Abstract
CUG-BP1 [CUG-binding protein 1 also called CELF (CUG-BP1 and ETR3 like factors) 1] is a human RNA-binding protein that has been implicated in the control of splicing and mRNA translation. The Xenopus homologue [EDEN-BP (embryo deadenylation element-binding protein)] is required for rapid deadenylation of certain maternal mRNAs just after fertilization. A variety of sequence elements have been described as target sites for these two proteins but their binding specificity is still controversial. Using a SELEX (systematic evolution of ligand by exponential enrichment) procedure and recombinant CUG-BP1 we selected two families of aptamers. Surface plasmon resonance and electrophoretic mobility-shift assays showed that these two families differed in their ability to bind CUG-BP1. Furthermore, the selected high-affinity aptamers form two complexes with CUG-BP1 in electrophoretic mobility assays whereas those that bind with low affinity only form one complex. The validity of the distinction between the two families of aptamers was confirmed by a functional in vivo deadenylation assay. Only those aptamers that bound CUG-BP1 with high affinity conferred deadenylation on a reporter mRNA. These high-affinity RNAs are characterized by a richness in UGU motifs. Using these binding site characteristics we identified the Xenopus maternal mRNA encoding the MAPK (mitogen-activated protein kinase) phosphatase (XCl100α) as a substrate for EDEN-BP. In conclusion, high-affinity CUG-BP1 binding sites are sequence elements at least 30 nucleotides in length that are enriched in combinations of U and G nucleotides and contain at least 4 UGU trinucleotide motifs. Such sequence elements are functionally competent to target an RNA for deadenylation in vivo.
Keywords: deadenylation, embryo deadenylation element-binding protein (EDEN-BP), mitogen-activated protein kinase (MAPK) phosphatase XCl100α, RNA-binding protein, systematic evolution of ligand by exponential enrichment (SELEX)
Abbreviations: ARE, AU-rich element; CUG-BP1, CUG-binding protein 1; CELF, CUG-BP1 and ETR3 like factors; DMPK, myotonic dystrophy protein kinase; EDEN, embryo deadenylation element; EDEN-BP, EDEN-binding protein; EMSA, electrophoretic mobility-shift assay; MAPK, mitogen-activated protein kinase; ORF, open reading frame; RU, resonance unit; SELEX, systematic evolution of ligand by exponential enrichment; SPR, surface plasmon resonance; UTR, untranslated region
INTRODUCTION
CUG-BP1 (CUG-binding protein 1) is a human RNA-binding protein that was first identified by its capacity to bind to a (CUG)8 probe [1,2]. It was thereby implicated in DM1 (type 1 myotonic dystrophy; MIM No. 160900), a neuromuscular disease associated with an unstable CUG triplet expansion in the 3′-UTR (3′-untranslated region) of the DMPK (myotonic dystrophy protein kinase) gene. A variety of functions have been described for CUG-BP1 (reviewed in [3]). In the nucleus it was demonstrated to control the alternative splicing of certain pre-mRNAs [4–10]. In the cytoplasm, it stimulates the translation of p21 mRNA [11] or controls the choice of translation initiation codon of C/EBP (CCAAT/enhancer-binding protein) mRNA [12]. The Xenopus equivalent of CUG-BP1, EDEN-BP (embryo deadenylation element-binding protein), is responsible for the rapid cytoplasmic deadenylation [poly(A) tail shortening] of certain maternal mRNAs [13], which is correlated with translational repression [14]. In Xenopus egg extracts the human CUG-BP1 and Xenopus EDEN-BP are interchangeable [15] and recently, CUG-BP1 was reported to act as a deadenylation factor in human cells by recruiting PARN deadenylase to target mRNA [16]. Lastly, CUG-BP1 is one of the founding members of the CELF (CUG-BP1 and ETR-3 like factors) family of RNA-binding proteins [17], key molecular factors which determine the fate of a large number of mRNAs (reviewed in [3]).
Despite the importance of CUG-BP1 and related proteins in the post-transcriptional regulation of gene expression, the sequence requirements for high-affinity CUG-BP1 binding are still ambiguous. This protein was initially reported to cross-link the DMPK 3′-UTR CUG expansion [1]. Other natural sequences, described for their capacity to bind to CUG-BP1, include U/G-rich intronic motifs [4,18,19], 3′-UTR U/G-rich motifs such as the c-jun ARE (AU-rich element) [20], c-mos EDEN [15], and GCN-rich coding regions [12,21,22]. However, the capacity of CUG-BP1 to bind to the CUG expansion has been questioned [23,24]. In two independent triple hybrid assays using synthetic probes, it was observed that CUG-BP1 had a low affinity for CUG repeats but interacted very efficiently with (UG)n probes [24,25]. In addition, CUG-BP1 crosslinks to a synthetic (UG)15 probe [26]. The sequence requirement for CUG-BP2/CELF2/ETR-3, that shares 75% identity with CUG-BP1 (and almost 100% identity in the consensus RNA binding domains [3]), is also ambiguous. RNA aptamers selected against this protein resulted in UG-rich sequences and in particular UGUU motifs [27], but this protein was also shown to interact with cycloxygenase-2 mRNA ARE [28]. This ARE is not UG-rich but rather contains several AUUUA motifs.
This variety in the sequences reported to be recognized by CUG-BP1, and the ambiguity of autonomous binding, highlights the need for a systematic, non-biased (without prior limitations), examination of the binding specificities of this protein. In this work, we have used a SELEX (systematic evolution of ligand by exponential enrichment) procedure [29,30] to select RNA aptamers from a library of random sequences on the basis only of their autonomous association with CUG-BP1. Analysis of the binding affinities and the functionality of the selected aptamers showed that a minimal high-affinity CUG-BP1 binding site could be defined as a relatively short RNA (around 36 nucleotides) highly enriched in UG repeats with a particular importance of the UGU motif. These criteria were used to identify a biologically important maternal Xenopus mRNA MAPK (mitogen-activated protein kinase phosphatase), XCl100α, that is a target for the Xenopus homologue of CUG-BP1.
EXPERIMENTAL
Recombinant protein production
The CUG-BP1 ORF (open reading frame) was cloned into the pTRC-His plasmid (Invitrogen). His-tagged protein was produced by overnight induction at 25 °C with 5 mM IPTG (isopropyl β-D-thiogalactoside) and purified on Ni-NTA (Ni2+-nitrilotriacetate) agarose beads (Qiagen) following the manufacturer's instructions. Denaturing electrophoresis and Coomassie Blue staining was used to check the purity of recombinant CUG-BP1 and to measure its concentration as compared with BSA.
Synthesis of the initial random RNA library
The initial random oligonucleotide was purchased from IBA GmbH/Göttingen. The sequence was 5′-CCACTAAACCAGCCTCAAGGGTACCGCTCTAGA(35N)GCTAGCGTATCTGCTCCTAATAAAAAGAGGATCCCC-3′ where N represents the randomized part and the underlined sequences are XbaI and NheI restriction sites respectively. This oligonucleotide was PCR amplified (4 cycles) using the forward (5′-AGTAATACGACTC-ACTATAGGGCCACTAAACCAGCCTCAAGG-3′) and reverse (5′-GGGGATCCTCTTTTTATTAGG-3′) primers to generate a double-stranded DNA template with a 5′-end T7 promoter (underlined in the forward primer). Each 100 μl PCR reaction mixture contained 8 pmol of random oligonucleotide template and 2.5 units of Amplitaq DNA polymerase (PerkinElmer). The PCR products were phenol/chloroform extracted, ethanol precipitated and further purified on Microcon YM30 columns (Amicon). The initial RNA pool was synthesized from 240 pmol of PCR DNA with 2400 units of T7 RNA polymerase (New England BioLabs) in a 6 ml reaction mixture supplemented with 24 units of inorganic pyrophosphatase (Sigma) and 300 units of porcine RNAse inhibitor (Amersham Biosciences). After 4 h at 37 °C, the reaction was treated (30 min at 37 °C) with 700 units of DNAse I (Invitrogen). The RNA mixture was extracted with phenol/chloroform, twice with chloroform, ethanol precipitated and further purified on Microcon YM30 columns. The RNA was eluted using the SELEX binding buffer [50 mM Tris/HCl (pH 7.5), 50 mM NaCl, 5 mM dithiothreitol and 0.1 mM CaCl2].
SELEX procedure
RNA (300 pmol) was refolded by heating at 85 °C and slowly cooled to room temperature (20 °C) before counter selection through a HAWP filter (Amicon). RNA was then incubated with CUG-BP1 (30 pmol) for 80 min (first round) or 30 min (subsequent rounds) in the presence of 1.2 nmol of tRNA (Ambion). Partition was achieved by filtration through a HAWP filter that was washed three times with 200 μl of binding buffer. The filter was incubated for 5 min at 85 °C in elution buffer (7 M urea, 100 mM sodium citrate and 3 mM EDTA) to recover the CUG-BP1-associated RNA species that were propan-1-ol precipitated. The RNA was refolded in binding buffer and subjected to a second counter selection as described above. The eluted RNA was ethanol-precipitated and resuspended in nuclease-free water.
After each round of selection, the RNA was hybridized with the reverse primer, reverse transcribed with 15 units of AMV reverse transcriptase (QBiogen) for 50 min at 42 °C and PCR amplified with 30 units of Amplitaq DNA polymerase as described for the initial RNA pool. The PCR products were purified on a Microcon YM30 column and 18 pmol of DNA were in vitro transcribed with 200 units of T7 RNA polymerase for 3 h at 37 °C. Finally, the RNA was treated with 140 units of DNAse I for 20 min at 37 °C, and then purified and quantified with Ribogreen (Molecular Probes).
Sequence analysis
Individual aptamers were cloned from a retro-transcribed RNA pool using a TA cloning procedure (Invitrogen) with the pCR2.1 TOPO plasmid as described by the manufacturer. The DNA of interest was PCR amplified from individual positive bacterial colonies using M13 forward and reverse primers and sequenced. The random sequences were extracted and analysed using the ClustalW algorithm [31] and Emboss web facility.
Evaluation of RNA affinity
Individual or pooled clones were PCR amplified using the forward (containing a T7 promoter) and reverse primers and Amplitaq DNA polymerase. The PCR products were purified and quantified. The RNAs were transcribed in the presence [for EMSA (electrophoretic mobility-shift assay)] or the absence [for SPR (surface plasmon resonance)] of [α-32P]UTP. The plasmids encoding the s3′Eg5 and s3′Eg5C6 RNAs have been described previously [13] and were used as transcription templates as above. EMSA was performed as described by Cosson et al. [32]. The SPR experiments were performed using a BIAcore 2000 biosensor system (BIA-core) at 25 °C according to the manufacturer's instruction. CUG-BP1 was immobilized directly on a CM5 sensor chip (BIAcore) using an amine coupling kit (BIAcore). This was preferred to attachment via a tag because two of the three RNA recognition motifs in CUG-BP1 are very close to the N- and C-termini and immobilization via an N- or C-terminal tag could interfere with the overall CUG-BP1 RNA recognition specificity. A blank flow cell (without protein) was used to evaluate non-specific interactions. RNA was diluted to 100 nM in running buffer [10 mM Hepes (pH 7.9), 150 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2 and 0.05% Tween 20] supplemented with 0.2 units/μl SuperasIn (Ambion), and injected for 2 min followed by a 2 min dissociation step at a flow rate of 20 μl/min. Regeneration was achieved with a 6 s pulse of 100% ethylene glycol at a flow rate of 50 μl/min. Each RNA sample was injected once before performing the second injection (serial duplicates procedure). The sensorgrams were analysed using the BIAevaluation software (Version 3.2, BIAcore).
In vivo deadenylation assay and analysis of maternal Xenopus mRNA
DNA corresponding to the RNA aptamers was digested by NheI and XbaI, and cloned into the pGbORF/mosEDEN plasmid in place of the minimal c-mos EDEN [13]. The 3′UTR of XCl100α maternal mRNA was amplified by PCR (forward primer 5′-CTAGCTAGCGGTACTGAGCAAACAGAC-3′; reverse primer 5′-CGGGATCCCAGTACAAATATCATAATTTA-3′) and cloned into the NheI and BamHI restriction sites of pGbORF [33]. The globin reading frame was then deleted by restriction with NheI and SacI, the NheI site was blunt ended, and religation. All constructs were sequenced.
Capped, polyadenylated radiolabelled RNA were microinjected in Xenopus 2-cell embryos and the deadenylation assays were performed as already described in [13]. Briefly, RNAs were extracted with Tri-Reagent (Euromedex) and separated by electrophoresis on 4% polyacrylamide gels containing urea. The dried gels were analysed using a Phosphoimager (STORM 840; Molecular Dynamics). The proportion of fully deadenylated RNA was quantified, using the ImageQuant software (Version 5.4, Molecular Dynamics), by determining the signal in a rectangle at the position of the fully deadenylated transcript (A)0 relative to the total signal of this transcript (fully adenylated, partially and totally deadenylated transcripts).
To analyse Xenopus maternal mRNAs, total RNA was extracted from embryos using Tri-Reagent (Euromedex) and separated into poly(A)+ and poly(A)− populations by oligo(dT)–cellulose chromatography (Promega). The RNAs were analysed by Northern blotting. XCl100α mRNA was revealed using 32P-labelled cDNA probes corresponding to the coding sequence (the plasmid was obtained from Dr J. E. Fenell Jr, Sanford, CA, U.S.A.; Accession number AJ320158). Immunoprecipitations and UV-induced cross-linking, using the indicated 32P-labelled RNAs, were performed as previously described [13]. The XCl100α RNA corresponded to the UGU rich region was amplified by PCR (forward primer, 5′-CTGGTACTGTAATTCCTGTG-3′; reverse primer, 5′-AAATCATAGCATAGCATACAA-3′).
RESULTS
The SELEX procedure against CUG-BP1
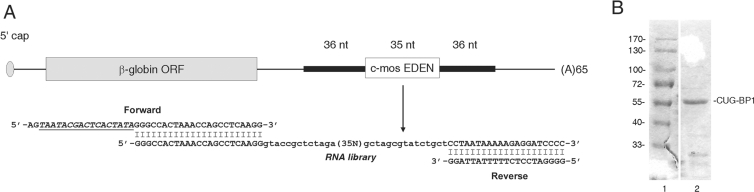
A SELEX procedure was developed that could generate fully functional CUG-BP1 RNA aptamers. An initial SELEX RNA library of 1.8×1014 distinct RNA sequences, randomized on 35 positions, was used (see Experimental section). The 35 nucleotide random sequence was flanked by invariant 5′ and 3′ regions derived from the β-globin 3′UTR (Figure 1A). After an initial counter selection to remove non-specific RNA species, the RNA pool was incubated with CUG-BP1 and loaded onto the filter. Bound RNA was eluted from the filter and submitted to a second counter selection. The RNA was then amplified by RT (reverse transcriptase)-PCR, transcribed into a new RNA library and then applied to the next round of selection/amplification. This selection/amplification procedure was repeated seven times but with a reduced incubation time to increase stringency. Throughout the experiments, the same preparation of recombinant, bacterially-produced CUG-BP1 protein was used. The integrity and purity of this protein were verified by SDS/PAGE (Figure 1B).
Figure 1. Overview of the SELEX procedure.
(A) The 36 nucleotide invariant sequences of the random RNA library were those flanking the c-mos EDEN in the GbORF/mosEDEN RNA. The forward and reverse primers, used for the PCR amplification steps, hybridize to approximately 20 nucleotides of these invariant sequences. The forward primer has a T7 promoter overhang (underlined). (B) Coomassie blue staining of a sample of the recombinant CUG-BP1 used in the experiments (lane 2). Lane 1, molecular mass markers, sizes in kDa are given on the left.
Evaluation of the enrichment using a SPR biosensor and EMSA
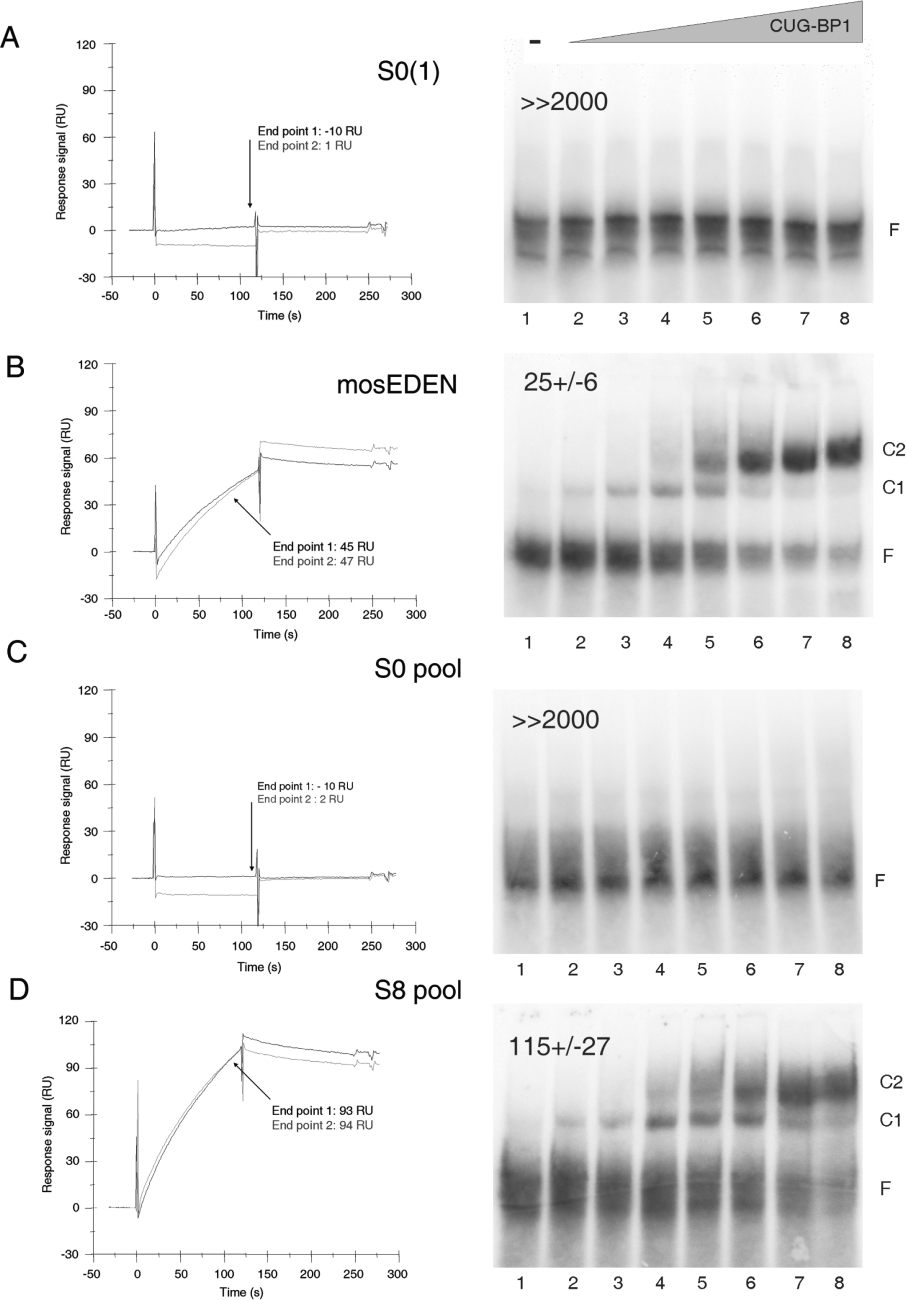
To evaluate the quality of the SELEX enrichment after eight rounds of selection, two analytical methods were used, an SPR assay using a BIAcore™ 2000 instrument and EMSA. The SPR assay was used for screening as it is rapid and can be automated. However, a high level of CUG-BP1 immobilization [approx. 10000 RU (resonance units)] was necessary to detect a clear association signal with c-mos EDEN RNA (positive control, see Figure 2B). A high immobilization level on the chip causes mass transport limitations in the proximity of the sensor chip surface [34] which prevents analysis of the kinetics of RNA–protein associations. Therefore we used a ranking analysis which consisted of comparing the overall qualitative shape of specific binding figures (sensorgrams) to determine the end-point association level (in RU) reached after injection of each analyte [35,36]. When all the samples are injected under the same conditions (concentration, volume, flow rate and temperature), the end-point value should be directly correlated with RNA affinity. The validity of this SPR method for the present case was demonstrated by the close correlation between the SPR and EMSA results (see below).
Figure 2. SPR and EMSA evaluations of SELEX enrichment.
The negative control [S0(1) RNA] (A), the positive control (c-mos EDEN RNA) (B), the initial random RNA population (S0 pool) (C) and the enriched RNA population (S8 pool) (D) were tested for their capacity to interact with recombinant CUG-BP1 by SPR (left-hand panels) and EMSA (right-hand panels). For SPR, RNAs were injected at 100 nM over a sensor chip containing approx. 11000 RU of immobilized CUG-BP1 (corresponding to 11 ng of CUG-BP1/mm2). The grey and black curves represent the first and the second injection of each RNA sample respectively. The end-point values (in RU) correspond to the response signal reached at the end of the RNA injections (t=120 s). For EMSA, uniformly labelled RNA probe was incubated with the following concentrations of CUG-BP1 (nM): lane 1, 0; lane 2, 0.5; lane 3, 2; lane 4, 8; lane 5, 31; lane 6, 125; lane 7, 500; lane 8, 2000. The free probe (F) and two complexes (C1 and C2) were resolved by native electrophoresis. The gel was dried and analysed using a phosphoimager and ImageQuant software. The concentrations (nM) of CUG-BP1 required to achieve 50% of shifted probe are indicated on the top of each gel (means±S.D. of three independent experiments).
First, the specificity and the sensitivity of both assays were assessed by running individual RNA sequences. Except where indicated, EMSA assays were performed with 4-fold serial dilutions of CUG-BP1, between 0.5 and 2000 nM. The S0(1) RNA was randomly taken from the cDNAs cloned from the starting pool of unselected aptamers. No association of this RNA with CUG-BP1 was detected by either the SPR assay or EMSA (Figure 2A). The c-mos EDEN, previously described as a good CUG-BP1 target in a UV cross-linking assay [15], was used as a positive control. This RNA presented a strong association in the SPR assay, reaching an end-point value of approx. 45 RU (Figure 2B, left-hand panel). In the EMSA (Figure 2B, right-hand panel) increasing amounts of CUG-BP1 shifted this RNA from the free state to a first (C1), and then a second (C2) complex. The affinity of CUG-BP1 for c-mos EDEN was estimated from the concentration of protein that bound 50% of the RNA probe. This value, which would be the Kd if only one complex was formed, was 25±6 nM (average of three independent experiments).
Pools of RNA were next tested. In both SPR and EMSA, the initial RNA library (S0 pool) behaved very similarly to the S0(1) RNA, demonstrating that the S0 pool had no overall affinity for CUG-BP1 (Figure 2C). In contrast, the enriched RNA pool (S8 pool) gave an association signal in the SPR assay, and this pool of RNA was shifted to two different complexes by increasing concentrations of CUG-BP1 in the EMSA (Figure 2D). The ‘fuzzy’ appearance of the shifted complexes in the EMSA using the S8 pool (Figure 2D, right-hand panel) may be due to the heterogeneity of the RNA population. The different sequences forming this pool may have different conformations and hence have different electrophoretic mobilities. This dispersion of the bands also reduces the precision of the quantification of the affinity of this pool of RNA for CUG-BP1 (115±27 nM). Irrespective of these considerations, the results in Figure 2 show that the SELEX procedure had successfully allowed the evolution of the large random RNA library into an enriched RNA population whose global affinity was similar to that of the c-mos EDEN. Four additional SELEX rounds did not increase the overall affinity of the RNA library (results not shown).
Characterization of individual aptamers after round eight
The cDNA libraries corresponding to RNA aptamers (S0 pool and S8 pool) were cloned and randomly selected clones were sequenced. As expected, no sequence similarity was detected in 41 clones from the S0 pool. The sequences of 101 clones from the S8 pool are given in the Supplementary data (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/400/bj4000291add.htm). For these S8 clones, although no consensus sequence or obvious conservation was detected, the Clustal algorithm (ClustalW) identified two families of aptamers. In family 1, containing 63 clones, the aptamers were enriched in uridine and depleted in adenosine and cytosine nucleotides relative to the 41 clones from the S0 pool. The nucleotide composition of the second family (38 clones) was not significantly different from that of the S0 pool. Within the 38 clones of family 2, a subpopulation of 10 clones containing six different sequences [S8(2) is present in 5 copies] displayed a conserved motif in the 3′region [GUUCUGCUAGUUU(U/C)GGUU; see Supplementary Figure 1 at http://www.BiochemJ.org/bj/400/bj4000291add.htm).
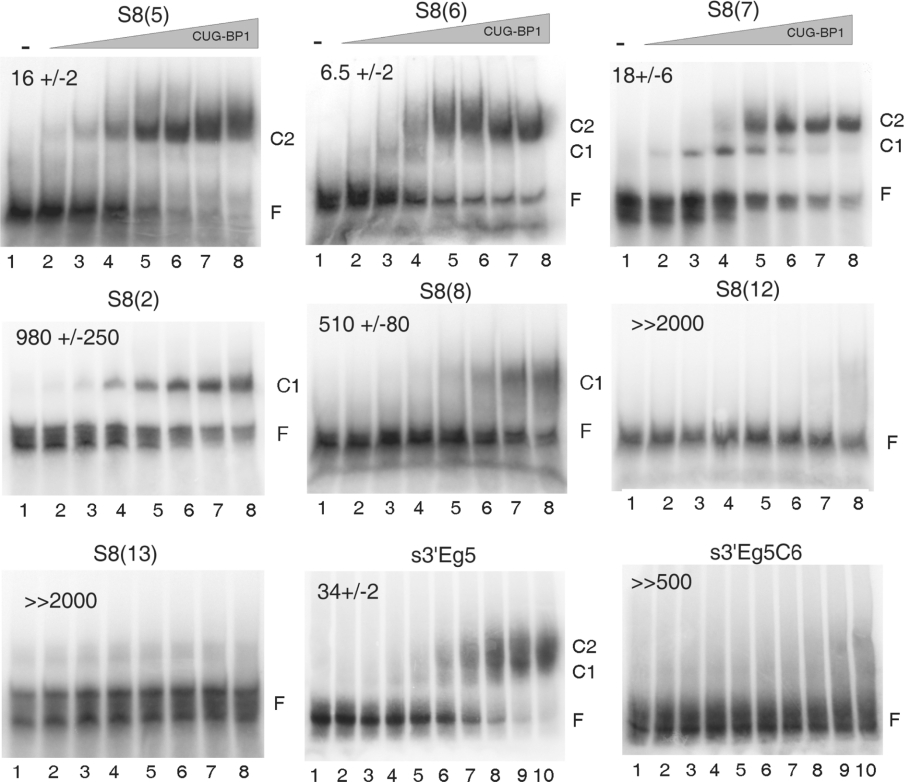
SPR and EMSA analyses were performed on seven individual aptamers from the S8 pool. In EMSA assays, two of them, S8(12) and S8(13), that both belong to family 2, were not shifted by CUG-BP1 (Figure 3). In the SPR assay, the same RNAs gave no association signal (see Table 1). In contrast, S8(5), S8(6), S8(7) and S8(8), that all belong to family 1 and S8(2) that belongs to family 2, showed association with CUG-BP1 in EMSA (Figure 3) and SPR (Table 1) assays. However, there was a noticeable difference between the EMSA patterns of these RNAs. S8(5), S8(6) and S8(7) produced the slower mobility complex, C2, (complex C1 was also observed at low CUB-BP1 concentrations) but only the fast mobility complex, C1, was observed for RNAs S8(2) and S8(8). These differences in EMSA pattern were reflected in the affinity of the individual RNAs for CUG-BP1 as estimated from the EMSA or the SPR assay (see Figure 3 and Table 1). The three RNAs that produced the C2 complex had an affinity for CUG-BP1 comparable with that of c-mos EDEN. For the two RNAs, S8(2) and S8(8), which only formed the C1 complex, at least 20-fold more protein was required to shift 50% of the probe.
Figure 3. EMSA analysis of individual aptamers.
The indicated aptamers were analysed by EMSA for their capacity to bind CUG-BP1 as described in Figure 2. For the EMSA with the s3′Eg5 and s3′Eg5C6 RNAs the protein concentrations were (nM): lane 1, 0; lane 2, 1; lane 3, 2; lane 4, 4; lane 5, 8; lane 6, 17; lane 7, 33; lane 8, 67; lane 9, 133 and lane 10, 266. The concentrations (nM) of CUG-BP1 required to achieve 50% of shifted probe are indicated at the top of each gel (means±S.D. of three independent experiments). The positions of the free probe (F) and the complexes C1 and C2 are indicated on the right of each panel.
Table 1. CUG-BP1 affinity of individual aptamers.
The indicated cloned RNA aptamers were analysed by SPR and EMSA assays as described in the legend to Figure 2. The sequence of the aptamers and the number of UGU trinucleotides are indicated. The aptamers are ordered according to the SPR values that are the averages from two separate injections. The results for the EMSA give the concentration of CUG-BP1 (nM) required to shift 50% of the RNA probe (averages from three separate experiments). The families are defined on the basis of sequence criteria. The Xenopus EDEN sequences and that from the S0 pool [S0(1)] are not clustered into a family. nd, not determined.
| Clone | Family | Sequence | SPR end-point | EMSA | Number of UGU |
|---|---|---|---|---|---|
| S8(7) | 1 | UAUGUUGUGUGUGUGUUGUUUUUGUGUUUUGUUUU | 203 | 18 | 10 |
| S8(6) | 1 | UUAUUAUUGUUUAUUUUGUGUUGUGUGUUUGUGUU | 151 | 6.4 | 8 |
| S8(5) | 1 | CCUGUGUCUUGUGUGUGUGUUUUGUUUCUGGUCUA | 104 | 16 | 8 |
| S8(10) | 1 | CUAGCAGUUUGUGUGUGUGUGUGUGUGUUAUGCUA | 70 | nd | 9 |
| c-mos EDEN | none | GUAUAUGUAUGUGUUGUUUUAUUGUGUGUGUGUGU | 51 | 25 | 10 |
| S8(9) | 1 | UGUGUGUGUGUUAUUUUGUUCGUGUUAUUGUGAUCUA | 48 | nd | 8 |
| s3′Eg5 | none | UAUAUAUGUGUGUCUAUCGUCACUUGUAUGUCAAAUAUU | nd | 34 | 5 |
| S8(2) | 2 | CCCGGUUACCGUGUUUGUUCUGCUAGAUUUUGGUU | 24 | 980 | 2 |
| S8(8) | 1 | AGCCGUCGCUAGUUUGAUUCCGUGUGUUAGUGGUU | 19 | 510 | 2 |
| S0(1) | none | AGUACUCGACGGGACGGCUGCUGUGAACGACUACG | 0 | >2000 | |
| S8(12) | 2 | GUGAUCCAAACCGUUUUUUCUGUCUCUCCAGUUUG | 0 | >2000 | 1 |
| S8(13) | 2 | UCGUAAAGGGCGUCGCUAAACUGAUUGCCGCUAGA | 0 | >2000 | 0 |
| S8(3) | 2 | GGCUGUACGCCUAUACGUUCUGCUAGAUUUCGGUU | 0 | nd | 1 |
| S8(4) | 2 | GGUUUUAACCUACAUUGUUCUGCUAGAUUUUGGUU | 0 | nd | 1 |
| S8(1) | 2 | UAAUCGUACUAUUUUUGUUCUGCUAGAUUUUGGUU | 0 | nd | 1 |
| S8(11) | 2 | UGAAGACGUAAACCGGUUCUUCACGAUGUGGGUGU | 0 | nd | 2 |
| S3′Eg5C6 | none | UAUAUAUCCCCCCUAUCGUCACUUGUAUGUCAAAUAUU | nd | >500 | 2 |
The results shown in Figures 2 and 3 and Table 1 demonstrate an excellent agreement between the SPR and EMSA results obtained with the individual RNAs. Hence six additional aptamers from the S8 pool were analysed by SPR alone; two were from family 1 and four from family 2 of which three came from the subpopulation with the conserved motif. Based on the end-point values, the two RNAs from family 1, S8(9) and S8(10), showed a high affinity for CUG-BP1 whereas none of the RNAs from family 2 displayed any binding to CUG-BP1 (see Table 1). This result indicates that the sequence present in several RNAs of family 2 is not implicated in binding to CUG-BP1 as determined by the assays used in the present study.
Functionality of SELEX aptamers
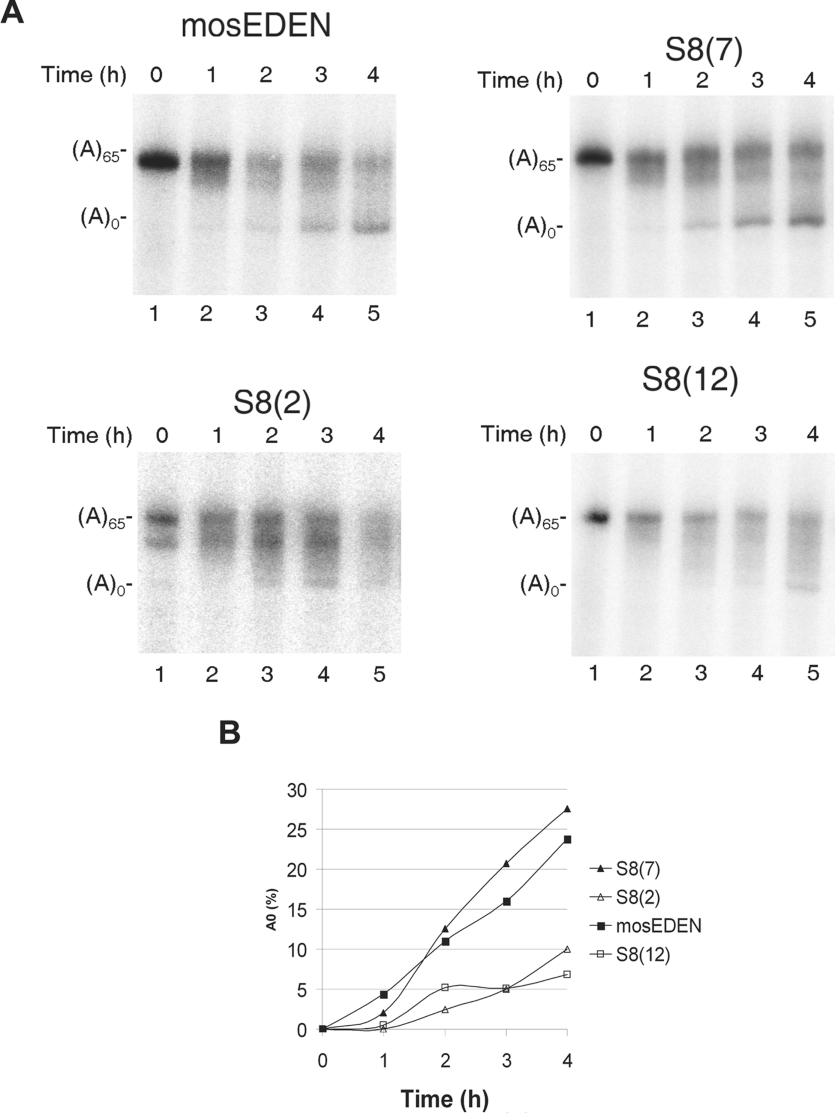
The SPR and EMSA assays allowed us to discriminate between the two aptamer families initially defined on a basis of ClustalW alignment. To confirm this discrimination biologically we used an in vivo test. In Xenopus embryos, EDEN-BP, the equivalent of human CUG-BP1, acts as a sequence-specific factor that directs target RNA for deadenylation [13]. For this process EDEN-BP can be replaced by the orthologous CUG-BP1 [15]. Therefore we used a deadenylation assay to evaluate aptamer functionality. The sequences to be tested were cloned in the 3′UTR of a β-globin cDNA upstream of a synthetic poly(A) tail (A)65. The corresponding radiolabelled RNA was injected into Xenopus embryos and samples were taken at hourly intervals to follow deadenylation kinetics. The deadenylation-targeting activities of the tested aptamers were compared with that of a similar mRNA but containing the c-mos EDEN in the 3′UTR.
As already reported, c-mos EDEN RNA (Figure 4A, top panel, left) was deadenylated by the EDEN pathway with the characteristic accumulation of the deadenylated [(A)0] mRNA within 1–2 h while fully adenylated [(A)65] mRNA persists [13]. In the experiment shown in the top panel of Figure 4(A), a completely deadenylated form of this transcript [(A)0] was detected as soon as 1 h after injection (lane 2) and, 4 h after injection, the (A)0 band was very abundant (lane 5). In Xenopus embryos, the (A)0 mRNAs are stable for at least 6 h [33,37]. The deadenylation pattern observed for the control c-mos EDEN RNA was compared with those conferred by one aptamer from family 1 that bound CUG-BP1 with high affinity, S8(7), one aptamer from family 2 with low affinity, S8(2) and one aptamer from family 2 for which no CUG-BP1 binding had been detected, S8(12) (See Table 1). The deadenylation of the RNA containing the S8(7) aptamer (Figure 4A, top panel, right) presented the characteristics of EDEN-dependent deadenylation, with an (A)0 band easily detecTable 2 h after injection and abundant after 4 h (lane 5). In contrast, the mRNA containing the S8(12) and S8(2) sequences (Figure 4A, two bottom panels) were not or only very slowly deadenylated. No deadenylated molecules (A)0 could be detected before 4 h of incubation (Figure 4A). Quantification of these experiments (Figure 4B) showed that the deadenylation kinetics of S8(7) and c-mos EDEN mRNAs were similar whereas that of S8(2) and S8(12) was about five times slower.
Figure 4. In vitro deadenylation assay of selected aptamers.
(A) SELEX sequences [S8(2), S8(7), and S8(12)] and c-mos EDEN (mosEDEN) were cloned upstream of a synthetic (A)65 tail in the 3′UTR of a β-globin reporter gene. Capped, in vitro synthesized radiolabelled transcripts were micro-injected into Xenopus embryos. At different time points, total RNA was extracted from batches of embryos and analysed by electrophoresis on a denaturing polyacrylamide gel. The positions of the fully adenylated (A)65 and the fully deadenylated (A)0 forms of the transcripts are indicated on the left of each panel. (B) Quantification of the experiments shown in (A). The percentage of (A)0 RNA at each time point was calculated as described in the Experimental section.
A threshold number of UGU is required for efficient CUG-BP1 binding
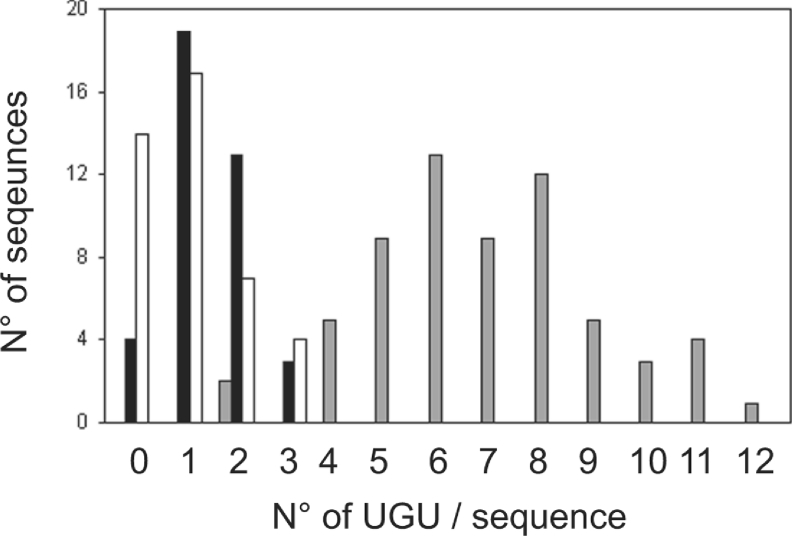
To better characterize the RNA sequence requirements for CUG-BP1 binding, the nucleotide composition of the sequenced RNAs from family 1 and 2 were analysed and compared with that of the 41 sequenced RNAs from the S0 pool. The graphical presentation of the proportions of mono-, di-, tri- and tetra-nucleotides in the sequenced RNAs from families 1 and 2 and the S0 pool are given in Supplementary Figure 2 (at http://www.BiochemJ.org/bj/400/bj4000291add.htm). The most notable features of these analyses were an enrichment in the family 1 RNAs of U mononucleotides and certain di- and tri-nucleotides containing different combinations of U and G nucleotides. In particular, UGU trinucleotides represented nearly 20% of all the trinucleotides in the family 1 RNAs.
In Table 1, the number of UGU trinucleotides for the RNAs tested in the EMSA and SPR experiments described above is reported. Sequences with 0–2 UGU do not bind, or bind only weakly, to CUG-BP1, whereas sequences with 8–10 motifs bind strongly. Analysis of the number of UGU in the individual RNA sequences (Figure 5) showed that the RNAs in the S0 pool and family 2 contain 2 or less UGU while those of family 1 contain 4 or more UGU. Importantly, more sequences are found with 6 UGU than with 4 UGU suggesting that aptamers with 6 UGU were more efficiently selected during the SELEX procedure than aptamers with 4 UGU. In contrast, more than 6 UGU does not appear to confer a selection advantage on an aptamer relative to those with 6 UGU. This suggests that efficient binding sites for CUG-BP1 consist of sequences containing at least 4–6 UGU. Accordingly, a sequence containing 5 UGU was tested. The EDEN motif in the 3′ portion of the Xenopus Eg5 mRNA (s3′Eg5) contains 5 UGU [13] (Table 1). EMSA analysis demonstrated that the affinity of the s3′Eg5 RNA is comparable with that of the c-mos, S8(5), S8(6) and S8(7) RNA (Figure 3 and Table 1), confirming that a threshold for efficient binding occurs between 4 and 5 UGUs. The binding affinity of a mutant RNA (s3′Eg5C6), in which three UGU trinucleotides were changed to cytosines, was next determined. This mutation reduces the number of UGUs from 5 to 2 (Table 1), which is below the proposed threshold. No retarded complex was observed for the s3′Eg5C6 RNA by EMSA, demonstrating a much reduced affinity (Figure 3 and Table 1). Together, these data indicate that the main sequence requirement for affinity binding to CUG-BP1 is the presence of at least 5 UGU within a 35 nucleotide window.
Figure 5. CUG-BP1 binding sites are enriched in UGU trinucleotides.
The number of UGU trinucleotides in the individual sequenced aptamers of the S0 pool and families 1 and 2 are shown as a histogram. Family 1, grey bars; family 2, black bars; S0 pool, white bars.
Identification of a maternal Xenopus mRNA containing a CUG-BP1/EDEN-BP binding site
In Xenopus embryos the majority of maternal mRNAs that are known functional targets for EDEN-BP encode kinases important for the cell cycle. Therefore to relate the sequence requirements for CUG-BP1/EDEN-BP binding to a biologically important event we searched 3′UTR sequences of Xenopus mRNAs encoding kinases or phosphatase for a potential novel target using the criteria defined above. This analysis identified the MAPK phosphatase XCl100 α [38] whose 3′UTR contains 12 UGU trinucleotides within a 35 nucleotide stretch (accession number AJ320158) making it a potential target for EDEN-BP, which would limit the expression of this protein after fertilization. This mRNA is of particular interest to the present paper as the over expression of XCl100α mRNA in embryos causes severe defects in gastrulation and posterior development ([39] and Y. Audic, unpublished work).
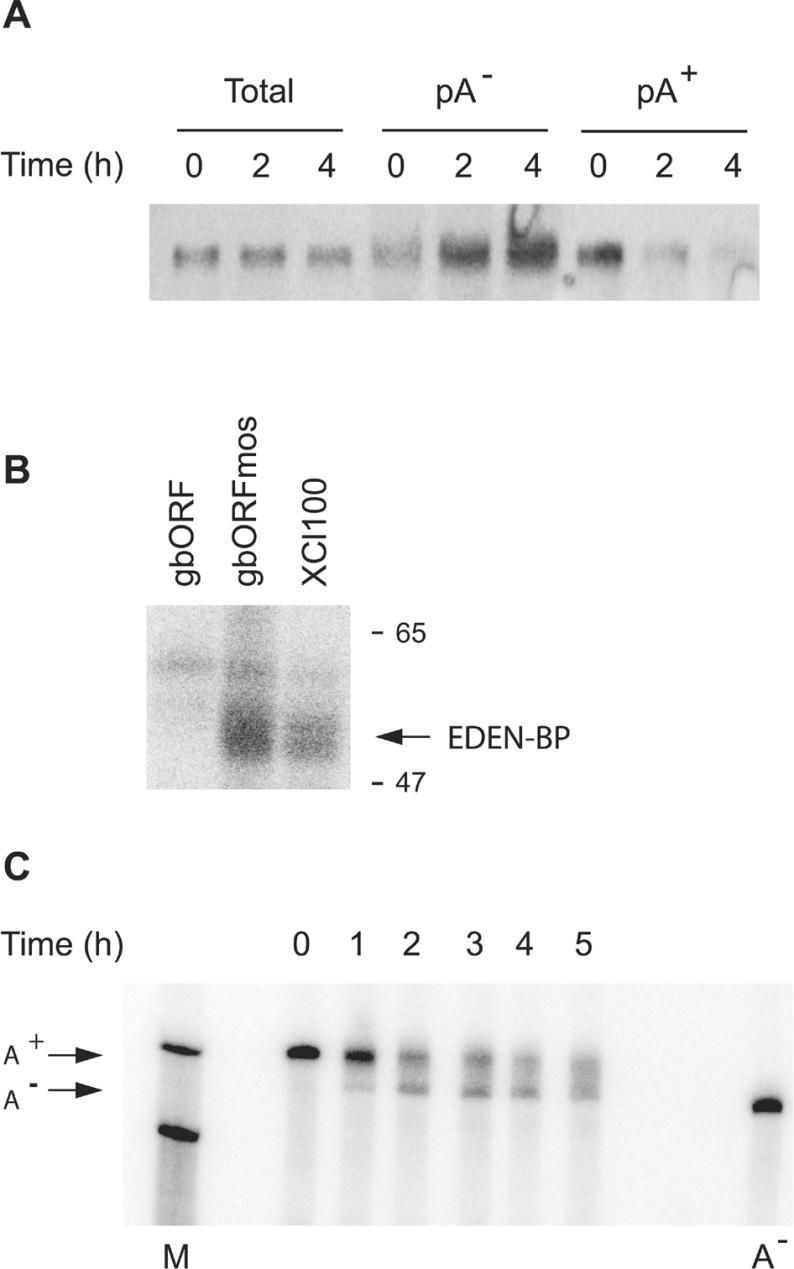
To ensure that the endogenous XCl100α mRNA is a potential target for CUG-BP1/EDEN-BP the adenylation behaviour of the maternal mRNA was determined. Total mRNA extracted from embryos at 0 h, 2 h and 4 h after fertilization were fractionated into poly(A)+ and poly(A)− populations and analysed by Northern blot (Figure 6A). As already observed for a number of maternal Xenopus mRNAs [40] the amount of XCl100α mRNA in the total population did not vary over this 4 h period. In contrast, there was a reciprocal exchange of this mRNA between the poly(A)+ and poly(A)− populations. The majority of XCl100α mRNA that is in the poly(A)+ population at fertilization (0 h) rapidly becomes deadenylated.
Figure 6. XCl100α maternal mRNA is a target for EDEN-BP and is deadenylated after fertilization.
(A) Maternal RNA isolated from unfertilized eggs (0 h) or 2 h and 4 h after fertilization was separated into poly(A+) and poly(A−) populations and analysed by Northern blotting. XCl100α was revealed using a 32P-labelled probe as described in the Experimental section. RNA loading was 1 and 11 embryo equivalents for total RNA and poly(A+) or poly(A−) RNAs respectively. (B) 32P-labelled RNAs containing the globin ORF with or without the c-mos EDEN, respectively gbORF and gbORFmos, and the UGU rich region of XCl100α 3′UTR were incubated in Xenopus egg extracts and processed for UV-induced cross-linking. After separation by SDS/PAGE the proteins that had become radiolabelled were revealed by phosphoimager analysis. The positions of molecular-mass markers (kDa) and of EDEN-BP are indicated on the right. (C) Xenopus 2-cell embryos were injected with a reporter mRNA containing the XCl100α 3′UTR. At the indicated times, samples were taken for extraction and analysis of the RNA. After separation by electrophoresis on denaturing gels the radiolabelled RNAs were revealed by phosphoimager analysis. The position of the fully adenylated (A+) and deadenylated (A−) RNAs are indicated on the left. Lane M, RNA markers of 888 and 715 nucleotides; lane A−, the reporter RNA containing the XCl100α 3′UTR synthesized devoid of a poly(A) tail.
Next, a radiolabelled RNA corresponding to the 180 nucleotide portion of the XCl100α 3′UTR that encompasses the UGU-rich motif was synthesized and used in a UV-induced cross-linking assay with Xenopus egg extracts (Figure 6B). In parallel with this RNA, the RNA GbORF and GbORF c-mos were included as negative and positive controls respectively. Both the XCl100α and GbORF c-mos RNA produced a strong radioactive signal for a protein migrating as a wide band between 54 and 56 kDa which is characteristic of Xenopus EDEN-BP [13]. The identity of this protein was confirmed by immunoprecipitation of the radiolabelled protein with anti-EDEN-BP antibodies (results not shown).
To verify that the 3′UTR of XCl100α could confer the deadenylation behaviour observed for the maternal mRNA we analysed the adenylation behaviour of a reporter mRNA containing this sequence after injection into Xenopus embryos (Figure 6C). This reporter mRNA was rapidly deadenylated with kinetics similar to those already described for the reporter mRNA containing the c-mos EDEN (compare Figure 4A with 6C). Together these results show that the 3′UTR of XCl100α targets an mRNA for rapid EDEN-dependent deadenylation in Xenopus embryos and therefore further validates the UGU-richness of the 3′UTR as a pertinent criterion to identify novel CUG-BP1/EDEN-BP mRNA targets.
DISCUSSION
A population of RNA (S8 pool) with a global affinity for CUG-BP1 equivalent to that of the c-mos EDEN was obtained after eight rounds of SELEX. Sequencing of 101 clones from this S8 pool did not identify any consensus motif, but allowed the clustering of these sequences into two families. The affinity of 13 individual aptamers was measured which showed that the majority (five out of six) of tested family 1 RNAs had a high affinity for CUG-BP1 and produced two shifted complexes (C1 and C2) in EMSA. Family 2 RNAs had no or a low (one out of seven) affinity for CUG-BP1 and the low affinity aptamers produced only the faster mobility complex (C1). In an in vivo deadenylation assay the family 1, but not the family 2, aptamers conferred efficient deadenylation on a reporter mRNA.
Analysis of the sequenced aptamers showed, for family 1, that UGU triplets represented approx. 20% of all the trinucleotides, and that at least four non-overlapping UGU are found in these aptamers. The number of UGU trinucleotides in the family 2 aptamers was not significantly different from that in the S0 pool. The importance of the UGU trinucleotide for CUG-BP1 binding was demonstrated by EMSA using the Eg5 EDEN and a mutant form that decreased the number of UGU trinucleotides from 5 to 2. This mutation drastically reduced the affinity of the RNA for CUG-BP1.
These data indicate that most functional CUG-BP1 binding sites will probably be sequences of more than 30 nucleotides containing at least four non-overlapping UGU motifs. Using this information we identified the Xenopus maternal mRNA encoding the MAPK phosphatase XCl100α as a substrate for the deadenylation process targeted by CUG-BP1/EDEN-BP. Gotoh et al. [39] have previously shown that the overexpression of XCl100α in Xenopus embryos caused severe defects during gastrulation and in posterior development. These data and data in the present paper imply that the post-fertilization deadenylation of the maternal mRNA, directed by the deadenylation factors CUG-BP1/EDEN-BP, is necessary in order to limit the amount of XCl100α expressed in the embryo. Furthermore, it shows that the criteria defined for autonomous binding of CUG-BP1 or the Xenopus homologue EDEN-BP can be used to identify functionally important mRNAs.
All but one of the sequenced aptamers of family 1 fulfil the criteria stated above (see Supplementary data at http://www.BiochemJ.org/bj/400/bj4000291add.htm) and this is also the case for natural sequences previously identified by their capacity to bind CUG-BP1. These include the cardiac troponin T (cTNT) [4] and the human muscle specific chloride channel (Clc-1) [19] mRNA precursors, the B recognition element [41] and the c-jun ARE [20]. Moreover, we noted that mutations in the cardiac troponin T, human muscle specific chloride channel and B recognition element sequences that abolished the interaction with CUG-BP1 [4,19,41] also disrupted these UGU motifs. CUG-BP1 was also shown to efficiently bind synthetic (UG)n repeats probes in UV cross-link experiments [26] and triple hybrid assays [24,25].
Although the SELEX procedure produced a population of aptamers, the S8 pool, that had overall affinity for CUG-BP1, this population contained a large proportion (approx. 38%) of aptamers with low or no affinity for this protein. It should be noted that the SELEX procedure and the SPR and EMSA analyses do not use equivalent criteria or conditions. The binding and selection steps in the SELEX procedure were performed in conditions that selected aptamers with a range of affinities and also a number of false positive aptamers. Although the SPR and EMSA analyses discriminated between the high and low affinity sites, we cannot formally exclude that the family 2 aptamers may represent binding sites that require auxiliary factors or are only used in certain cellular contexts. The family 1 UGU-rich aptamers, tested for their ability to interact with CUG-BP1 [S8(5), S8(6), S8(7) and c-mos EDEN] form two complexes, C1 and C2, in EMSA. In contrast, the two aptamers with a low affinity for CUG-BP1 that were tested, S8(2) and S8(8), only formed the first complex, C1. The different complexes could correspond to different conformations of the RNA probes associated with a single CUG-BP1 protein. However, this explanation would mean that, as the CUG-BP1 concentration increased, all of the family 1 RNAs tested (including those in the S8 pool), but not the RNAs from family 2, would be able to take up two different conformations; we consider this unlikely. An alternative explanation is that the protein components of the two complexes are different. Although we cannot exclude that the minor contaminants in the recombinant CUG-BP1 preparation partake in these complexes we think that this is improbable. We have previously shown that EDEN-BP could form a homodimer in a yeast 2-hybrid assay [42] and we have now demonstrated that this can occur in vivo [32]. Therefore we prefer the explanation that the two complexes correspond to different oligomerization states of CUG-BP1 bound to the RNA probe.
What are the RNA binding specificities of proteins close to human CUG-BP1? In humans, CUG-BP1 is a member of the BRUNOL (Bruno-like) or CELF family of RNA-binding proteins ([17,41] and reviewed in [3]). The best known member of this family, with CUG-BP1, is ETR-3 (also named CUG-BP2 or BRUNOL3), that shares 75% identity with CUG-BP1. This protein was shown to bind to an AU-rich element in cyclooxygenase-2 mRNA that is not enriched in UGU trinucleotide [28], which suggests a different binding specificity between CUG-BP1 and ETR-3. However, an ETR-3 SELEX selected sequences enriched in UG and UGUU [27], which is closer to the sequence requirement for CUG-BP1. In addition, Suzuki et al. [7] showed using UV-crosslinking that CUG-BP1 and ETR-3 could bind to UG-rich sequences. Binding to motifs enriched in UGU may therefore be a general property of at least these two BRUNOL/CELF proteins.
The proportion of CUG and GCN sequences decreased during the SELEX procedure (see Supplementary data at http://www.BiochemJ.org/bj/400/bj4000291add.htm) in contradiction with previously published data for CUG-BP1 targets [2,12,21,22]. Other authors also failed to find any affinity between CUG-BP1 and these sequences in three-hybrid assays [24,25]. Moreover, CUG expansions form double-stranded RNA that is unable to bind CUG-BP1 [23]. This does not exclude that CUG-BP1 could bind to non-UGU rich elements when associated with particular co-factors. For instance, the association of CUG-BP1 with another RNA-BP (RNA-binding protein) could significantly modify the characteristics of the element to which the complex is bound. For example, hnRNPE1 is involved both in the silencing of 15-lipoxygenase mRNA and the stabilization of β-globin mRNA during erythroid differentiation, and these properties are mediated by binding to different sequences [43,44]. Also, hnRNPD/AUF1 [ARE/poly(U)-binding/degradation factor] is involved in AU-rich element mediated mRNA degradation [45,46] but belongs to stabilizing complexes that are associated with the coding region of c-fos mRNA [47] or the 3′UTR of α-globin mRNA [48]. Again, the binding sites for AUF1 in these opposing functions are different.
Online data
Acknowledgments
We thank D. To Van (Biacore) for advice and helpful discussions during the SPR experiments, Agnès Méreau (UMR 6061) for help in designing bandshifts, and Valérie Allo (Généthon) who provided technical help for the constructs. This work was supported by the Association Française contre les Myopathies (to C.L.B.), by grants from ARC 4791 and Ministère de la Jeunesse, de l'Education Nationale et de la Recherche ACI BCMS314 (to L.P.), European Union DG XII biotechnology program contract QLK3-CT-2000-00721 (to H.B.O.). J.M. was financed by doctoral fellowships from the CNRS and Généthon and B.C. by postdoctoral fellowships from the CNRS and Rennes Métropole.
References
- 1.Timchenko L. T., Miller J. W., Timchenko N. A., DeVore D. R., Datar K. V., Lin L., Roberts R., Caskey C. T., Swanson M. S. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Timchenko L. T., Timchenko N. A., Caskey C. T., Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum. Mol. Genet. 1996;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- 3.Barreau C., Paillard L., Mereau A., Osborne H. B. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Philips A. V., Timchenko L. T., Cooper T. A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 5.Ho T. H., Bundman D., Armstrong D. L., Cooper T. A. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 6.Ladd A. N., Taffet G., Hartley C., Kearney D. L., Cooper T. A. Cardiac tissue-specific repression of celf activity disrupts alternative splicing and causes cardiomyopathy. Mol. Cell. Biol. 2005;25:6267–6278. doi: 10.1128/MCB.25.14.6267-6278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki H., Jin Y., Otani H., Yasuda K., Inoue K. Regulation of alternative splicing of α-actinin transcript by Bruno-like proteins. Genes Cells. 2002;7:133–141. doi: 10.1046/j.1356-9597.2001.00506.x. [DOI] [PubMed] [Google Scholar]
- 8.Gromak N., Matlin A. J., Cooper T. A., Smith C. W. Antagonistic regulation of α-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA. 2003;9:443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mankodi A., Takahashi M. P., Jiang H., Beck C. L., Bowers W. J., Moxley R. T., Cannon S. C., Thornton C. A. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 10.Poleev A., Hartmann A., Stamm S. A trans-acting factor, isolated by the three-hybrid system, that influences alternative splicing of the amyloid precursor protein minigene. Eur. J. Biochem. 2000;267:4002–4010. doi: 10.1046/j.1432-1327.2000.01431.x. [DOI] [PubMed] [Google Scholar]
- 11.Timchenko N. A., Iakova P., Cai Z. J., Smith J. R., Timchenko L. T. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell. Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timchenko N. A., Welm A. L., Lu X., Timchenko L. T. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPβ mRNA and regulates translation of C/EBPβ isoforms. Nucleic Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paillard L., Omilli F., Legagneux V., Bassez T., Maniey D., Osborne H. B. EDEN and EDEN-BP, a cis element and an associated factor that mediates sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ezzeddine N., Paillard L., Capri M., Maniey D., Bassez T., Aït-Ahmed O., Osborne B. EDEN dependent translational repression of maternal mRNAs is conserved between Xenopus and Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2002;99:257–262. doi: 10.1073/pnas.012555499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paillard L., Legagneux V., Osborne H. B. A functional deadenylation assay identifies human CUG-BP as a deadenylation factor. Biol. Cell. 2003;95:107–113. doi: 10.1016/s0248-4900(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 16.Moraes K. C., Wilusz C. J., Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladd A. N., Charlet-B N., Cooper T. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savkur R. S., Philips A. V., Cooper T. A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 19.Charlet B. N., Logan P., Singh G., Cooper T. A. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 20.Paillard L., Legagneux V., Maniey D., Osborne H. B. c-Jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J. Biol. Chem. 2002;277:3232–3235. doi: 10.1074/jbc.M109362200. [DOI] [PubMed] [Google Scholar]
- 21.Timchenko N. A., Cai Z. J., Welm A. L., Reddy S., Ashizawa T., Timchenko L. T. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 22.Timchenko N. A., Patel R., Iakova P., Cai Z. J., Quan L., Timchenko L. T. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 23.Michalowski S., Miller J. W., Urbinati C. R., Paliouras M., Swanson M. S., Griffith J. Visualization of double-stranded RNAs from the myotonic dystrophy protein kinase gene and interactions with CUG-binding protein. Nucleic Acids Res. 1999;27:3534–3542. doi: 10.1093/nar/27.17.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi N., Sasagawa N., Suzuki K., Ishiura S. The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem. Biophys. Res. Commun. 2000;277:518–523. doi: 10.1006/bbrc.2000.3694. [DOI] [PubMed] [Google Scholar]
- 25.Kino Y., Mori D., Oma Y., Takeshita Y., Sasagawa N., Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 26.Delaunay J., Le Mee G., Ezzeddine N., Labesse G., Terzian C., Capri M., Ait-Ahmed O. The Drosophila Bruno paralogue Bru-3 specifically binds the EDEN translational repression element. Nucleic Acids Res. 2004;32:3070–3082. doi: 10.1093/nar/gkh627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faustino N. A., Cooper T. A. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol. Cell. Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukhopadhyay D., Houchen C. W., Kennedy S., Dieckgraefe B. K., Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 29.Klug S. J., Famulok M. All you wanted to know about SELEX. Mol. Biol. Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 30.Gold L., Polisky B., Uhlenbeck O., Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 31.Thompson J. D., Higgins D. G., Gibson T. J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cosson B., Gautier-Courteille C., Maniey D., Ait-Ahmed O., Lesimple M., Osborne H. B., Paillard L. Oligomerization of EDEN-BP is required for specific mRNA deadenylation and binding. Biol. Cell. 2006;98:651–662. doi: 10.1042/BC20060054. [DOI] [PubMed] [Google Scholar]
- 33.Audic Y., Omilli F., Osborne H. B. Postfertilization deadenylation of mRNAs in Xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Mol. Cell. Biol. 1997;17:209–218. doi: 10.1128/mcb.17.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myszka D. G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr. Opin. Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 35.Shen B., Shimmon S., Smith M. M., Ghosh P. Biosensor analysis of the molecular interactions of pentosan polysulfate and of sulfated glycosaminoglycans with immobilized elastase, hyaluronidase and lysozyme using surface plasmon resonance (SPR) technology. J. Pharm. Biomed. Anal. 2003;31:83–93. doi: 10.1016/s0731-7085(02)00606-4. [DOI] [PubMed] [Google Scholar]
- 36.Karlsson R., Kullman-Magnusson M., Hamalainen M. D., Remaeus A., Andersson K., Borg P., Gyzander E., Deinum J. Biosensor analysis of drug-target interactions: direct and competitive binding assays for investigation of interactions between thrombin and thrombin inhibitors. Anal. Biochem. 2000;278:1–13. doi: 10.1006/abio.1999.4406. [DOI] [PubMed] [Google Scholar]
- 37.Voeltz G. K., Steitz J. A. AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol. Cell. Biol. 1998;18:7537–7545. doi: 10.1128/mcb.18.12.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis T., Groom L. A., Sneddon A. A., Smythe C., Keyse S. M. XCL100, an inducible nuclear MAP kinase phosphatase from Xenopus laevis: its role in MAP kinase inactivation in differentiated cells and its expression during early development. J. Cell Sci. 1995;108:2885–2896. doi: 10.1242/jcs.108.8.2885. [DOI] [PubMed] [Google Scholar]
- 39.Gotoh Y., Masuyama N., Suzuki A., Ueno N., Nishida E. Involvement of the MAP kinase cascade in Xenopus mesoderm induction. EMBO J. 1995;14:2491–2498. doi: 10.1002/j.1460-2075.1995.tb07246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paris J., Osborne H. B., Couturier A., LeGuellec R., Philippe M. Changes in the polyadenylation of specific stable RNAs during the early development of Xenopus laevis. Gene. 1988;72:169–176. doi: 10.1016/0378-1119(88)90139-4. [DOI] [PubMed] [Google Scholar]
- 41.Good P., Chen O., Warner S., Herring D. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J. Biol. Chem. 2000;275:28583–28592. doi: 10.1074/jbc.M003083200. [DOI] [PubMed] [Google Scholar]
- 42.Bonnet-Corven S., Audic Y., Omilli F., Osborne H. B. An analysis of the sequence requirements of EDEN-BP for specific RNA binding. Nucleic Acids Res. 2002;30:4667–4674. doi: 10.1093/nar/gkf586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostareck-Lederer A., Ostareck D. H., Hentze M. W. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 44.Ostareck-Lederer A., Ostareck D. H. Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biol. Cell. 2004;96:407–411. doi: 10.1016/j.biolcel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 45.DeMaria C. T., Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 46.Loflin P., Chen C. Y., Shyu A. B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grosset C., Chen C. Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A. B. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 48.Kiledjian M., DeMaria C. T., Brewer G., Novick K. Identification of AUF1 (heterogeneous nuclear riboprotein d) as a component of the α-globin mRNA stability complex. Mol. Cell. Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.