Abstract
The ARE (AU-rich element) is a post-transcriptional element controlling both mRNA turnover and translation initiation by primarily inducing poly(A) tail shortening. The mechanisms by which the ARE-associated proteins induce deadenylation are still obscure. One possibility among others would be that an ARE–ARE-BP (ARE-binding protein) complex intervenes in the PABP [poly(A)-binding protein]–poly(A) tail association and facilitates poly(A) tail accessibility to deadenylases. Here, we show by several experimental approaches that AUF1 (AU-rich element RNA-binding protein 1)/hnRNP (heterogeneous nuclear ribonucleoprotein) D, an mRNA-destabilizing ARE-BP, can bind poly(A) sequence in vitro. First, endogenous AUF1 proteins from HeLa cells specifically bound poly(A), independently of PABP. Secondly, using polyadenylated RNA probes, we showed that (i) the four recombinant AUF1 isoforms bind poly(A) as efficiently as PABP, (ii) the AUF1 binding to poly(A) does not change when the polyadenylated probe contains the GM-CSF (granulocyte/macrophage-colony stimulating factor) ARE, suggesting that, in vitro, the AUF1–poly(A) association was independent of the ARE sequence itself. In vitro, the binding of AUF1 isoforms to poly(A) displayed oligomeric and co-operative properties and AUF1 efficiently displaced PABP from the poly(A). Finally, the AUF1 molar concentration in HeLa cytoplasm was only 2-fold lower than that of PABP, whereas in the nucleus, its molar concentration was similar to that of PABP. These in vitro results suggest that, in vivo, AUF1 could compete with PABP for the binding to poly(A). Altogether, our results may suggest a role for AUF1 in controlling PABP–poly(A) tail association.
Keywords: AU-rich element, granulocyte/macrophage-colony stimulating factor, heterogeneous nuclear ribonucleoprotein (hnRNP) D/AU-rich element RNA-binding protein 1 (AUF1), mRNA, poly(A)-binding protein (PABP), post-transcriptional regulation
Abbreviations: ARE, AU-rich element; ARE-BP, ARE-binding protein; AUF1, AU-rich element RNA-binding protein 1; BB, binding buffer; DTT, dithiothreitol; eIF, eukaryotic initiation factor; GM-CSF, granulocyte/macrophage-colony stimulating factor; GMSA, gel mobility-shift assay; GST, glutathione S-transferase; HuR, Hu-antigen R; HRP, horseradish peroxidase; NP40, Nonidet P40; PABP, poly(A)-binding protein; PAIP-1, PABP-interacting protein-1; PARN, poly(A) ribonuclease; RRM, RNA recognition motif; RNP, ribonucleoprotein; hnRNP, heterogeneous nuclear RNP; siRNA, small interfering RNA; U1 snRNP70k, U1 small nuclear RNP-70 k
INTRODUCTION
The ARE (AU-rich element) is a powerful cis-regulatory determinant found in the 3′-untranslated region of many unstable mRNAs [1] that controls both mRNA turnover and translation [2–5]. In mammalian cells, the ARE induces mRNA decay by triggering deadenylation as a first and necessary step; deadenylation is then immediately followed by decapping and exonucleic digestions [4,6]. We recently reported that the ARE intervenes in the binding of PABP [poly(A)-binding protein] to poly(A) [7]. The ARE-mediated PABP release may deprotect the poly(A) tail, opening access to deadenylases and allowing rapid deadenylation. Direct consequences of this deadenylation may be a break of the protective interactions mediated by the mRNA extremities, leading to a decircularization of the transcript, translational inhibition and mRNA body degradation [4,8]. Therefore the PABP–poly(A) tail association may be the primary target of the ARE-mediated mechanisms.
Many ARE-BPs (ARE-binding proteins) have been described [2,4,5]. They participate in ARE functions by favouring or impeding either decay or translation. Controversial data have been reported pertaining to the role of the protein AUF1 (AU-rich element RNA-binding protein 1)/hnRNP [heterogeneous nuclear RNP (ribonucleoprotein)] D in ARE functions [9]. Whereas AUF1 triggers rapid decay of many ARE-containing mRNAs in various cellular environments [2,4,5,10], its ectopic expression stabilizes mRNAs containing either various AREs or the mCRD (major coding region determinant of instability) present in the c-fos mRNA [11]. The high AUF1 expression levels obtained in the latter report are far from physiological and could account for this disparity [12]. Therefore AUF1 is mostly considered as a destabilizing factor and this is supported by its ability to interact with the exosome machinery responsible for the 3′ to 5′ RNA body degradation [13]. However, investigations on the role of AUF1 in ARE functions have been obscured by the facts that (i) four AUF1 isoforms of 37, 40, 42 and 45 kDa, generated by alternative splicing, are present simultaneously [10], (ii) these may have opposite roles in ARE-mediated decay [5,14], (iii) hundreds of mRNAs are bound by AUF1, among which some do not contain obvious ARE [15,16], and (iv) a non-ARE mRNA destabilizing complex contains AUF1 [17].
One model to explain the ARE-mediated deadenylation process postulates that the ARE complex, comprising the ARE and its associated ARE-BPs, induces the removing of the PABP from the poly(A) tail, exposing the naked poly(A) tail to the degradation by either the deadenylase PARN [poly(A) ribonuclease] or the exosome [4,18]. In this model, the PABP–poly(A) tail dissociation is mediated by the ARE–ARE-BP complex [4]. Our recent results support such a model [7]. However, it is noteworthy that (i) PABP can bind to the ARE [19,20], and (ii) both AUF1 and PABP participate in ARE [21] and non-ARE [17] destabilizing complexes. Thus, alternatively, the ARE-BPs could control the activation status of the deadenylases or the decapping enzymes and/or their access to their RNA targets. Supporting this hypothesis, a direct interaction between the ARE-associated destabilizing factor KSRP (K homology RNA-binding protein) and the deadenylase PARN has been reported [22]. Moreover, the ARE-BP tristetraprolin was shown to recruit and activate mRNA decay enzymes by two internal domains [23]. Thus the molecular mechanism of the ARE-mediated deadenylation is far from being clear and the role of AUF1 in this step remains fully obscure.
Here, we show by several experimental approaches that the endogenous and recombinant AUF1 proteins can bind poly(A) in an oligomeric fashion and that AUF1 can efficiently displace PABP from the poly(A) by molecular competition, thus supporting the model described in [4].
EXPERIMENTAL
Plasmids and siRNA (small interfering RNA)
The pBAD/His-AUF1 plasmids were kindly provided by Gary Brewer (Piscataway, NJ, U.S.A.). The FLAG-AUF1 plasmids were kindly provided by Robert J. Schneider (New York, U.S.A.). GC-UTRp(A+), AU-UTRp(A+), pGEM-GC and pGEM-AT vectors were kindly provided by Dr Joan A. Steitz (New Haven, CT, U.S.A.). The pTrcHisB-PABP vector was kindly provided by Richard E. Lloyd (Houston, TX, U.S.A.). A cocktail of four siRNAs directed against PABP was purchased from Dharmacon. The control scramble siRNA was purchased from Eurogentec. The siRNAs were transfected in HeLa cells with the Transmessenger Reagent (Qiagen).
Cell culture and cytoplasmic extract preparations
HeLa cells were cultured in Dulbecco's modified Eagle's medium and 10% (v/v) fetal bovine serum. Cytoplasmic lysates were prepared from HeLa cells as described in [7].
Agarose poly(A)-binding assay
Poly(A)–agarose or poly(C)–agarose beads (Sigma) were washed three times in BB (binding buffer: 25 mM Hepes, pH 7.9, 150 mM KCl, 2 mM EDTA, pH 8.0, 1 mM DTT (dithiothreitol), 5% glycerol and 0.5% NP40 (Nonidet P40)], incubated with 400 μg of whole HeLa cytoplasmic proteins for 4 h at +4 °C under rotation and finally washed four times in BB. The bead pellet was resuspended in Laemmli buffer, boiled for 3 min and resolved on an SDS/10% polyacrylamide gel. The amount of beads was determined to get 7.5 μg of polynucleotide in each reaction. When indicated, the protein extract was treated in the absence or presence of micrococcal nuclease (0.7 unit/μl) in Nuclear buffer, corresponding to the protein extract in BB supplemented first with 1mM CaCl2 and then 15 min later with 4 mM EGTA to stop the digestion reaction.
Antibodies and Western-blot analyses
Western-blot analyses were carried out as described in [7,17]. Antibodies against hnRNP K, GST (glutathione S-transferase) [HRP (horseradish peroxidase)-labelled] and U1 snRNP70k (U1 small nuclear RNP-70 k) were obtained from Santa Cruz Biotechnology. Antibody against AUF1 (5B9) was from Euromedex/Upstate Biotechnology. Antibodies against actin and α-tubulin were from Sigma. The mouse monoclonal 10E10 antibody against PABP was kindly provided by Dr Gideon Dreyfuss (Philadelphia, NJ, U.S.A.). In Western blotting experiments, the protein amount was determined by a 5 min incubation of the membrane with the Supersignal West Dura Extended Duration Substrate (Pierce) and subsequent measure of chemiluminescence using a VersaDoc Imaging system (Bio-Rad). In the poly(A)–agarose experiments, 100% corresponds to the amount of either PABP or AUF1 selected on the poly(A)–agarose column when using the control extracts.
Recombinant proteins
His6–AUF1 proteins
The pBAD/HisB plasmids (Invitrogen) containing the whole coding region of the 37, 40, 42 or 45 kDa AUF1 isoforms were each introduced into TOP10 bacteria (Invitrogen). AUF1 expression was induced by adding 0.0002% arabinose in culture media for 5 h [24]. After induction, each histidine-tagged protein was purified from bacteria pellet using the Talon purification kit (BD Biosciences, Clontech) under non-denaturing conditions. The purification was completed by high-performance gel-filtration chromatography using AKTA purifier system (Amersham Biosciences). An aliquot of each isoform (200 μl) was submitted to chromatography on a Superdex 75 HR 10/30 column equilibrated with the TGN buffer (10 mM Tris/HCl, pH 7.4, 150 mM NaCl and 10% glycerol) at a flow rate of 0.2 ml/min. Proteins, monitored by absorbance at 280 nm, were recovered in 250 μl fractions. Selected fractions were electrophoresed and the protein profile was assessed by silver staining. Fractions of interest were pooled and concentrated using an Amicon Ultra-4 centrifugal filter device (Millipore, Molsheim, France). Concentrates were aliquoted and stored at –80 °C in TGN buffer. Purified recombinant AUF1 concentration was determined by colorimetry using the BCA (bicinchoninic acid) protein assay kit (Pierce) and using Coomassie Blue-stained SDS/polyacrylamide gels.
His6–PABP
The pTrcHisB plasmid (Invitrogen) containing the whole coding region of the PABP [25] was introduced into TOP10 bacteria and the PABP expression was induced by adding 1 mM isopropyl β-D-thiogalactoside in culture media for 5 h. After induction, the His6–PABP was purified from bacteria pellet using the Talon purification kit (BD Biosciences, Clontech) as above. Protein purity was checked on a Coomassie Blue-stained SDS/10% polyacrylamide gel. The protein was then concentrated and stored as described above.
GST–PAIP2 (PABP-interacting protein-2)
The GST–PAIP2 protein was kindly provided by Dr Nahum Sonenberg (Montreal, QC, Canada).
Preparation of RNA probes
In vitro transcription reactions were performed using T7 RNA polymerase (Promega) and either the SacI-linearized AU-UTRp(A)+ and GC-UTRp(A)+ plasmids (polyadenylated RNA probes) or XbaI-linearized AU-UTRpGEM and GC-UTRpGEM (non-adenylated RNA probes) plasmids as templates [26]. Labelled RNA transcripts were produced by including [α-32P]ATP (3000 Ci/mmol; PerkinElmer) in the reaction mixtures. Non-adenylated RNA probes were purified on polyacrylamide–urea gel before use.
Analysis of RNP interactions with purified proteins
The binding reactions were performed in binding buffer A (10 mM Hepes, pH 7.6, 40 mM KCl, 5% glycerol, 0.5% NP40, 3 mM MgCl2, 2 mM DTT, 0.3 mg/ml heparin, 0.2 mg/ml tRNA and 150 nM BSA) as described previously [7]. The volume of each reaction mixture was 10 μl and incubations were done at room temperature. Each purified protein (concentrations as indicated) was incubated for 15 min with 1 nM of the indicated 32P-labelled RNA probe. When using the polyadenylated RNA probes, mixtures were further treated with RNase T1 (5 units/tube) for 20 min prior to loading on to a non-denaturing low-ionic-strength 6% polyacrylamide gel. Finally, the gel was dried, scanned using an Instant imager (Packard) and exposed to a film.
Protein affinity for the non-adenylated probes was determined by measuring the amount of the unbound RNA probes (counts). The amount of unbound RNA versus log His6–protein concentration was plotted using GraphPad Prism 3.0 computer software. The apparent Kd was determined as the EC50, i.e. the protein concentration at which 50% of RNA was bound.
Protein affinity for the poly(A) tail was determined by measuring the amount of poly(A)-specific RNP complexes formed with the polyadenylated probes (counts). Under all conditions, a concentration of 360 nM in His6–protein was sufficient to reach the maximal binding on polyadenylated RNA probes. The amount of His6–protein–RNA complex versus log His6–protein concentration was plotted using GraphPad Prism 3.0 computer software. The apparent Kd was determined as the EC50, which is the protein concentration corresponding to the half-maximal binding of the His6–protein to the poly(A) part of the RNA probes.
RESULTS
Endogenous AUF1 proteins specifically bind to poly(A) in vitro
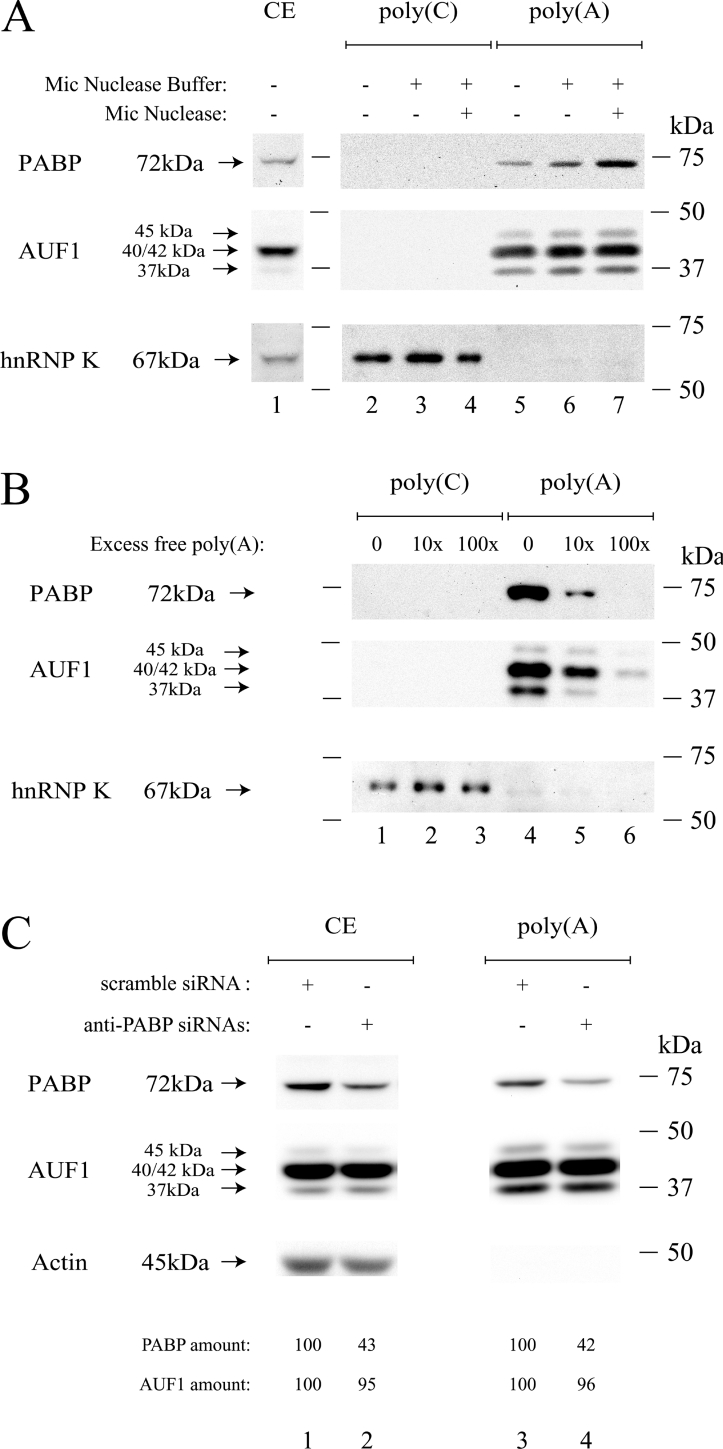
We recently showed that the GM-CSF (granulocyte/macrophage-colony stimulating factor) ARE–ARE-BP complex intervenes in the PABP–poly(A) association [7]. This led us to hypothesize that some ARE-BPs may be involved in the PABP removal from the poly(A) tail. We hypothesized that this removal could be mediated by protein competition for the poly(A). Following the report of Moraes et al. [27] showing that AUF1 had structural and binding similarities to PABP we investigated whether AUF1 could specifically associate with the poly(A) in vitro. To do so, we performed affinity chromatography using a poly(A)–agarose resin and HeLa cytoplasmic extracts followed by Western-blot analyses. As control, a similar affinity chromatography using a poly(C)–agarose resin was performed in parallel and the specific binding of either hnRNP K [28] to the poly(C) (Figures 1A and 1B, lower panel) or PABP to the poly(A) (Figure 1A) was verified. These results validated specific selection of our protein of interest when using the chosen resins. Blotting with the polyclonal anti-AUF1 antibody clearly showed that three bands, corresponding to the 37, 40+42 and 45 kDa AUF1 isoforms [12,14,29] respectively, were strictly selected on the poly(A)–agarose (Figures 1A, middle panel). The Western-blot data did not enable us to determine whether the middle band contained the 40 kDa, the 42 kDa or both isoforms, raising the possibility that one of them was not selected under our experimental conditions. However, all recombinant AUF1 isoforms were able to bind poly(A) in vitro (see below) and each ectopically expressed FLAG-tagged AUF1 isoform was specifically selected on the poly(A)–agarose column, whereas HuR (Hu-antigen R), another ARE-BP [2,4,16], was not (results not shown). Treatment of the HeLa cytoplasmic extract with the micrococcal nuclease prior to selection on agarose beads increased the amount of PABP molecules selected on poly(A)–agarose resin (Figure 1A, upper panel, compare lanes 5 and 7), likely due to an increase in the amount of free PABP following mRNA digestion. The Micrococcal nuclease treatment did not decrease the amount of AUF1 isoforms bound to poly(A) (Figure 1A, middle panel, compare lanes 5 and 7), indicating that the AUF1 isoforms were not selected on poly(A)–agarose resin by mRNA tethering. To support the binding specificity on the poly(A)–agarose, we added increasing amounts of free poly(A) during our affinity- chromatography selections. Whereas excess of poly(A) did not alter the binding of the hnRNP K to the poly(C) resin (Figure 1B, lower panel), it strongly competed with the binding of both PABP and AUF1 isoforms on the poly(A) resin (Figure 1B). Finally, we checked whether AUF1-specific binding to poly(A)–agarose could be consecutive to a co-purification with the PABP, since given RNP complexes were shown to contain both AUF1 and PABP [17,21]. To do so, PABP expression in HeLa cells was knocked-down by transfection with a cocktail of four specific siRNAs directed against the human PABPC1 mRNA (GenBank® accession number NM_002568). Similar experiments were performed with a non-specific scramble siRNA as control. In each case, PABP expression was followed by Western blotting during the next 3 days (results not shown). The amount of either PABP or AUF1 in the cytoplasmic extracts (Figure 1C, ‘CE’) was calculated and normalized by comparison with the amount of actin used as protein-loading control. Semi-quantitative experiments (Figure 1C, lanes 1 and 2) showed that PABP expression was reduced to 43% in cells transfected with the cocktail of anti-PABP siRNAs in comparison with the cells transfected with the control siRNA (Figure 1C, compare lanes 1 and 2). Under the same conditions, the amount of AUF1 did not change (Figure 1C, compare lanes 1 and 2). Poly(A)-affinity chromatography was then performed using 400 μg of cytoplasmic extracts prepared from HeLa cells grown for 3 days after siRNA transfections (Figure 1C, ‘CE’). The amount of PABP selected on the poly(A)–agarose resin was clearly reduced to 42% in cytoplasmic extracts from cells transfected with the anti-PABP siRNAs as compared with control cell extracts (Figure 1C, compare lanes 3 and 4). Under the same conditions, the amount of the AUF1 proteins selected on the resin did not change (Figure 1C, compare lanes 3 and 4). Altogether, these results demonstrated that the binding of the different cytoplasmic AUF1 isoforms to the poly(A) is specific and independent of the amount of PABP in the cytoplasm.
Figure 1. Endogenous AUF1 proteins bind to poly(A).
HeLa cytoplasmic proteins (400 μg) were incubated with either poly(A)–agarose or poly(C)–agarose beads as described in the Experimental section. Proteins from HeLa cytoplasmic lysates (CE; A, lane 1, 10 μg of total proteins; C, lanes 1 and 2, 20 μg of total proteins) or selected on either poly(C)–agarose (A, lanes 2–4; B, lanes 1–3) or poly(A)–agarose resin (A, lanes 5–7; B, lanes 4–6; C, lanes 3 and 4) were resolved on a 10% polyacrylamide gel and analysed by Western blotting using either an anti-PABP (A–C, upper panels), anti-AUF1 (A–C, middle panels), anti-hnRNP K (A, B, lower panels) or anti-actin (C, lower panel) antibody. (A) Prior to selection on the columns, cell extracts were either untreated (lanes 2 and 5) or incubated in nuclease buffer without (lanes 3 and 6) or with (lanes 4 and 7) micrococcal nuclease. (B) Prior to selection on the columns, cell extracts were added with no (lanes 1 and 4), 10-fold (lanes 2 and 5) or 100-fold (lanes 3 and 6) excess amount of free poly(A). (C) HeLa cytoplasmic extracts (CE), prepared from cells transfected with either a control siRNA (lanes 1 and 3) or an anti-PABP siRNA cocktail (lanes 2 and 4), were resolved on gel before (lanes 1 and 2) or after (lanes 3 and 4) selection on poly(A)–agarose resin. The amount of either PABP or AUF1 (means for two independent experiments) was calculated as described in the Experimental section. The name and size of the identified proteins are as shown on the left. Molecular-mass markers are as shown on the right.
Recombinant AUF1 proteins specifically bind to poly(A) in vitro
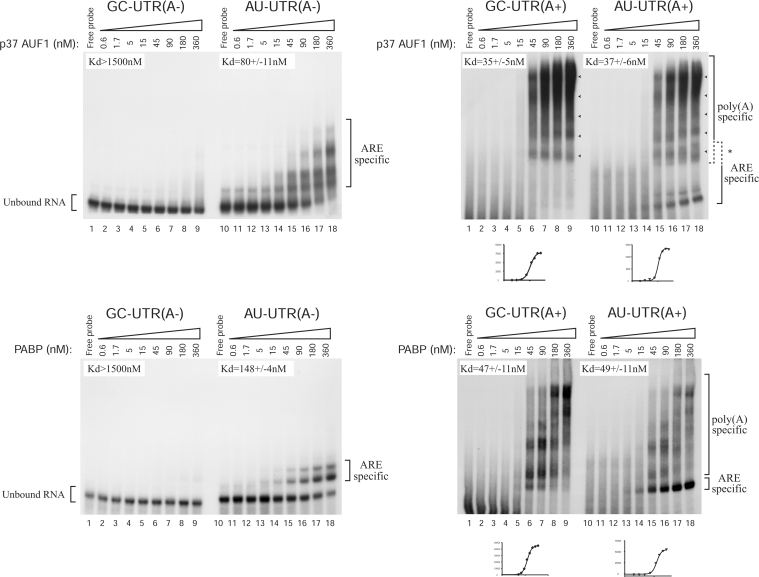
To study the AUF1–poly(A) tail association further, we performed gel retardation assays with several RNA probes and increasing amounts of purified histidine-tagged proteins. The in vitro-transcribed RNA probes used here were as previously described [7,26]. We first evaluated the ability of the purified p37AUF1 to bind to the non-adenylated RNA probes. As shown (Figure 2), p37AUF1 specifically bound to the AU-UTR(A–), the GM-CSF-ARE-containing probe, in an oligomeric fashion while its concentration increased (Figure 2, upper left panel, Kd=80±11 nM), but not to its non-ARE mutated counterpart on to which no cytoplasmic proteins bound, the GC-UTR(A–) probe (Figure 2, upper left panel, Kd>1500 nM). Because p37AUF1 binding to AU-UTR(A–) resulted in the formation of multiple protein–RNA complexes, the Kd determined in these experiments is only indicative and shows the specific binding of p37AUF1 to the ARE. It should be noted that this result is consistent with previous results showing a sequential association of AUF1 dimers with the ARE [29,30]. When performing control experiments with a purified histidine-tagged PABP, we observed a clear and specific binding of PABP to the ARE (Figure 2, lower left panel, Kd=148±4 nM), with a 2-fold lower affinity than p37AUF1, supporting previously reported results [19,20]. The PABP binding to the GM-CSF-ARE-containing AU-UTR(A–) seemed less oligomeric in comparison with p37AUF1. Here again, since two protein–RNA complexes formed when the PABP amount increased, the Kd values were only indicative and show a specific binding of PABP to the ARE and not to the GC-UTR(A–) probe. In our hands, affinities of these two proteins for the GM-CSF ARE were higher than those described in previous reports [20,29,30], which may be explained by the use of various types of AREs or different GMSA (gel mobility-shift assay) conditions.
Figure 2. Recombinant p37AUF1 and PABP bind to the GM-CSF ARE and poly(A) in vitro.
The AU-UTR(A–) probe contains the GM-CSF ARE. The GC-UTR(A–) probe is the non-ARE mutated counterpart of the AU-UTR(A–) probe on to which no cytoplasmic proteins bound [7]. The GC-UTR(A+) probe contains the non-ARE GC-UTR(A–) probe sequence followed by a 100 nt-long poly(A) sequence. As the GC-UTR part does not bind any proteins, the GC-UTR(A+) probe can be considered as a poly(A) sequence. The AU-UTR(A+) probe contains both the ARE sequence from the AU-UTR(A–) probe and a 100 nt-long poly(A) sequence. All gel retardations described in the present study were done in the presence of 150 nM BSA as an irrelevant protein and tRNA as non-specific RNA competitor. Purified His6–p37AUF1 (upper panels) or His6–PABP (lower panels) (concentrations as indicated) was mixed with 1 nM 32P-labelled GC-UTR(A–), AU-UTR(A–), GC-UTR(A+) or AU-UTR(A+) RNA probe as indicated at the top of the panels. When using GC-UTR(A+) or AU-UTR(A+) RNA probe, the reaction mixtures were treated with RNase T1 prior to loading on to gel. The unbound RNA and the ARE- or poly(A)-specific RNP complexes are shown in brackets. Arrowheads show the ladder of the p37AUF1–poly(A) complexes. Kd values were calculated from three independent experiments as described in the Experimental section. A representative curve corresponding to the binding of each His6–protein to the poly(A) sequence of the two polyadenylated RNA probes according to the increasing protein concentration is shown below the corresponding gel. The asterisk shows our inability to distinguish, in the spanned area, between the upper-migrating ARE-specific complexes and the lower-migrating poly(A)-specific complexes.
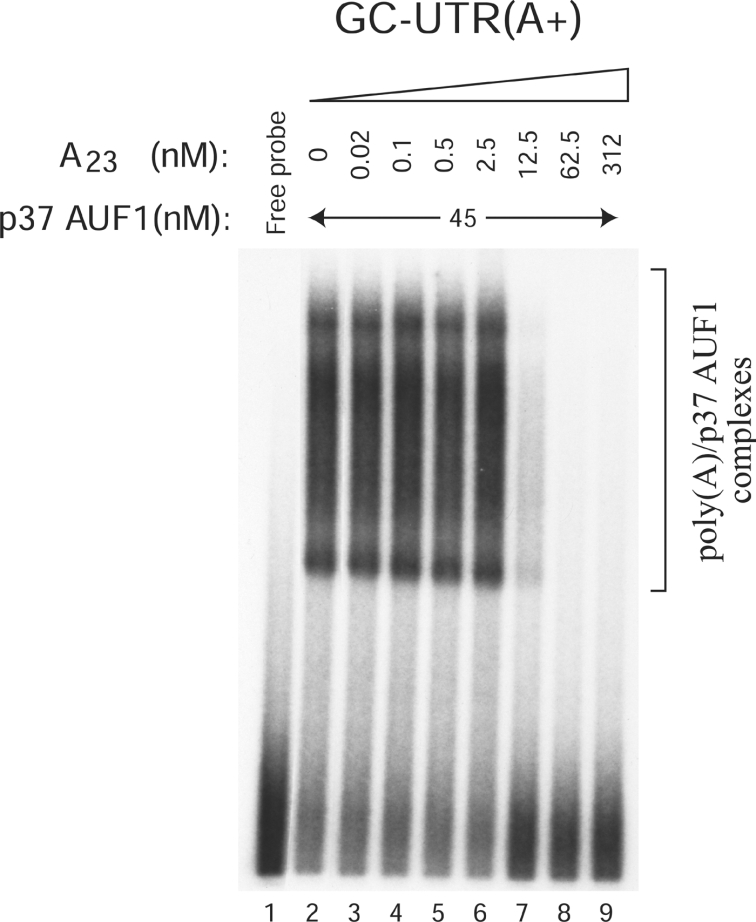
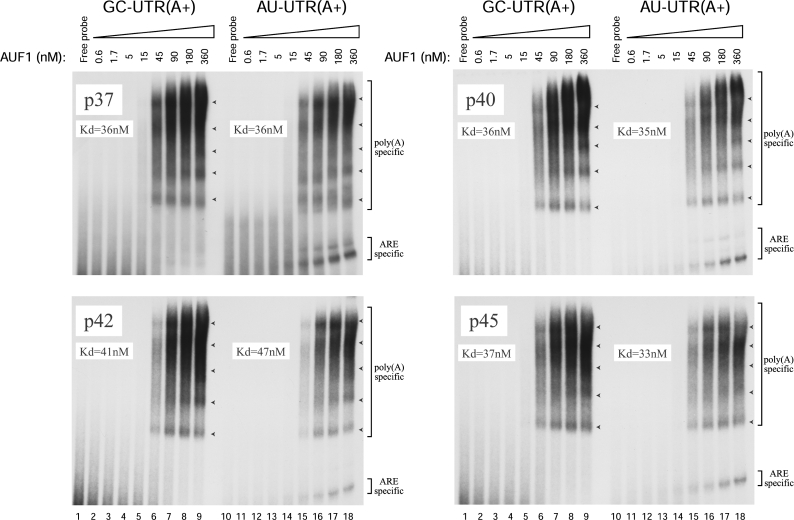
We then investigated the ability of p37AUF1 to bind to the polyadenylated RNA probes AU-UTR(A+) and GC-UTR(A+) constructed from AU-UTR(A–) and GC-UTR(A–) respectively, to which a 100 nt-long poly(A) tail had been added. Note that as GC-UTR(A–) does not bind any protein, GC-UTR(A+) can be considered as a poly(A) sequence. When loaded on to a non-denaturing low-ionic-strength 6% polyacrylamide gel, the undigested polyadenylated RNA probes were smearing, whereas they showed mostly a single band when loaded on to a 6% urea-containing polyacrylamide gel (results not shown). As this smear was not observed with the deadenylated GC-UTR(A–) or AU-UTR(A–) probe (see Figure 2, left panels), it is likely due to the poly(A) sequence. In accordance with this observation, similar results were obtained in non-denaturing low-ionic-strength polyacrylamide gel by other authors when using a 100 nt-long RNA probe containing a 85 nt-long poly(A) sequence [31]. Finally, in order to distinguish between the ARE- and the poly(A)-specific RNP complexes, we treated the binding mixtures with RNase T1 prior to loading on to gel. As shown in Figure 2 (upper right panel), poly(A)-specific RNP complexes were easily detectable with 45 nM of p37AUF1 and 1 nM of either GC-UTR(A+) or AU-UTR(A+) probe (lanes 6 and 15 respectively). Here again, the binding pattern of p37AUF1 displayed an oligomeric pattern with a ladder of RNP complexes (arrowheads). Moreover, the intensity of the higher oligomeric complexes dose-dependently increased, while AUF1 concentration increased. Because the binding samples were treated with RNase T1, the apparent affinity of p37AUF1 for the poly(A) (Kd=35±5 nM) was evaluated through the amount of RNP complexes formed with the poly(A) part of each polyadenylated probe and cannot be compared with the affinity obtained with the ARE, where the Kd values were calculated with the remaining free RNA probe (see the Experimental section for details). Affinity of p37AUF1 for the poly(A) tail was similar in the presence [GC-UTR(A+)] or the absence [AU-UTR(A+)] of the GM-CSF ARE (Figure 2, upper right panel). Control GMSA performed with PABP and the GC-UTR(A+) probe showed, as expected, that the PABP associated in vitro with its natural ligand (Figure 2, lower right panel, Kd=47±11 nM) with an affinity comparable with that of p37AUF1. Here again, the presence of the ARE did not notably change the PABP-binding affinity. Interestingly, the binding pattern of PABP was different from that obtained with p37AUF1. Indeed, as noted above, p37AUF1 binding to the poly(A) part of GC-UTR(A+) was oligomeric (Figure 2, upper right panel). In the case of PABP, a similar degree of oligomerization was obtained with the poly(A) at a concentration approximately 4 times higher than with p37AUF1 (Figure 2, lower right panel). It is noteworthy that an increase of 0.92 log units in p37AUF1 concentration was required to increase the fraction of p37AUF1–poly(A) complex from 10 to 90%, indicating a possible co-operativity in the binding of p37AUF1 to poly(A) [32]. To support further the specificity of the AUF1–poly(A) tail association, we performed competitive experiments using 1 nM of GC-UTR(A+) probe, 45 nM p37AUF1 and an increasing amount of an unlabelled 23-mer oligo(A). As shown in Figure 3, a 12.5-fold excess of unlabelled 23-mer oligo(A) very efficiently abolished the binding of p37AUF1 to the poly(A), demonstrating the specificity of the interaction between p37AUF1 and the poly(A). It is noteworthy that when the unlabelled competitor was in a 2.5-fold excess, the binding of p37AUF1 to GC-UTR(A+) remained mainly unchanged. However, when the competitor was in 12.5-fold excess, the binding of p37AUF1 to GC-UTR(A+) was suddenly lost. These results are consistent with the results of Figure 2 and support the idea that in vitro, both association and dissociation of p37AUF1 with or from the 100 nt-long poly(A) are co-operative. The ability of a unlabelled 23-mer oligo(A) to compete efficiently with the binding of p37AUF1 to a 100 nt-long poly(A) also suggests that the binding site of AUF1 for poly(A) spans 23 adenine bases or less. Since both GC-UTR(A+) and AU-UTR(A+) RNA probes are 100 nt long, these probes may potentially contain at least four AUF1-binding sites. This hypothesis is supported by the fact that the oligomeric pattern of AUF1 binding to the poly(A) sequence of either GC-UTR(A+) or AU-UTR(A+) showed five potential protein–RNA complexes (arrowheads in Figure 2, upper right panel). Finally, we compared the in vitro binding affinity of the four different AUF1 isoforms for the poly(A). Our results (Figure 4) showed that the four isoforms bound to the poly(A) in an oligomeric and co-operative way as efficiently as shown for p37AUF1 and with a ladder of RNP complexes corresponding to five potential binding sites on the 100 nt-long poly(A) (arrowheads). Here again, the ARE did not notably change the binding affinity of the four AUF1 isoforms for the poly(A) tail (Figure 4).
Figure 3. The binding of p37AUF1 to the poly(A) is specific.
Purified His6–p37AUF1 (45 nM) was mixed with 1 nM of 32P-labelled GC-UTR(A+) and an increasing amount of unlabelled oligo(A)23 (concentrations as indicated). Mixtures were treated by RNase T1 prior to loading on to gel.
Figure 4. In vitro, the four AUF1 isoforms bind similarly to poly(A).
Purified His6–p37AUF1 (upper left panel), His6–p40AUF1 (upper right panel), His6–p42AUF1 (lower left panel) or His6–p45AUF1 (lower right panel) (concentrations as indicated) was mixed with 1 nM of either 32P-labelled GC-UTR(A+) or AU-UTR(A+) as indicated above the panels. The reaction mixtures were treated with RNase T1 prior to loading on to gel. Arrowheads show the ladder of the AUF1 protein–poly(A) complexes. Kd values were calculated from the presented experiments.
Recombinant AUF1 proteins efficiently compete with PABP for the binding to poly(A)
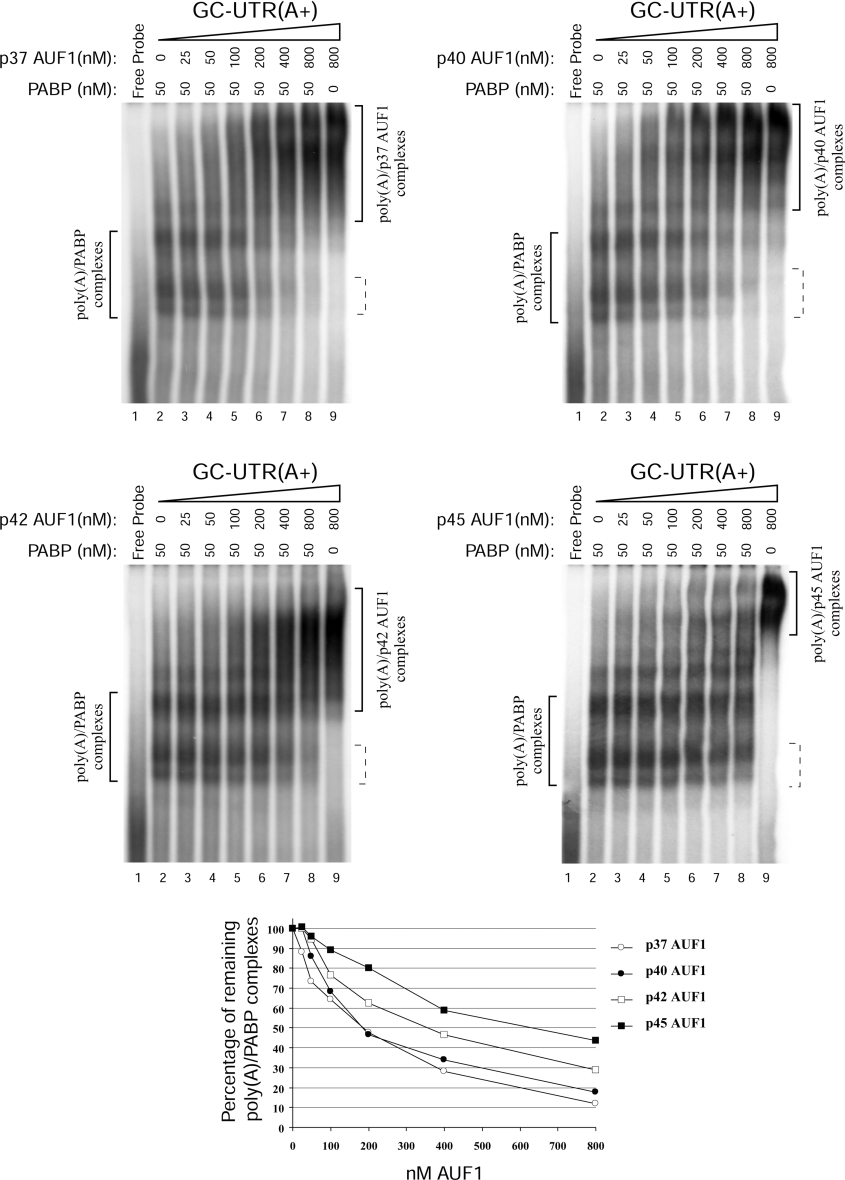
The observation of AUF1 binding to poly(A) led us to investigate whether this could play a role in the ARE-mediated removal of PABP from the poly(A) tail [7]. Since p37AUF1 and PABP had a similar affinity for the poly(A) (Figure 2), we hypothesized that the interference with the PABP–poly(A) tail association by the ARE–ARE-BP complex [7] and the destabilizing effect of AUF1 in mRNA decay could be due to a competition between AUF1 and PABP for the poly(A). We first assessed that the formation of the PABP–poly(A) complexes under our GMSA conditions could be efficiently abrogated in vitro by a specific inhibitor. This was done by performing competition binding assays using 1 nM GC-UTR(A+) probe, 50 nM PABP and an increasing amount of purified PAIP2, a PAIP known to inhibit the binding of PABP to poly(A) [33]. As expected, PAIP2 specifically inhibited the binding of PABP to poly(A), whereas it was unable to dislodge p37AUF1 from the poly(A) (results not shown). Secondly, we checked whether or not AUF1 was able to interact with PABP under our GMSA conditions. PAIP2 was used as a control for our interaction assays. Either 200 nM of GST–PAIP2 (PABP has two PAIP2-binding sites [33]) or 100 nM of p37AUF1 was resolved on a native polyacrylamide gel in the presence or absence of 100 nM of recombinant PABP (and in the presence of 150 nM BSA). As shown in Figure 5, when the mixture of PABP and PAIP2 was resolved on the native gel, a retardation of the migration of both proteins was observed (lane 6, arrowheads). Unexpectedly, PABP was also partially detected by the 5B9 polyclonal anti-AUF1 antibody (Figure 5, upper panel, lane 2). This cross-immunodetection allowed us to observe the retardation of PABP migration in the presence of PAIP2 (Figure 5, upper panel, lane 6, asterisk). Taken together, these results clearly demonstrated that PAIP2 and PABP interacted with each other under our GMSA conditions. Surprisingly, the detection of PABP by immunoblotting was strongly decreased in the presence of PAIP2 (Figure 5, middle panels, compare lanes 2 and 4 with lane 6). It should be noted that the electrophoresis and protein transfer were made under non-denaturing conditions. Therefore this observation may suggest that the antigenic epitope recognized by the 10E10 anti-PABP antibody was masked by the interaction of PAIP2 with PABP and is localized in one of the two PAIP2-binding sites for PABP. In the presence of 100 nM of p37AUF1, no retardation in the migration of either PABP (Figure 5, middle panels, compare lanes 2 and 4) or PAIP2 (Figure 5, bottom panel, compare lanes 3 and 5) was observed, demonstrating that under our GMSA conditions, p37AUF1 did not interact with either PABP or PAIP2. Similar results were obtained with 100 nM of the three other AUF1 isoforms (results not shown). We then investigated whether the different AUF1 isoforms could compete with PABP for the poly(A) in vitro. Competition assays were first performed using 1 nM GC-UTR(A+) probe, 50 nM PABP and an increasing amount of each recombinant AUF1 isoform. That concentration of PABP was chosen as it is close to the Kd of PABP for the poly(A) and because, at this concentration, the PABP–poly(A) complexes displayed a low degree of oligomerization (Figure 2, lower right panel). Results in Figure 6 showed that increasing concentrations of AUF1 dose-dependently reduced the formation of the PABP–poly(A) complexes, while there was a progressive increase in the formation of the AUF1–poly(A) complexes. Approximately 50% reduction in the formation of the PABP–poly(A) complexes was obtained using a 4-fold molar excess of either p37AUF1 or p40AUF1 (Figure 6, see graph). A similar percentage reduction in the formation of the PABP–poly(A) complexes was obtained using an 8-fold molar excess of p42AUF1 or a 12-fold molar excess of p45AUF1. These results demonstrated that the ability of the different AUF1 isoforms to displace the PABP from the poly(A) was not equivalent and that p37AUF1 and p40AUF1 were the most efficient competitors, whereas p45AUF1 was the least efficient. As appearance of a new complex containing PABP, AUF1 and the polyadenylated probe was not observed in our assays, we concluded that under our conditions, AUF1 and PABP were unable to interact with each other in the presence of the poly(A). Altogether, these results allowed us to conclude that AUF1 isoforms (in this order p37AUF1=p40AUF1>p42AUF1>p45AUF1) can efficiently displace the PABP from the poly(A) in vitro. In converse experiments, the reduction of the AUF1–poly(A) complexes could not be easily observed because of the co-migration of the PABP–poly(A) complexes when the PABP molar concentration increased. However, we observed a dose-dependent reduction in the formation of a part of the p37AUF1–poly(A) and p40AUF1–poly(A) complexes when PABP molar concentrations increased (results not shown). But because of this co-migration, we were unable to estimate the reduction of the p37AUF1–poly(A) or p40AUF1–poly(A) complexes when the concentration of PABP increased. From these results, we can conclude that, at least for p37AUF1 and p40AUF1, AUF1 and PABP can alternatively displace each other from the poly(A).
Figure 5. AUF1 does not interact with PABP under our GMSA conditions, whereas PABP and PAIP2 do.
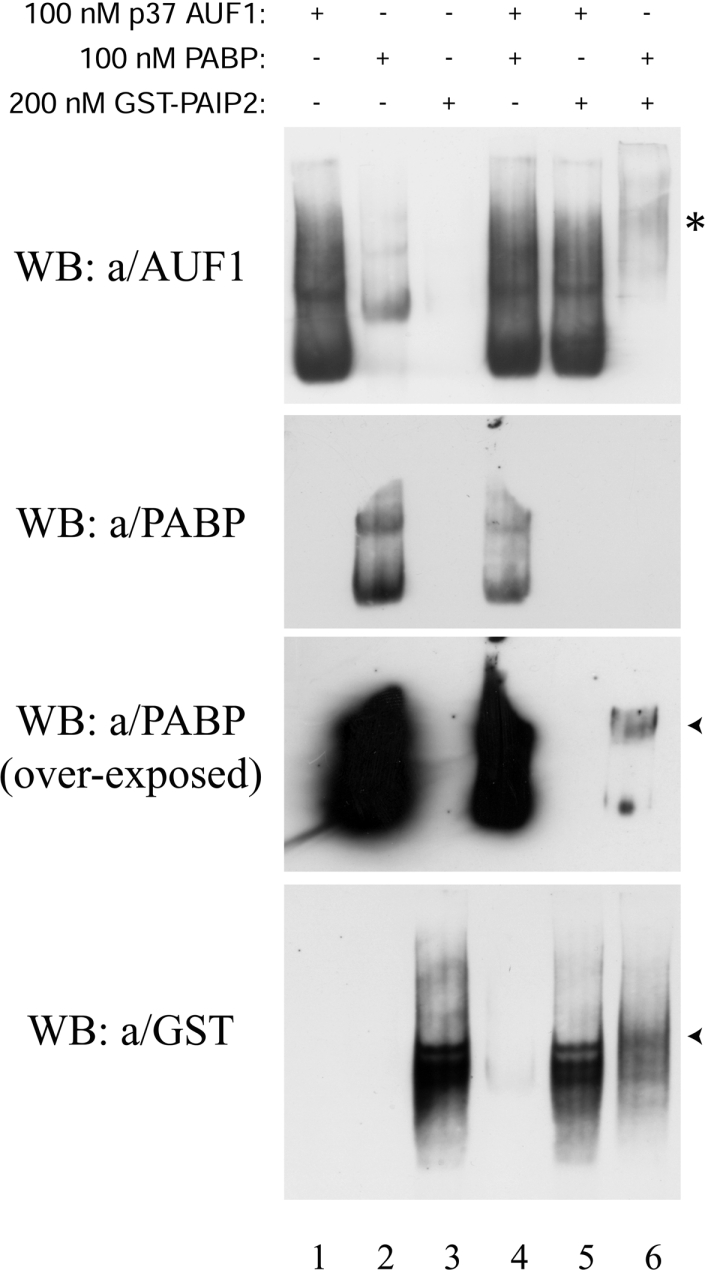
Purified His6–p37AUF1 (100 nM), His6–PABP (100 nM) or GST–PAIP2 (200 nM) were incubated for 15 min at room temperature in binding buffer A alone or together as indicated. The reaction mixtures were then resolved on a non-denaturing low-ionic-strength 6% polyacrylamide gel. After transfer in 0.5×TBE buffer (1×TBE=45 mM Tris/borate and 1 mM EDTA), the proteins were detected by Western blotting using the 5B9 anti-AUF1, 10E10 anti-PABP and anti-GST HRP-labelled antibodies respectively. Arrowheads show the retardation of the migration of either PABP or PAIP2. The asterisk shows the retardation of the migration of PABP revealed by the cross-reactivity using the 5B9 polyclonal anti-AUF1 antibody.
Figure 6. AUF1 proteins compete with PABP for the binding to poly(A).
Purified His6–p37AUF1 (upper left panel), His6–p40AUF1 (upper right panel), His6–p42AUF1 (middle left panel) or His6–p45AUF1 (middle right panel) (concentrations as indicated) was mixed with 1 nM of 32P-labelled GC-UTR(A+) and 50 nM of His6–PABP (except in lanes 9). The reaction mixtures were treated with RNase T1 prior to loading on to gel. The percentages of the remaining poly(A)–PABP complexes (shown in dashed brackets) were plotted against the concentration of each recombinant AUF1 protein; 100% corresponds to the amount of poly(A)–PABP complex formed from the mixture of 1 nM of 32P-labelled GC-UTR(A+) and 50 nM of His6–PABP (lane 2) in the absence of recombinant AUF1 proteins. The results are summarized in the graph at the bottom of the panel. Results are representative of three independent experiments.
AUF1 is an abundant protein in HeLa cytoplasm and nucleus
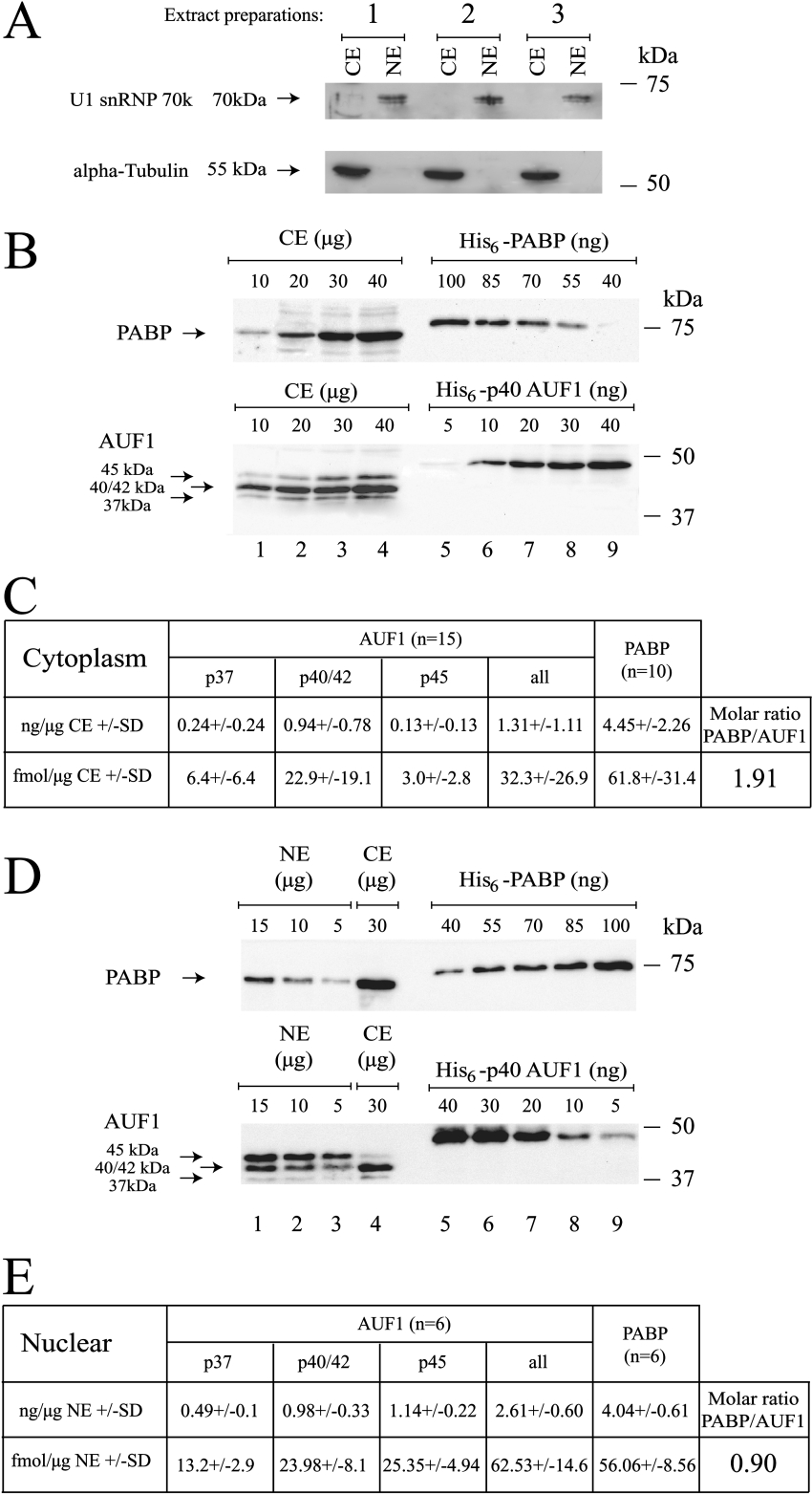
On the basis of these results, we wondered whether the respective amounts of AUF1 and PABP in the cytoplasm and nuclear compartments may allow such competition to occur in vivo. To address this question, AUF1 and PABP were quantified in HeLa cytoplasmic and nuclear extracts using a semi-quantitative Western blotting procedure. We first assessed the purity of three different cytoplasmic and nuclear HeLa extracts by Western blotting. α-Tubulin and U1 snRNP70k, which are respectively specific to the cytoplasm and the nucleus, were only detected in their specific cellular compartment (Figure 7A). We thus proceeded to the determination of AUF1 and PABP amounts in these extracts. A standard curve was created for each protein by plotting the amounts of recombinant protein against the chemiluminescence signal. Several quantities of extract were tested in order to obtain a chemiluminescence signal in the linear range of the standard curve. One representative experiment is shown in Figure 7(B) with the cytoplasmic extracts and one representative experiment is shown in Figure 7(D) with the nuclear extracts. By this strategy, we found that the amounts of AUF1 and PABP in the cytoplasm were respectively 32.3±26.9 and 61.8±31.4 fmol/μg of cytoplasmic proteins (Figure 7C; this difference was statistically significant by using a t test, P<0.03). In the nucleus, the amounts of AUF1 and PABP were respectively 62.53±14.60 and 56.06±8.56 fmol/μg of nuclear proteins (Figure 7E). Because the PABP/AUF1 molar ratio is only 1.91 in HeLa cell cytoplasm and 0.90 in the nucleus, these results show that AUF1 is abundant in both HeLa cytoplasm and nucleus, and support the idea that AUF1 and PABP proteins could compete in vivo for the poly(A).
Figure 7. AUF1 is an abundant protein in HeLa cytoplasm and nucleus.
(A) Cytoplasmic extracts (CE) (20 μg) and 5 μg of nuclear extracts (NE) from three distinct preparations were resolved on a 10% polyacrylamide gel and analysed by Western blotting using either an anti-α-tubulin or anti-U1 snRNP70k antibody as indicated. (B) Various amounts of HeLa cytoplasmic proteins (CE, lanes 1–4, indicated in μg), His6–PABP (lanes 5–9, top panel, indicated in ng) or His6–p40AUF1 (lanes 5–9, bottom panel, indicated in ng) were resolved on an 8% polyacrylamide gel and analysed by Western blotting using either an anti-PABP (upper panel) or anti-AUF1 (bottom panel) antibody. (C) The Table summarizes the results from independent experiments (number as indicated) using the three distinct HeLa cytoplasmic extracts shown in (A). Results are expressed in either ng or fmol of corresponding protein per μg of HeLa cytoplasmic extract±S.D. (D) Various amounts of HeLa nuclear proteins (NE, lanes 1–3, indicated in μg) and cytoplasmic proteins (CE, lane 4, indicated in μg), His6–PABP (lanes 5–9, upper panel, indicated in ng) or His6–p40AUF1 (lanes 5–9, bottom panel, indicated in ng) were resolved on an 8% polyacrylamide gel and analysed by Western blotting using either an anti-PABP (upper panel) or anti-AUF1 (lower panel) antibody. (E) The Table summarizes the results from independent experiments (number as indicated) using the three distinct HeLa nuclear extracts shown in (A). Results are expressed in either ng or fmol of corresponding protein per μg of HeLa cytoplasmic extract±S.D.
DISCUSSION
Since we previously showed that the GM-CSF ARE–ARE-BP complex intervenes in the binding of PABP to poly(A) and knowing that HuR and TIAR [TIA-1 (T-cell-restricted intracellular antigen)-related protein] do not bind to the poly(A) part of our polyadenylated RNA probes [7], we investigated the hypothesis that other ARE-BPs could bind to poly(A) and intervene in the PABP–poly(A) association. We focused our investigations on AUF1, since its RRM-2 (RNA recognition motif-2) had been shown to share structural and binding similarities with that of PABP [27]. Results from our affinity-chromatography experiments confirmed that like PABP, AUF1 can specifically associate with the poly(A) sequence (Figure 1). Previous reports have shown by co-immunoprecipitation that AUF1 and PABP can belong to the same multimolecular RNP complex [17,21]. Here, we showed that the AUF1–poly(A) association was independent of the PABP amount in the cytoplasmic extract (Figure 1C). This suggests that, in vivo, AUF1 and PABP do not directly or stably interact with each other. This is in accordance with the fact that results of the investigations done with the aim to identify the proteic partners of either AUF1 or PABP [34–36] failed to show a direct interaction between these two proteins. An exception to this concerns the recent report by Lu et al. [37] showing that AUF1, eIF4G (eukaryotic initiation factor 4G) and PABP could directly interact with each other. However, the display of these interactions required the use of a chemical cross-linking agent, suggesting their weakness or instability. An interesting point, however, is the observation that the weak interaction observed between AUF1 and PABP is abrogated by the binding of AUF1 to the ARE [37], a property that may help AUF1 to act later as a competitor for the poly(A). In our interaction assays (Figure 5), whereas PABP directly and firmly interacted with PAIP2, we were unable to show any interaction of AUF1 with either PABP or PAIP2. It is therefore likely that AUF1 and PABP participate in the same regulatory processes, since they belong to the same multimolecular RNP complex [17,21] and bind to and compete for the poly(A) (the present study). However, our results do not support the idea that these two proteins directly or firmly interact with each other.
Whereas association of AUF1 with the ARE is widely documented [2,4], its association with the poly(A) had never been investigated. We therefore further investigated the AUF1–poly(A) association with recombinant histidine-purified AUF1 proteins. Our in vitro results showed that the four AUF1 isoforms bound to the poly(A) sequence with an affinity comparable with that of PABP. Complementary experiments showed that this binding did not depend on the GM-CSF ARE sequence itself. The binding pattern of the four AUF1 proteins to our 100 nt-long poly(A) sequence displayed a clear oligomeric pattern (Figures 2 and 4) showing some property of a co-operative binding [32]. Similar results were obtained when AUF1 dissociated from our poly(A) (Figure 3). AUF1, which is dimeric in solution, can bind to AREs in the form of tetramers or hexamers [10,29,30]. It is assumed that such a large structure might help in the packaging of the RNA or facilitate its interaction with other regulatory proteins [10]. It is therefore likely that AUF1 binds to poly(A) with similar or even higher degrees of oligomerization than previously reported. This is supported by the binding pattern of the AUF1 proteins to the poly(A) showing a ladder likely to correspond to a successive association of AUF1 dimers. When using the PABP as positive control for the binding to our polyadenylated RNA probes, an oligomeric pattern was also observed with the poly(A) sequence. However, higher concentrations of PABP were required in order to display an oligomerization amount comparable with that of p37AUF1. Therefore not only AUF1 specifically bound in vitro to the poly(A) tail with an affinity close to that obtained with the PABP, but it could also generate, at a given concentration, higher ranks of oligomerization with the poly(A) than PABP. This feature could help AUF1 to efficiently prevent the PABP–poly(A) tail association and/or provoke the PABP–poly(A) dissociation. The ability of AUF1 to remove the PABP from the poly(A) was demonstrated in our competitive experiments (Figure 6) where an increasing amount of AUF1 dose-dependently displaced the PABP from the poly(A) by generating poly(A)–AUF1 complexes of higher molecular mass. In our hands, the four AUF1 isoforms efficiently compete with PABP for the poly-(A) in this order p37AUF1=p40AUF1>p42AUF1>p45AUF1. Because in vitro the different AUF1 isoforms specifically bind to poly(A) in an oligomeric fashion with a binding affinity close to that of PABP and compete with PABP, AUF1 could be considered as an efficient PABP competitor in vivo.
Some published results indirectly suggested an interaction of AUF1 with non-ARE parts of the mRNA. First, the different AUF1 isoforms were shown to reduce the half-life of a non-ARE reporter mRNA and p37AUF1 was shown to interact directly with this reporter mRNA [12]. Secondly, a hepatocyte mRNA population significantly reduced the interaction between AUF1 and the GM-CSF ARE in the presence of tRNAs used as non-specific competitors [15]. The identification of RNP–RNA interactions in vivo showed that AUF1 bound to the 5′ and 3′ non-translated regions of the ankylosis mRNA into cortical neurons [38]. Cellular factors, including AUF1, were shown to associate with the non-translated region of the hepatitis C virus RNA (+) genome [39]. Finally, two different strategies led to the identification of hundreds of AUF1-interacting mRNAs, of which some do not contain obvious AREs [15,16].
Concerning the interaction of AUF1 with the poly(A), here again, published results supported such an interaction. By comparative molecular modelling, RRM-2 of AUF1 was shown to have structural similarities to the RRM-2 of PABP, and to be able to elaborate either base-specific or non-base-specific interactions with the adenine residues through hydrogen bond or stacking interaction respectively [27]. In the same report, GMSA experiments with a purified His6-tagged p37AUF1 showed that AUF1 had comparable affinities for various RNA probes constituted by the wild-type TNF-α (tumour necrosis factor-α) ARE, several mutants of this ARE or a 25 nt-long poly(A) [27]. Another report corresponding to the original discovery of AUF1 [31] showed that a 7 S complex from the human erythroleukaemia K562 post-ribosomal S130 supernatant containing AUF1 and displaying an mRNA decay activity could bind to the c-myc ARE. The binding of this complex to the c-myc ARE was displaced by either a poly(U) or a poly(A) competitor. However, AUF1 binding was not detected when using a radiolabelled poly(A) probe [31].
Immunofluorescence experiments have shown that PABP is mainly cytoplasmic [40], whereas AUF1 is largely but not exclusively localized in the nucleus [12]. Because it is presumed that PABP is more abundant than AUF1 in the cytoplasm, it is difficult to understand how AUF1 could compete in vivo with PABP in this compartment. To assess this point precisely, we determined by Western blotting the amount of each protein in HeLa cytoplasm and nucleus. Our results showed that, like PABP, AUF1 is an abundant protein in HeLa cells, being roughly 2-fold less abundant than PABP in the cytoplasm and similarly abundant in the nucleus. Because AUF1 can bind the poly(A) and is an abundant protein, this raises the idea that AUF1 may compete with PABP in vivo.
It is therefore possible that in the nucleus, AUF1, with the help of other factors and/or depending on its phosphorylation status, may associate with both the ARE and poly(A) tail of the same mRNA before its export to the cytoplasm. Once in the cytoplasm, since PABP is (two times) more concentrated than AUF1 in this compartment, it may compete with AUF1 for the poly(A) and maybe also for the ARE (since PABP binds to the ARE [19,20]; the present study) to allow mRNA translation. Therefore the balance between the association of either AUF1 or PABP with the poly(A) and/or the ARE may control the fate of the ARE-containing mRNAs by triggering either their decay or their translation. Such a model is compatible with the interesting observations that (i) AUF1 activity in mRNA decay depends on its nuclear import [41], (ii) AUF1 is associated with cytosolic fraction [16], whereas PABP is found in polysome-bound fraction, (iii) AUF1-induced mRNA decay is linked to the exosome [13] and the ubiquitin–proteasome pathway [21].
In conclusion, from the in vitro results presented in the current study, we propose that the destabilizing ARE-BP AUF1 could play a major role in the ARE-mediated dissociation or non-association of PABP with the poly(A) tail. The direct consequences of this non-association would be an activation of the poly(A) shortening, decapping and of mRNA body degradation [4,5,18]. In vivo studies are currently under way to investigate the physiological relevance of the AUF1 binding to the poly(A) in the ARE-mediated processes.
Acknowledgments
We thank Gary Brewer (Department of Molecular Genetics, Microbiology and Immunology, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, Piscataway, NJ, U.S.A.), Gideon Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, Philadelphia, PA, U.S.A.), Richard E. Lloyd (Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX, U.S.A.), Robert J. Schneider (Department of Microbiology, New York University School of Medicine, New York, NY, U.S.A.), Nahum Sonenberg (Department of Biochemistry and McGill Cancer Center, McGill University, Montreal, Quebec, Canada) and Joan A. Steitz (Department of Molecular Biophysics and Biochemistry, and Howard Hughes Medical Institute, Yale University School of Medicine, New Haven, CT, U.S.A.) for kindly providing materials. We thank Sonia Dheur, Michel Hugues, Agnès Méreau and Luc Paillard for helpful discussions. This work was supported by the INSERM (Paris, France), the Université Victor Segalen Bordeaux 2, the Conseil Régional d'Aquitaine, the Ligue Régionale contre le Cancer – Comités Aquitaine – Dordogne (Bordeaux, France), and the Association pour la Recherche contre le Cancer (Paris, France).
References
- 1.Bakheet T., Frevel M., Williams B. R., Greer W., Khabar K. S. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guhaniyogi J., Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 3.Kruys V., Huez G. Translational control of cytokine expression by 3′ UA-rich sequences. Biochimie. 1994;76:862–866. doi: 10.1016/0300-9084(94)90188-0. [DOI] [PubMed] [Google Scholar]
- 4.Wilusz C. J., Wormington M., Peltz S. W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 5.Barreau C., Paillard L., Osborne H. B. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoecklin G., Mayo T., Anderson P. ARE-mRNA degradation requires the 5′–3′ decay pathway. EMBO Rep. 2005;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grosset C., Boniface R., Duchez P., Solanilla A., Cosson B., Ripoche J. In vivo studies of translational repression mediated by the granulocyte-macrophage colony-stimulating factor AU-rich element. J. Biol. Chem. 2004;279:13354–13362. doi: 10.1074/jbc.M308003200. [DOI] [PubMed] [Google Scholar]
- 8.Gallie D. R. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 9.Dean J. L., Sully G., Clark A. R., Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell. Signalling. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Wilson G. M., Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 11.Xu N., Chen C. Y., Shyu A. B. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar B., Xi Q., He C., Schneider R. J. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 2003;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C. Y., Gherzi R., Ong S. E., Chan E. L., Raijmakers R., Pruijn G. J., Stoecklin G., Moroni C., Mann M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 14.Raineri I., Wegmueller D., Gross B., Certa U., Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya S., Giordano T., Brewer G., Malter J. S. Identification of AUF-1 ligands reveals vast diversity of early response gene mRNAs. Nucleic Acids Res. 1999;27:1464–1472. doi: 10.1093/nar/27.6.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lal A., Mazan-Mamczarz K., Kawai T., Yang X., Martindale J. L., Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosset C., Chen C. Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A. B. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 18.Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 19.Bollig F., Winzen R., Gaestel M., Kostka S., Resch K., Holtmann H. Affinity purification of ARE-binding proteins identifies polyA-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem. Biophys. Res. Commun. 2003;301:665–670. doi: 10.1016/s0006-291x(03)00015-9. [DOI] [PubMed] [Google Scholar]
- 20.Sladic R. T., Lagnado C. A., Bagley C. J., Goodall G. J. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur. J. Biochem. 2004;271:450–457. doi: 10.1046/j.1432-1033.2003.03945.x. [DOI] [PubMed] [Google Scholar]
- 21.Laroia G., Cuesta R., Brewer G., Schneider R. J. Control of mRNA decay by heat shock–ubiquitin–proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 22.Gherzi R., Lee K. Y., Briata P., Wegmuller D., Moroni C., Karin M., Chen C. Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Lykke-Andersen J., Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson G. M., Sutphen K., Chuang K., Brewer G. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J. Biol. Chem. 2001;276:8695–8704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]
- 25.Joachims M., Van Breugel P. C., Lloyd R. E. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voeltz G. K., Ongkasuwan J., Standart N., Steitz J. A. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moraes K. C., Lee W. H., Kobarg J. Analysis of the structural determinants for RNA binding of the human protein AUF1/hnRNP D. Biol. Chem. 2002;383:831–837. doi: 10.1515/BC.2002.087. [DOI] [PubMed] [Google Scholar]
- 28.Makeyev A. V., Liebhaber S. A. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson G. M., Sun Y., Lu H., Brewer G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J. Biol. Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- 30.DeMaria C. T., Sun Y., Wagner B. J., Long L., Brewer G. A. Structural determination in AUF1 required for high affinity binding to A+U-rich elements. Nucleic Acids Symp. Ser. 1997;36:12–14. [PubMed] [Google Scholar]
- 31.Brewer G. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 33.Khaleghpour K., Kahvejian A., De Crescenzo G., Roy G., Svitkin Y. V., Imataka H., O'Connor-McCourt M., Sonenberg N. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 2001;21:5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craig A. W., Haghighat A., Yu A. T., Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 35.Khaleghpour K., Svitkin Y. V., Craig A. W., DeMaria C. T., Deo R. C., Burley S. K., Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 36.Moraes K. C., Quaresma A. J., Maehnss K., Kobarg J. Identification and characterization of proteins that selectively interact with isoforms of the mRNA binding protein AUF1 (hnRNP D) Biol. Chem. 2003;384:25–37. doi: 10.1515/BC.2003.004. [DOI] [PubMed] [Google Scholar]
- 37.Lu J. Y., Bergman N., Sadri N., Schneider R. J. Assembly of AUF1 with eIF4G–poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12:883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zielinski J., Kilk K., Peritz T., Kannanayakal T., Miyashiro K. Y., Eiriksdottir E., Jochems J., Langel U., Eberwine J. In vivo identification of ribonucleoprotein–RNA interactions. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1557–1562. doi: 10.1073/pnas.0510611103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris D., Zhang Z., Chaubey B., Pandey V. N. Identification of cellular factors associated with the 3′ nontranslated region of the hepatitis C virus genome. Mol. Cell. Proteomics. 2006;5:1006–1018. doi: 10.1074/mcp.M500429-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Gorlach M., Burd C. G., Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp. Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 41.Chen C. Y., Xu N., Zhu W., Shyu A. B. Functional dissection of hnRNP D suggests that nuclear import is required before hnRNP D can modulate mRNA turnover in the cytoplasm. RNA. 2004;10:669–680. doi: 10.1261/rna.5269304. [DOI] [PMC free article] [PubMed] [Google Scholar]