner to that seen in Figure 1(C); however, HNF1α and actin levels remained constant and Sp1 slightly increased (Figure 2A), indicating that the effect of doxorubicin on HNF4α1/2 was not due to a global decrease in transcription. HNF6α protein levels also decreased but only after 25 h (Figure 2A, panel labelled HNF6α), which could be due to the fact that HNF4α regulates the HNF6α gene [34]. Taken together, these results indicated that the decrease in HNF4α1/2 protein levels was not due to lower levels of the transcription factors that up-regulate the P1 promoter.
Figure 2. High levels of endogenous p53 protein induced by DNA damaging agents are associated with decreased HNF4α1/2 protein and mRNA levels.
(A and B) IB (immunoblot) as in Figure 1 (C) of HepG2 cells treated with doxorubicin (0.5 μg/ml) for the times indicated. Protein levels were analysed using antibodies against HNF4α (α-445), HNF6α (H-100), HNF1α, Sp1, p53 (DO-1) or β-actin. (C) IB analysis as in (A) of whole cell extracts from HepG2 cells harvested after exposure to UV (75 J/m2 or 150J/m2) at the times indicated.
The effect of doxorubicin was reversible as at 48 h p53 protein levels decreased and HNF4α1/2 protein levels increased, and both returned to near normal levels by 72 h (Figure 2B). UV, another DNA damaging agent known to increase p53 levels, also decreased the HNF4α1/2 protein levels (Figure 2C), further confirming that high levels of endogenous, stress-induced p53 protein correlate with a decrease in HNF4α1/2 protein and mRNA levels.
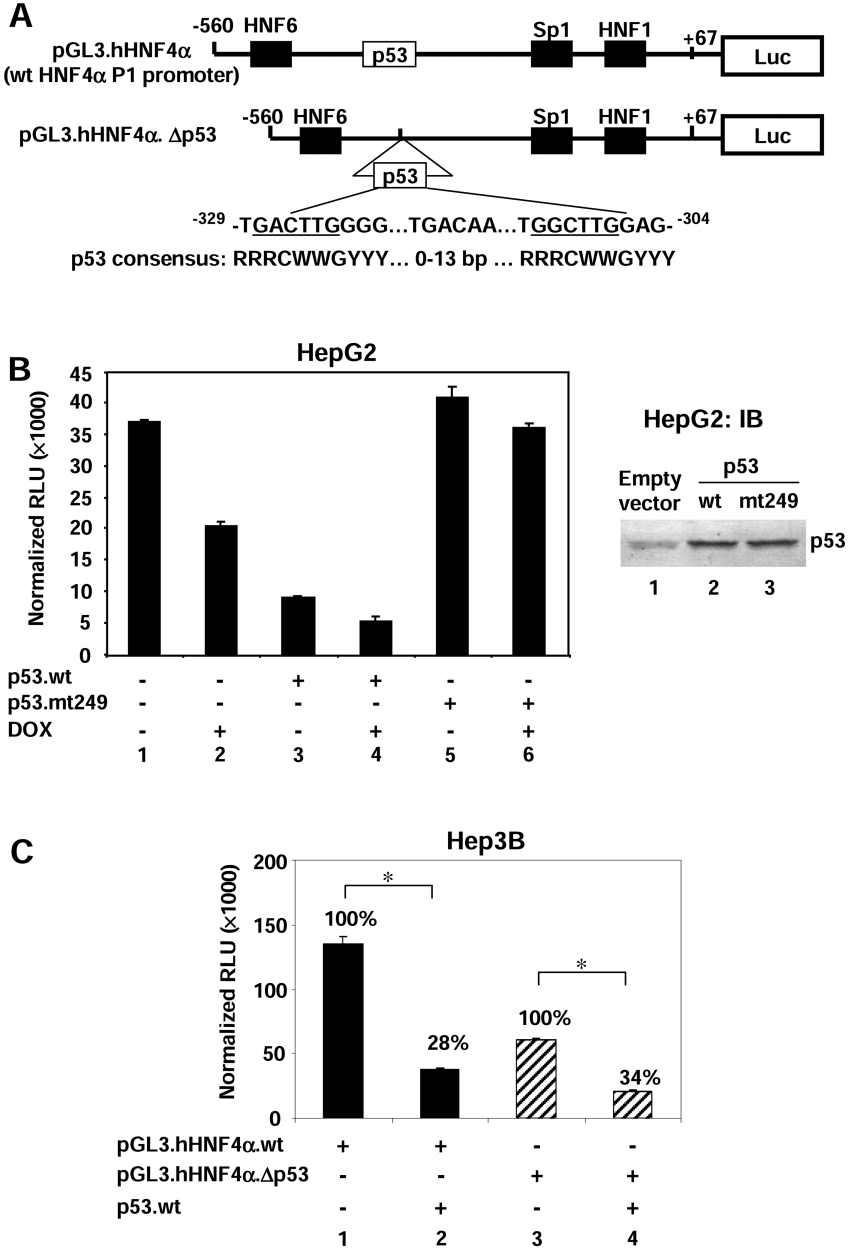
Wild-type but not mutant p53 down-regulates the HNF4α P1 promoter
To determine whether p53 acts directly on the HNF4α P1 promoter, we performed transient transfection analysis with a luciferase reporter construct containing a fragment of the human HNF4α P1 promoter (pGL3.hHNF4α) (Figure 3A). The P1 promoter has been shown previously to be responsible for much of the expression of the HNF4α gene in HepG2 cells [20]. Upon cotransfection into HepG2 cells, a wild-type p53 expression vector repressed the HNF4α P1 promoter activity (Figure 3B, lane 3 compared with lane 1) but a construct containing a DNA-binding domain mutant of p53 often found in liver cancer cells (R249S, p53mt249) did not (Figure 3B, lane 5). Immunoblot analysis indicated that both the wild-type and mutant p53 proteins were expressed at comparable levels (Figure 3B, right-hand panel). Doxorubicin treatment also decreased P1 promoter activity (Figure 3B, lane 2) and enhanced the repression of p53 (Figure 3B, lane 4), possibly due to increased expression of the endogenous p53. In contrast, doxorubicin treatment did not substantially repress HNF4α P1 promoter activity in the presence of mutant p53 (Figure 3B, lane 6). p53 mutants such as R249S are known to display dominant-negative behaviour because incorporation of a single mutant protein into the p53 tetramer is sufficient to inhibit DNA binding [35]. Thus the prevention of doxorubicin-mediated down-regulation of the HNF4α P1 promoter by the mutant p53 might be due to a dominant-negative effect of the transfected mutant p53 on the endogenous wild-type p53 protein, and suggests that the binding of p53 to the HNF4α P1 promoter may be involved.
Figure 3. p53 represses the activity of the human HNF4α P1 promoter.
(A) Schematic diagram of the reporter construct of the wild-type human HNF4α P1 promoter driving luciferase (Luc) (pGL3.hHNF4α). A potential p53 binding site at −329 to −304 was deleted in pGL3.hHNF4α.Δp53. The sequence of this site loosely fits the p53 consensus sequence, given in IUBMB nomenclature (R=G or A, W=A or T,Y=T or C). (B) Left-hand panel: transient transfection into HepG2 cells of pGL3.hHNF4α (2 μg), CMV.β-gal (2 μg), pcDNA3.p53wt (3 μg) or pcDNA3.p53mt249 (3 μg) in the absence or presence of doxorubicin (DOX; 0.5 μg/ml) as indicated. Cells were harvested at 24 h after transfection. Results are means±S.D. of one representative experiment performed in triplicate. Right-hand panel: IB analysis was performed using the colour reaction of whole cell extracts from HepG2 cells (70 μg of total protein per lane) transfected with pcDNA3 empty vector, pcDNA3.p53wt or pcDNA3.p53mt249 (3 μg each) as indicated using a p53 antibody (DO-1) to show equivalent expression of the wild-type and mutant p53 proteins. (C) Hep3B cells were cotransfected with pGL3.hHNF4α (2 μg), pGL3.hHNF4α.Δp53 (2 μg), CMV.β-gal (2 μg) or pcDNA3.p53wt (3 μg) as indicated. Percentages relative to pGL3.hHNF4α or pGL3.hHNF4α.Δp53 alone are shown. Results are means±S.D. of one representative experiment performed in triplicate. *P<0.05. Similar results were obtained in HepG2 cells (results not shown).
We identified a potential p53 binding element (−329 to −304) in the human HNF4α P1 promoter that nominally matches the p53 consensus site (Figure 3A). To determine whether p53 represses HNF4α gene expression specifically via this potential binding element, we deleted nucleotides −330 to −305 from the promoter (pGL3.hHNF4α.Δp53). The reporter assay revealed that wild-type p53 protein still repressed the promoter activity (Figure 3C, lane 4) even though the basal activity of pGL3.hHNF4α.Δp53 was much lower than the wild-type promoter (Figure 3C, compare lane 3 with lane 1). This result suggests that either p53 suppresses HNF4α gene expression via a different response element in the P1 promoter, or that it represses transcription via a mechanism that does not require DNA binding. It should be noted that the R249S mutation not only inhibits the ability of p53 to bind DNA but also alters the local three-dimensional structure of the DNA binding domain [36]. Therefore, it is also possible that the lack of repression by R249S is due not only to the lack of DNA binding but also to some other, as yet unidentified, function of the DNA-binding domain.
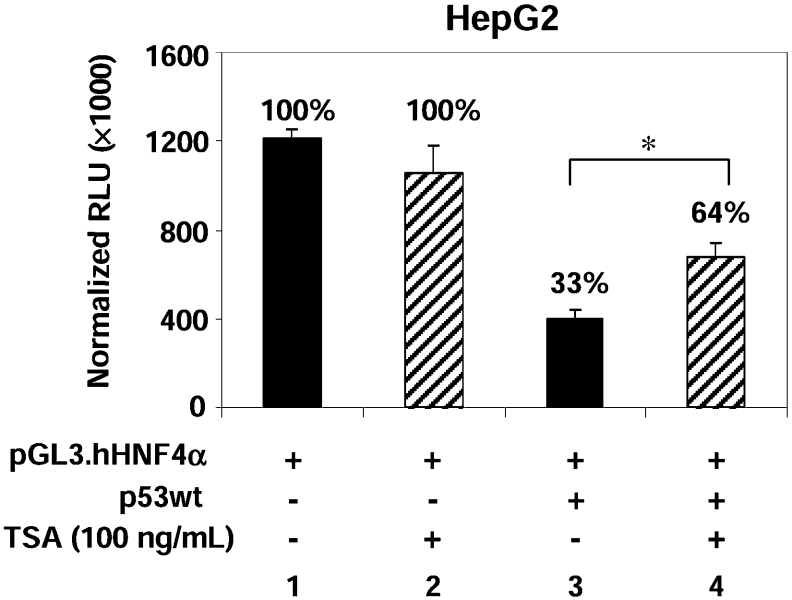
An HDAC inhibitor, TSA (trichostatin A), partially recovers p53-mediated repression of the HNF4α P1 promoter
Modifications of histones, especially acetylation, often play a critical role in regulating gene expression. Therefore to determine whether p53 represses HNF4α P1 promoter activity by recruitment of HDAC activity, we examined the effect of the HDAC inhibitor TSA in the luciferase reporter assay (Figure 4). Although TSA did not have a significant effect on the basal promoter activity (Figure 4, lane 2), it did cause a significant, although not complete, recovery of promoter activity in the p53-treated cells (Figure 4, lane 4). This suggests that the recruitment of HDAC activity plays a partial role in p53-mediated repression of the HNF4α P1 promoter.
Figure 4. HDAC inhibitor TSA partially recovers the repression of the HNF4α P1 promoter by p53.
HepG2 cells were co-transfected with pGL3.hHNF4α (0.5 μg), pcDNA3.p53wt (0.5 μg) or pcDNA3 empty vector (0.5 μg), and CMV.β-gal (0.2 μg) as indicated. Cells were treated with 100 ng/ml TSA 8 h before harvesting (20 h after addition of DNA). Percentages relative to the reporter alone are given. Results are means±S.D. of one representative experiment performed in triplicate. *P<0.05.
Potential involvement of HNF6α in the p53-mediated repression of the HNF4α P1 promoter
To determine whether p53 inhibits HNF4α gene expression by affecting the binding of other transcriptional activators to the HNF4α promoter, EMSA was performed using probes corresponding to binding sites for HNF1α, Sp1 and HNF6α (FP1, FP2 and FP4 respectively [20], Figure 5A). HNF1α and Sp1 binding were not significantly affected by a 15 h doxorubicin treatment of HepG2 cells (Figure 5B, panel labelled FP1 and FP2, compare lane 3 with lane 2). However, HNF6α DNA binding was somewhat decreased (Figure 5B, panel labelled FP4, compare lane 3 with lane 2). Furthermore, when EMSA was performed with extracts from untreated cells in the presence of purified p53 protein, the binding of HNF6α was nearly completely abolished, but that of HNF1α or Sp1 was not affected (Figure 5B, all panels, lane 5). This suggests that p53 specifically inhibits the ability of HNF6α to bind DNA in vitro.
Figure 5. p53 interacts with HNF6α in vivo and in vitro and inhibits its binding to the HNF4α P1 promoter in vitro.
(A) Schematic diagram of HNF4α P1 promoter adapted from [20]. (B) DNA binding activities analysed by EMSA performed with HepG2 nuclear extracts (700 ng each) from control cells (C) or cells treated with doxorubicin (D; 0.5 μg/ml) for 15 h in the presence or absence (+ or −) of purified wild-type p53 protein and 32P-labelled probes of HNF1α, Sp1 and HNF6α response elements of the HNF4α P1 promoter as indicated (FP1, FP2 and FP4). Shift complexes (hatched arrows) were verified by supershift (open arrows) using antibodies against HNF1α or HNF6α for the FP1 and FP4 probes respectively. A non-specific complex (arrowhead) serves as a control for loading and provides evidence of the specificity of the inhibition of HNF6α binding by p53 (right-hand panel, lane 5). Shown are autoradiograms of the shift gels. (C) Co-IP assay using untransfected (Mock) and HNF6α-transfected COS-7 cells. The immunoprecipitation was performed using the mouse monoclonal antibody that recognizes p53 (DO-1; lanes 3 and 6) and mouse IgG as a control (lanes 2 and 5). The immunoblot was performed using anti-HNF6 N-terminus antibody and ECL® detection. Faint bands in the Mock Co-IP (lanes 2 and 3) were most likely caused by cross-reaction with the heavy chain of the antibodies for immunoprecipitation. (D) GST pull-down assay was performed with in vitro-synthesized 35S-labelled HNF6α, 35S-labelled p53.wt or p53.mt249 and immobilized GST, GST.p53.wt or GST.HNF6α proteins as indicated. The autoradiogram of the SDS-gel transferred to Immobilon is shown. Coomassie stain shows the amount of GST proteins loaded in each lane.
p53 alone did not bind any of the probes (Figure 5B, all panels, lane 6), suggesting that p53 may inhibit HNF6α DNA binding by interacting with the HNF6α protein. To investigate a potential protein–protein interaction between p53 and HNF6α, we performed a Co-IP analysis with COS-7 cells transfected with an HNF6α expression vector. Immunoprecipitation of endogenous p53 resulted in a substantial co-precipitation of HNF6α protein (Figure 5C, lane 6). A direct interaction between HNF6α and wild-type p53 was further verified in a pulldown assay using in vitro synthesized 35S-labelled HNF6α and bacterially expressed GST.p53.wt (Figure 5D, left-hand panel). To determine whether the interaction with HNF6α was critical for the repression by p53, we repeated the pulldown assay with 35S-labelled p53 proteins (wild-type and R249S) and GST.HNF6α and found that, surprisingly, both the wild-type and the mutant p53 bound HNF6α in vitro. Since the mutant p53 did not repress the P1 promoter (Figure 3B), these results suggest that the interaction between p53 and HNF6α may not be sufficient for the repression. However, it is still possible that the interaction is necessary.
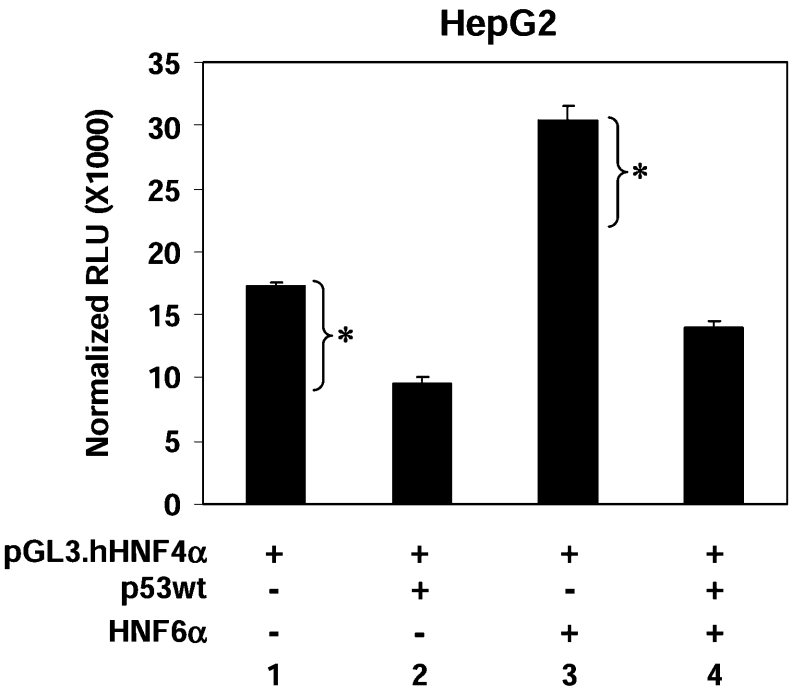
To determine whether p53 inhibits the ability of HNF6α to activate transcription of the HNF4α promoter in vivo, the P1 promoter construct was co-transfected into HepG2 cells with HNF6α and wild-type p53 expression vectors. As previously reported [20], exogenously expressed HNF6α stimulated the P1 promoter above the basal level of expression seen in HepG2 cells (Figure 6, lane 3). Transfection with p53 alone repressed the P1 promoter activity below the basal level, as seen previously (Figure 6, lane 2). Importantly, co-transfection of both HNF6α and p53 also resulted in P1 activity below the basal level (Figure 6, lane 4), indicating that p53 blocked the HNF6α-mediated stimulation of the HNF4α P1 promoter. This observation is qualitatively different than the result expected if p53 did not act on the P1 promoter via HNF6α. If p53 repressed the HNF4α P1 promoter by a mechanism not involving HNF6α, the level of luciferase activity in lane 4 would be that of lane 3 minus the basal repression (indicated on Figure 6 as a bracket with asterisk, the difference between lanes 1 and 2, resulting in approx. 22000 RLU). However, the luciferase activity is much lower (approx. 14000 RLU), suggesting that p53 represses the transcriptional activity of the exogenously expressed HNF6α and supports the notion that p53-mediated repression of the P1 promoter acts in part via inhibition of HNF6α activity.
Figure 6. Co-transfection of p53 and HNF6α suggests that p53 inhibits HNF6α transcriptional activation of the HNF4α P1 promoter.
HepG2 cells were transfected with pGL3.hHNF4α promoter (2 μg), CMV.β-gal (2 μg), pcDNA3.p53wt (1 μg) and pcDNA3.HNF6α (1 μg) as indicated. Cells were harvested at 30 h after transfection. Results are means±S.D. of one representative experiment performed in triplicate. The bracket with asterisk indicates the amount of basal repression by p53 (difference between lane 1 and lane 2). Results indicate that p53 repressed the transcriptional activity of the exogenously expressed HNF6α (see text for details).
p53 is recruited to the HNF4α P1 promoter under conditions of cellular stress
To further investigate whether p53 represses HNF4α1/2 expression via binding the P1 promoter, the ChIP assay using anti-p53 and anti-HNF6α antibodies was performed on HepG2 cells treated with doxorubicin for 0 to 24 h (Figure 7). Immunoblot analysis verified that under the conditions used for the ChIP analysis, p53, HNF6α and HNF4α protein levels exhibited the same trends as in previous experiments (Figure 7A). As the p53 protein levels increased, there was a concomitant increase in the amount of p53 bound to the HNF4α P1 promoter, reaching a peak of a 17.2-fold increase at 20 h (Figure 7B, panel a). Surprisingly, HNF6α was observed on the promoter at all times, with only a marginal decrease at 24 h when the total HNF6α protein levels were lower (Figure 7B, panel b). Additionally, ChIP analysis indicated that Sp1 also remained on the P1 promoter after doxorubicin treatment (Figure 7C). More than three independent time-course ChIP assays were performed with similar results. These data may be interpreted in two different ways. One is that p53 does not inhibit HNF6α from binding DNA in vivo, i.e. both p53 and HNF6α can occupy the same promoter at the same time. The alternative explanation is that since the ChIP assay examines the whole population of P1 promoters, some promoters have p53 but not HNF6α bound, whereas others have HNF6α but not p53 bound. The current experiments cannot distinguish between these two possibilities.
Figure 7. p53 and HNF6α bind the human HNF4α P1 promoter in vivo after doxorubicin treatment.
(A) IB (immunoblot) analysis using ECL® verifying an increase in p53 and a decrease in HNF4α, but no substantial change in HNF6α protein levels in HepG2 cells after doxorubicin treatment (1 μg/ml) over 24 h. Protein levels were analysed using antibodies against HNF4α (α-445), HNF6α (N-terminus), p53 (FL-393) and β-actin. The solid line separates two gels loaded with identical samples. (B) Upper panel: schematic diagram of the human HNF4α P1 proximal promoter showing the relative position of the PCR primers used in the ChIP assay. Lower panel: ChIP assay of HepG2 cells treated with doxorubicin (1 μg/ml) for 0–24 h using anti-p53 (FL-393, a), anti-HNF6α (H-100, b), control mouse IgG (c) and 10% input (d). The amount of p53 bound to the P1 promoter increased after doxorubicin treatment but there was no substantial change in the amount of HNF6α bound to the promoter. Shown are the ethidium bromide-stained agarose gels. The bands were quantified using the NIH (National Institute of Health) Image J program and the fold-change relative to 0 h (control, non-treated) was calculated and normalized to the input signal. Results are from one representative experiment of at least three that were performed. (C) ChIP assay as in (B) using an anti-Sp1 (PEP2) antibody.
DISCUSSION
The results in the present study demonstrate that in two human liver cancer cell lines ectopically expressed p53 and elevated levels of endogenous p53 result in substantially lower levels of HNF4α1/2 protein and mRNA (Figures 1 and 2). Transient transfection assays show that expression of exogenous p53 is by itself sufficient to decrease transcriptional activity of the HNF4α P1 promoter (Figures 3 and 4). The results also indicate several potential mechanisms by which p53 may down-regulate the HNF4α P1 promoter (Figures 5–7).
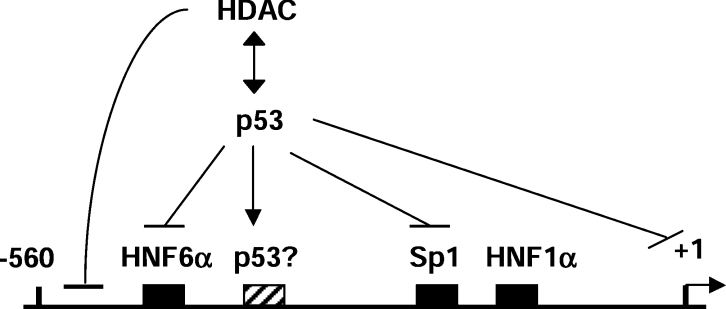
p53 represses the HNF4α P1 promoter through multiple mechanisms
The mechanisms by which p53 represses transcription are known to be varied and complex [3]. The situation with the HNF4α promoter is apparently no different and is summarized in Figure 8. In the present study we show that p53 mutated in the DNA-binding domain, although still able to interact with HNF6α (Figure 5D), does not repress the P1 promoter (Figure 3B). This suggests that the DNA binding activity, or at least an intact DNA-binding domain, of p53 is important for the repression. This is consistent with the results from the ChIP assay indicating that p53 is recruited to the P1 promoter in vivo (Figure 7). However, when a site that loosely fits the p53 consensus binding element was deleted from the P1 promoter, repression by p53 was not eliminated in Hep3B cells (Figure 3C) or HepG2 cells (results not shown). This indicates that there may be other more divergent p53 binding sites in the promoter that mediate repression. Others have noted that the p53 response elements involved in transcriptional repression do not fit the consensus as well as those involved in activation [3,37].
Figure 8. Model showing the multiple mechanisms by which p53 may mediate repression of the HNF4α P1 promoter (see text for details).
Another possibility is that p53 acts on the P1 promoter via binding to other transcription factors, such as HNF6α. The ChIP assay showed that HNF6α remains on the promoter during doxorubicin treatment (Figure 7B) and the GST pulldown and Co-IP assays indicated that p53 interacts directly with HNF6α (Figures 5C and 5D). Furthermore, the transient transfection assay demonstrated that p53 represses the transactivation activity of HNF6α (Figure 6). These results, in combination with the partial reversal of repression by the HDAC inhibitor TSA (Figure 4), suggest that p53 could be recruited to the HNF4α promoter via interaction with HNF6α, and then subsequently recruit HDAC activity. The only results that appear to be at odds with this mechanism are the ability of p53 to repress HNF6α DNA binding in vitro (Figure 5B) and the ability of the mutant p53 to interact with HNF6α (Figure 5D). However, there are possible explanations for both of these results as well. The former could be explained by differences in DNA binding in vitro compared with in vivo, the in vitro binding was on naked DNA whereas in vivo there is chromatin and other transcription factors to consider. The latter result could be explained if there were a requirement for both DNA binding and interaction with HNF6α for the repression.
There are also other potential factors that might be involved. One possibility is that Sp1, a transcription factor previously shown to interact with p53 [38], could be recruiting p53 to the P1 promoter. This is consistent with our ChIP results indicating that, like HNF6α, Sp1 remains on the promoter during doxorubicin treatment (Figure 7C). Another possibility is that repression is mediated via interaction with TBP (TATA binding protein) as others have observed on other promoters [39–41]. In summary, we propose that the mechanism by which p53 represses the HNF4α P1 promoter is a complex one that may involve multiple factors, including direct DNA binding to as yet unidentified sites and/or recruitment by other transcription factors, such as HNF6α.
Function of p53-mediated HNF4α repression
Although the mechanism by which p53 represses the HNF4α gene is multi-faceted and complex, the effect of stress on HNF4α gene expression is clear and specific. Our results indicate that the decreased expression of HNF4α in response to doxorubicin treatment is not a general effect of stress or DNA damage; the treated cells continue to express normal levels of actin and other transcription factors such as Sp1 and HNF1α. The down-regulation of HNF4α was also observed in unstressed cells expressing exogenous p53 (Figures 1 and 2). The specificity of this response and the fact that there are apparently multiple mechanisms by which p53 inhibits HNF4α activity, i.e. direct inhibition of protein function [10] and inhibition of gene expression (the present study), suggest that repression of HNF4α is an important function of p53 in the liver.
A number of other studies have demonstrated that cellular stress conditions can regulate the expression of HNF4α. For example, hypoxia [42] and arsenic trioxide [43] have both been shown to down-regulate HNF4α gene expression, and both are also known to up-regulate p53 protein levels [44,45]. Hypoxia in particular is associated with increasing transcriptional repression, as opposed to activation, by p53 [46]. Therefore it appears that the cell uses p53 to down-regulate HNF4α gene expression and protein function in response to stress. Additional studies are underway to determine the purpose of such a down-regulation.
Effect of chronic stress on the liver
The results in the present study shed additional light on the observation that long-term clinical use of anti-cancer treatments that chronically induce p53, such as doxorubicin, often lead to liver damage and liver toxicity [47–50]. Doxorubicin causes hepatocellular stress by generating semiquinon free radicals and superoxide anion radicals [49]. In response to this stress, p53 protein levels are increased, resulting in the classical blockage of the cell cycle and cell death via apoptosis. It is also known that p53 itself causes liver injury [51–54]. Since HNF4α function is known to be critical for the differentiated liver phenotype [55–57], the down-regulation of HNF4α by activated p53 that we demonstrate in the present study could, in the chronic situation, lead to decreased liver differentiation and hence decreased liver function. This could explain some of the liver toxicity of doxorubicin, and other DNA damaging agents that increase p53 protein levels.
Acknowledgments
This work was supported by NIH (National Institute of Health) Grants R01DK053892 to F. M. S., R01CA075180 to X. L. and R01CA052457 to L. B. O., fellowships from the Japan Society for the Promotion of Science (to Y. M.) and UC Toxic Substances Research and Teaching Program (to Y. M. and W. W. H.-V). We thank D. Lee and A. G. Li for advice on ChIP analysis and E. Martinez for thoughtful discussion.
References
- 1.Bargonetti J., Manfredi J. J. Multiple roles of the tumor suppressor p53. Curr. Opin. Oncol. 2002;14:86–91. doi: 10.1097/00001622-200201000-00015. [DOI] [PubMed] [Google Scholar]
- 2.Esteve P. O., Chin H. G., Pradhan S. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1000–1005. doi: 10.1073/pnas.0407729102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho J., Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 4.Ko L. J., Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 5.Mirza A., Wu Q., Wang L., McClanahan T., Bishop W. R., Gheyas F., Ding W., Hutchins B., Hockenberry T., Kirschmeier P., et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene. 2003;22:3645–3654. doi: 10.1038/sj.onc.1206477. [DOI] [PubMed] [Google Scholar]
- 6.Murphy M., Ahn J., Walker K. K., Hoffman W. H., Evans R. M., Levine A. J., George D. L. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu G., Schwartz J. A., Brooks S. C. p53 down-regulates ER-responsive genes by interfering with the binding of ER to ERE. Biochem. Biophys. Res. Commun. 1999;264:359–364. doi: 10.1006/bbrc.1999.1525. [DOI] [PubMed] [Google Scholar]
- 8.Maiyar A. C., Phu P. T., Huang A. J., Firestone G. L. Repression of glucocorticoid receptor transactivation and DNA binding of a glucocorticoid response element within the serum/glucocorticoid-inducible protein kinase (sgk) gene promoter by the p53 tumor suppressor protein. Mol. Endocrinol. 1997;11:312–329. doi: 10.1210/mend.11.3.9893. [DOI] [PubMed] [Google Scholar]
- 9.Yap N., Yu C. L., Cheng S. Y. Modulation of the transcriptional activity of thyroid hormone receptors by the tumor suppressor p53. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4273–4277. doi: 10.1073/pnas.93.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda Y., Seidel S. D., Wei G., Liu X., Sladek F. M. Repression of hepatocyte nuclear factor 4α tumor suppressor p53: involvement of the ligand-binding domain and histone deacetylase activity. Mol. Endocrinol. 2002;16:402–410. doi: 10.1210/mend.16.2.0769. [DOI] [PubMed] [Google Scholar]
- 11.Sladek F. M., Seidel S. D. Hepatocyte Nuclear Factor 4α. In: Burris T. P., McCabe E. R. B., editors. Nuclear Receptors and Genetic Diseases. London: Academic Press; 2001. pp. 309–361. [Google Scholar]
- 12.Darlington G. J. Molecular mechanisms of liver development and differentiation. Curr. Opin. Cell Biol. 1999;11:678–682. doi: 10.1016/s0955-0674(99)00035-6. [DOI] [PubMed] [Google Scholar]
- 13.Zaret K. S. Regulatory phases of early liver development: paradigms of organogenesis. Nat. Rev. Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 14.Zhao R., Duncan S. A. Embryonic development of the liver. Hepatology. 2005;41:956–967. doi: 10.1002/hep.20691. [DOI] [PubMed] [Google Scholar]
- 15.Ryffel G. U. Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol. 2001;27:11–29. doi: 10.1677/jme.0.0270011. [DOI] [PubMed] [Google Scholar]
- 16.Briancon N., Bailly A., Clotman F., Jacquemin P., Lemaigre F. P., Weiss M. C. Expression of the α7 isoform of hepatocyte nuclear factor (HNF) 4 is activated by HNF6/OC-2 and HNF1 and repressed by HNF4α1 in the liver. J. Biol. Chem. 2004;279:33398–33408. doi: 10.1074/jbc.M405312200. [DOI] [PubMed] [Google Scholar]
- 17.Eeckhoute J., Moerman E., Bouckenooghe T., Lukoviak B., Pattou F., Formstecher P., Kerr-Conte J., Vandewalle B., Laine B. Hepatocyte nuclear factor 4α isoforms originated from the P1 promoter are expressed in human pancreatic β-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology. 2003;144:1686–1694. doi: 10.1210/en.2002-0024. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka T., Jiang S., Hotta H., Takano K., Iwanari H., Sumi K., Daigo K., Ohashi R., Sugai M., Ikegame C., et al. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4α in the pathogenesis of human cancer. J. Pathol. 2006;208:662–672. doi: 10.1002/path.1928. [DOI] [PubMed] [Google Scholar]
- 19.Torres-Padilla M. E., Fougere-Deschatrette C., Weiss M. C. Expression of HNF4α isoforms in mouse liver development is regulated by sequential promoter usage and constitutive 3′ end splicing. Mech. Dev. 2001;109:183–193. doi: 10.1016/s0925-4773(01)00521-4. [DOI] [PubMed] [Google Scholar]
- 20.Hatzis P., Talianidis I. Regulatory mechanisms controlling human hepatocyte nuclear factor 4α gene expression. Mol. Cell Biol. 2001;21:7320–7330. doi: 10.1128/MCB.21.21.7320-7330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatzis P., Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 22.Lannoy V. J., Burglin T. R., Rousseau G. G., Lemaigre F. P. Isoforms of hepatocyte nuclear factor-6 differ in DNA-binding properties, contain a bifunctional homeodomain, and define the new ONECUT class of homeodomain proteins. J. Biol. Chem. 1998;273:13552–13562. doi: 10.1074/jbc.273.22.13552. [DOI] [PubMed] [Google Scholar]
- 23.Lannoy V. J., Rodolosse A., Pierreux C. E., Rousseau G. G., Lemaigre F. P. Transcriptional stimulation by hepatocyte nuclear factor-6. Target-specific recruitment of either CREB-binding protein (CBP) or p300/CBP-associated factor (p/CAF) J. Biol. Chem. 2000;275:22098–22103. doi: 10.1074/jbc.M000855200. [DOI] [PubMed] [Google Scholar]
- 24.Sladek F. M., Zhong W. M., Lai E., Darnell J. E., Jr Liver-enriched transcription factor HNF4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 25.Wang B., Cai S. R., Gao C., Sladek F. M., Ponder K. P. Lipopolysaccharide results in a marked decrease in hepatocyte nuclear factor 4 α in rat liver. Hepatology. 2001;34:979–989. doi: 10.1053/jhep.2001.28885. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W. W., Fang X., Mazur W., French B. A., Georges R. N., Roth J. A. High-efficiency gene transfer and high-level expression of wild-type p53 in human lung cancer cells mediated by recombinant adenovirus. Cancer Gene Ther. 1994;1:5–13. [PubMed] [Google Scholar]
- 27.Jiang G., Nepomuceno L., Hopkins K., Sladek F. M. Exclusive homodimerization of the orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Mol. Cell Biol. 1995;15:5131–5143. doi: 10.1128/mcb.15.9.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie Y., Li H. H., Bula C. M., Liu X. Stimulation of p53 DNA binding by c-Abl requires the p53 C-terminus and tetramerization. Mol. Cell Biol. 2000;20:741–748. doi: 10.1128/mcb.20.3.741-748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yew P. R., Liu X., Berk A. J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 30.Liu X., Miller C. W., Koeffler P. H., Berk A. J. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol. Cell Biol. 1993;13:3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda Y., Rachez C., Hawel L., 3rd, Byus C. V., Freedman L. P., Sladek F. M. Polyamines modulate the interaction between nuclear receptors and vitamin D receptor-interacting protein 205. Mol. Endocrinol. 2002;16:1502–1510. doi: 10.1210/mend.16.7.0883. [DOI] [PubMed] [Google Scholar]
- 32.Lee T. K., Lau T. C., Ng I. O. Doxorubicin-induced apoptosis and chemosensitivity in hepatoma cell lines. Cancer Chemother. Pharmacol. 2002;49:78–86. doi: 10.1007/s00280-001-0376-4. [DOI] [PubMed] [Google Scholar]
- 33.Jiang G., Nepomuceno L., Yang Q., Sladek F. M. Serine/threonine phosphorylation of orphan receptor hepatocyte nuclear factor 4. Arch. Biochem. Biophys. 1997;340:1–9. doi: 10.1006/abbi.1997.9914. [DOI] [PubMed] [Google Scholar]
- 34.Lahuna O., Rastegar M., Maiter D., Thissen J. P., Lemaigre F. P., Rousseau G. G. Involvement of STAT5 (signal transducer and activator of transcription 5) and HNF4 (hepatocyte nuclear factor 4) in the transcriptional control of the hnf6 gene by growth hormone. Mol. Endocrinol. 2000;14:285–294. doi: 10.1210/mend.14.2.0423. [DOI] [PubMed] [Google Scholar]
- 35.Guimaraes D. P., Hainaut P. TP53: a key gene in human cancer. Biochimie. 2002;84:83–93. doi: 10.1016/s0300-9084(01)01356-6. [DOI] [PubMed] [Google Scholar]
- 36.Bullock A. N., Fersht A. R. Rescuing the function of mutant p53. Nat. Rev. Cancer. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 37.Hearnes J. M., Mays D. J., Schavolt K. L., Tang L., Jiang X., Pietenpol J. A. Chromatin immunoprecipitation-based screen to identify functional genomic binding sites for sequence-specific transactivators. Mol. Cell Biol. 2005;25:10148–10158. doi: 10.1128/MCB.25.22.10148-10158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Innocente S. A., Lee J. M. p53 is a NF-Y- and p21-independent, Sp1-dependent repressor of cyclin B1 transcription. FEBS Lett. 2005;579:1001–1007. doi: 10.1016/j.febslet.2004.12.073. [DOI] [PubMed] [Google Scholar]
- 39.Crighton D., Woiwode A., Zhang C., Mandavia N., Morton J. P., Warnock L. J., Milner J., White R. J., Johnson D. L. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. Embo J. 2003;22:2810–2820. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farmer G., Friedlander P., Colgan J., Manley J. L., Prives C. Transcriptional repression by p53 involves molecular interactions distinct from those with the TATA box binding protein. Nucleic Acids Res. 1996;24:4281–4288. doi: 10.1093/nar/24.21.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seto E., Usheva A., Zambetti G. P., Momand J., Horikoshi N., Weinmann R., Levine A. J., Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc. Natl. Acad. Sci. U.S.A. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazure N. M., Nguyen T. L., Danan J. L. Severe hypoxia specifically downregulates hepatocyte nuclear factor-4 gene expression in HepG2 human hepatoma cells. Tumour Biol. 2001;22:310–317. doi: 10.1159/000050632. [DOI] [PubMed] [Google Scholar]
- 43.Yu D., Wang Z. H., Cheng S. B., Li H. K., Chan H. B., Chew E. C. The effect of arsenic trioxide on the expression of Hsc and HNF4 in nuclear matrix proteins in HepG2 cells. Anticancer Res. 2001;21:2553–2559. [PubMed] [Google Scholar]
- 44.Filippova M., Duerksen-Hughes P. J. Inorganic and dimethylated arsenic species induce cellular p53. Chem. Res. Toxicol. 2003;16:423–431. doi: 10.1021/tx025606a. [DOI] [PubMed] [Google Scholar]
- 45.Pluquet O., Hainaut P. Genotoxic and non-genotoxic pathways of p53 induction. Cancer Lett. 2001;174:1–15. doi: 10.1016/s0304-3835(01)00698-x. [DOI] [PubMed] [Google Scholar]
- 46.Koumenis C., Alarcon R., Hammond E., Sutphin P., Hoffman W., Murphy M., Derr J., Taya Y., Lowe S. W., Kastan M., Giaccia A. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol. Cell Biol. 2001;21:1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bagchi D., Bagchi M., Hassoun E. A., Kelly J., Stohs S. J. Adriamycin-induced hepatic and myocardial lipid peroxidation and DNA damage, and enhanced excretion of urinary lipid metabolites in rats. Toxicology. 1995;95:1–9. doi: 10.1016/0300-483x(94)02867-t. [DOI] [PubMed] [Google Scholar]
- 48.Ganey P. E., Kauffman F. C., Thurman R. G. Oxygen-dependent hepatotoxicity due to doxorubicin: role of reducing equivalent supply in perfused rat liver. Mol. Pharmacol. 1988;34:695–701. [PubMed] [Google Scholar]
- 49.Kalender Y., Yel M., Kalender S. Doxorubicin hepatotoxicity and hepatic free radical metabolism in rats. The effects of vitamin E and catechin. Toxicology. 2005;209:39–45. doi: 10.1016/j.tox.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Kimura T., Fujita I., Itoh N., Muto N., Nakanishi T., Takahashi K., Azuma J., Tanaka K. Metallothionein acts as a cytoprotectant against doxorubicin toxicity. J. Pharmacol. Exp. Ther. 2000;292:299–302. [PubMed] [Google Scholar]
- 51.Jaeschke H., Gores G. J., Cederbaum A. I., Hinson J. A., Pessayre D., Lemasters J. J. Mechanisms of hepatotoxicity. Toxicol. Sci. 2002;65:166–176. doi: 10.1093/toxsci/65.2.166. [DOI] [PubMed] [Google Scholar]
- 52.Schafer T., Scheuer C., Roemer K., Menger M. D., Vollmar B. Inhibition of p53 protects liver tissue against endotoxin-induced apoptotic and necrotic cell death. FASEB J. 2003;17:660–667. doi: 10.1096/fj.02-0774com. [DOI] [PubMed] [Google Scholar]
- 53.Sola S., Ma X., Castro R. E., Kren B. T., Steer C. J., Rodrigues C. M. Ursodeoxycholic acid modulates E2F-1 and p53 expression through a caspase-independent mechanism in transforming growth factor β1-induced apoptosis of rat hepatocytes. J. Biol. Chem. 2003;278:48831–48838. doi: 10.1074/jbc.M300468200. [DOI] [PubMed] [Google Scholar]
- 54.Yahagi N., Shimano H., Matsuzaka T., Sekiya M., Najima Y., Okazaki S., Okazaki H., Tamura Y., Iizuka Y., Inoue N., et al. p53 involvement in the pathogenesis of fatty liver disease. J. Biol. Chem. 2004;279:20571–20575. doi: 10.1074/jbc.M400884200. [DOI] [PubMed] [Google Scholar]
- 55.Hayhurst G. P., Lee Y. H., Lambert G., Ward J. M., Gonzalez F. J. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J., Ning G., Duncan S. A. Mammalian hepatocyte differentiation requires the transcription factor HNF4α. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 57.Spath G. F., Weiss M. C. Hepatocyte nuclear factor 4 expression overcomes repression of the hepatic phenotype in dedifferentiated hepatoma cells. Mol. Cell Biol. 1997;17:1913–1922. doi: 10.1128/mcb.17.4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]