Abstract
RNA-binding activity of IRP1 (iron regulatory protein 1) is regulated by the insertion/extrusion of a [4Fe-4S] cluster into/from the IRP1 molecule. NO (nitic oxide), whose ability to activate IRP1 by removing its [4Fe-4S] cluster is well known, has also been shown to down-regulate expression of the IRP1 gene. In the present study, we examine whether this regulation occurs at the transcriptional level. Analysis of the mouse IRP1 promoter sequence revealed two conserved putative binding sites for transcription factor(s) regulated by NO and/or changes in intracellular iron level: Sp1 (promoter-selective transcription factor 1) and MTF1 (metal transcription factor 1), plus GAS (interferon-γ-activated sequence), a binding site for STAT (signal transducer and activator of transcription) proteins. In order to define the functional activity of these sequences, reporter constructs were generated through the insertion of overlapping fragments of the mouse IRP1 promoter upstream of the luciferase gene. Transient expression assays following transfection of HuH7 cells with these plasmids revealed that while both the Sp1 and GAS sequences are involved in basal transcriptional activity of the IRP1 promoter, the role of the latter is predominant. Analysis of protein binding to these sequences in EMSAs (electrophoretic mobility-shift assays) using nuclear extracts from mouse RAW 264.7 macrophages stimulated to synthesize NO showed a significant decrease in the formation of Sp1–DNA and STAT–DNA complexes, compared with controls. We have also demonstrated that the GAS sequence is involved in NO-dependent down-regulation of IRP1 transcription. Further analysis revealed that levels of STAT5a and STAT5b in the nucleus and cytosol of NO-producing macrophages are substantially lower than in control cells. These findings provide evidence that STAT5 proteins play a role in NO-mediated down-regulation of IRP1 gene expression.
Keywords: iron metabolism, iron regulatory protein 1 (IRP1), nitric oxide, promoter regulation, signal transducer and activator of transcription (STAT), transcription factor
Abbreviations: 1400W, N-[3-(aminonethyl)benzoyl]acetamide; DETA/NO, diethylentriamine NONOate (diazeniumdiolate); DFO, desferrioxamine®; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; FAC, ferric ammonium citrate; FCS, fetal calf serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IFN-γ, interferon-γ; GAS, IFN-γ-activated sequence; IRE, iron-responsive element; IRP, iron regulatory protein; KO, knockout; LIP, labile iron pool; L-NMMA, L-NG-monomethyl-L-arginine; LPS, lipopolysaccharide; MRE, metal responsive element; MTF1, metal transcription factor 1; NOS2, nitric oxide synthase 2; ONPG, o-nitrophenyl-β-D-galactopyranoside; RT, reverse transcriptase; Sp1, promoter-selective transcription factor 1; STAT, signal transducer and activator of transcription; SV40, simian virus 40; TF, transcription factor
INTRODUCTION
In mammals, cellular iron homoeostasis is largely co-ordinated at the post-transcriptional level through the action of two cytoplasmic IRPs (iron regulatory proteins) (IRP1 and IRP2). They function by binding to RNA stem–loop structures, known as IREs (iron-responsive elements), located in the 5′- or 3′-untranslated regions respectively of specific mRNAs that code for ferritin and transferrin receptors. The activity of IRPs is modulated by changes in intracellular iron concentrations, becoming activated as translational repressors and/or molecules conferring mRNA stability by low iron and inactivated by high levels of iron in the LIP (labile iron pool) [1]. The inactive IRP1 has a secondary function as cytosolic aconitase when co-ordinating a [4Fe-4S] cluster, which masks the IRE-binding site. It has long been held that IRP1 is a major post-transcriptional regulator that controls iron metabolism in response to fluctuations in intracellular iron. However, recent studies on KO (knockout) IRP1 [2] and KO IRP2 mice [3,4] strongly suggest that IRP2, rather than IRP1, is responsible for maintenance of the constitutive body iron balance. The new concept of IRP1 as a molecular target responding preferentially to stimuli other than iron-related challenges is now under debate [5,6]. Indeed, the two activities of IRP1 have been shown to be modulated by NO (nitric oxide) [7,8], oxidative stress [9,10], phosphorylation [11] and hypoxia/reoxygenation [12].
NO and/or NO-derived species appear to be chemical messengers able to induce a broad spectrum of post-translational modifications of the IRP1 molecule. First, NO may act directly on the [4Fe-4S] cluster of IRP1, inducing its gradual disassembly and removal, favouring the IRE-binding conformation of the protein [13,14]. Secondly, nitration of IRP1, reported in vitro following its exposure to peroxynitrite [14] and in NO- and superoxide (O2•−)-producing macrophages [15], has been shown to prevent IRP1 activation. Lastly, NO is also responsible for the down-regulation of IRP1 expression at both mRNA and protein levels [16].
Needless to say, modulation of IRP1 function is an important element but not the sole contributor in a complex network of interactions between NO and intracellular iron (reviewed in [17,18]). Besides IRP1, IRP2 has been also shown to be regulated by NO [19,20]. It is also important to recall that one of the most apparent hallmarks of NO action originally described by Hibbs et al. [21] is depletion of intracellular iron. Recently, a mechanism of NO-induced iron loss involving glutathione and multidrug resistance-associated protein 1 has been proposed [22]. Another common feature of cells exposed to NO is the formation of iron–nitrosyl complexes, with multiple haem and non-haem iron proteins inhibiting key enzymes in DNA synthesis and mitochondrial respiration (reviewed in 23,24]). Finally, NO has been reported to be a strong inducer of haem oxygenase expression at both the transcriptional [25] and post-transcriptional level [26].
In the present study, we focused on NO-mediated down-regulation of IRP1, an effect that may be of considerable biological importance, especially under conditions of sustained NO synthesis, because it could reduce the amount of the protein available for post-translational modification. The biological mechanisms underlying the NO-induced decrease in IRP1 expression are unknown, although it has been suggested that NO can modify transcriptional activity of the IRP1 gene [16]. Indeed, accumulating evidence suggests that NO affects gene expression at the transcriptional level [27] by modulating the activities of various TFs (transcription factors) such as NF-κB (nuclear factor κB) [28], AP-1 [29], Sp1 [30], HIF-1 (hypoxia-inducible factor-1) [31] and STAT (signal transducer and activator of transcription) [32]. Here, we sought to locate putative binding sites for TFs regulated by NO and/or changes in intercellular iron level. Through examination of the nucleotide sequence of the mouse IRP1 promoter (GenBank® accession no. AJ427344), we identified the following conserved motifs: one Sp1 sequence, one MRE (metal responsive element) and one binding site for STAT family proteins {GAS [IFN-γ (interferon-γ)-activated sequence]}. Functional analysis of the IRP1 promoter region and analysis of these putative TF-binding sites by EMSA (electrophoretic mobility-shift assay) using nuclear extracts from NO-producing RAW 264.7 macrophages indicate that the GAS is an important regulatory element responsible for down-regulation of IRP1 expression by NO.
EXPERIMENTAL
Materials
DMEM (Dulbecco's modified Eagle's medium) was obtained from Gibco-BRL. Low endotoxin FCS (fetal calf serum), FAC (ferric ammonium citrate), DFO (desferrioxamine®) and LPS (lipopolysaccharide) from Escherichia coli (serotype O55:B5) were purchased from Sigma (St. Louis, MO, U.S.A.). Murine recombinant IFN-γ was from RandD Systems. 1400W {N-[3-(aminomethyl)benzoyl]acetamide}, L-NMMA (L-NG-monomethyl-L-arginine) and DETA/NO [diethylentriamine NONOate (diazeniumdiolate)] were purchased from Cayman Chemical Co. (Ann Arbor, MI, U.S.A.). Restriction and modification enzymes were from Invitrogen (Carlsbad, CA, U.S.A.). All oligonucleotides were synthesized by Invitrogen. Competent E. coli DH5α™ cells, ONPG (o-nitrophenyl β-D-galactopyranoside) and Lipofectamine™ were from Invitrogen. Luciferin was from Euromedex. All other chemicals were obtained from Sigma.
Cell culture and treatment
RAW 264.7 murine macrophages, a cell line established from a tumour induced by Abelson murine leukaemia virus, were obtained from the American Type Culture Collection (Rockville, MD, U.S.A.). Cells were cultured in DMEM containing 5% (v/v) FCS and gentamicin (50 μg/ml) in 100 cm2 plastic culture flasks (Nunc) in a humidified atmosphere of 95% air and 5% CO2 at 37 °C. For NOS2 (nitric oxide synthase 2) induction, RAW 264.7 macrophages were stimulated for 16 h with 20 units/ml IFN-γ and 50 ng/ml LPS. In other experiments these cells were incubated with 250 μM DFO and/or 100 μM FAC for 24 h. The human hepatoma cell line HuH7 was maintained in DMEM supplemented with 10% (v/v) FCS, 50 units/ml penicillin and 50 μg/ml streptomycin.
Nitrite measurement
NO production was determined by quantifying nitrite, the stable end-product of NO, spectrophotometrically by a colorimetric assay. Briefly, 200 μl of sample medium was reacted with 800 μl of Griess reagent (1% sulfanilamide and 0.1% naphthylethylenediamine), and the absorbance was measured at 543 nm. Sodium nitrite was used as a standard.
Plasmid constructs
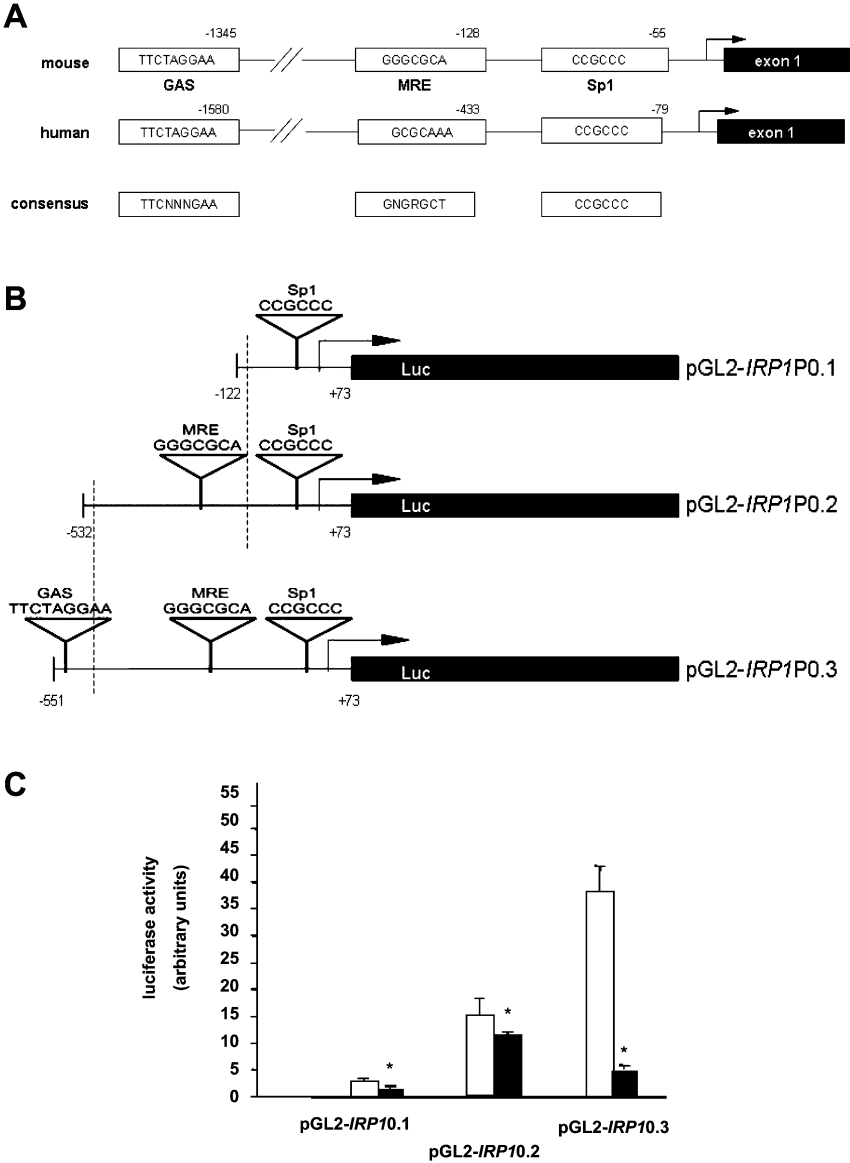
Two fragments from the IRP1 promoter region were PCR amplified using two different forward primers: promIRP1F1 5′-CCGACGCGTCCCGACTAGACTGCAAGCTTCAGCC-ATC-3′ (for the fragment encompassing −122 to +73 bp) and promIRP1F2 5′-CCGACGCGTAGAAATCGGCTCTACACTTCCTGGGACTGAG (for the fragment encompassing −532 to +73 bp) and the same reverse primer promIRP1R 3′-CCGCTCGAGCCGACTCCGCGCTCCTCCCACTGA–5′. The two PCR fragments of respectively 195 and 605 bp were digested with XhoI/MluI and ligated to the XhoI/MluI-cleaved pGL2-basic vector (Promega, Madison, WI, U.S.A.) to create plasmids pGL2-IRP1P0.1 and pGL2-IRP1P0.2 in which the IRP1 promoter sequences were transcriptionally fused in the correct orientation to the luciferase reporter gene. To generate a third construct, pGL2-IRP1P0.3, a double-stranded oligonucleotide containing the upstream GAS sequence from the IRP1 promoter (boldface) as well as 5′ KpnI (underlined) and a 3′ MluI (underlined) restriction site 5′-CCGGGTACCAGGCATTTCTAGGAAAATAAGAACGCGTGCC-3′ was inserted into the pGL2-IRP1P0.2 plasmid following MluI/KpnI digestion to place it upstream of the 605 bp fragment. The orientation and the integrity of all plasmids were verified by sequencing. Figure 1(B) shows a schematic representation of the luciferase reporter constructs used in the present study.
Figure 1. Functional studies of the IRP1 promoter region.
(A) The location of putative binding sites for the TFs regulated by NO and/or changes in intracellular iron level in the mouse IRP1 promoter. The −2790 to +90 bp fragment of the mouse IRP1 gene (GenBank® accession no. AJ427344) was analysed using TESS software and the TRANSFAC database. The indicated sequences located at positions −61 to −55 bp, −135 to −128 bp and −1354 to −1345 bp upstream of the transcription initiation site, represent three potential cis-acting regulatory elements corresponding to Sp1, MRE and the GAS sequence respectively. The consensus sequences and the position of the above-mentioned putative binding sites in human IRP1 gene sequence (AL161783) are indicated. (B) Structure of reporter gene constructs used in transient transfection assays. All constructs were prepared using the pGL2-basic vector. Two overlapping fragments of the mouse IRP1 promoter sequence extending from −122 to +73 bp and −532 to +73 bp relative to the transcription start were amplified and cloned in front of the coding region of the luciferase gene to generate two constructs: pGL2-IRP1P0.1 and pGL2-IRP1P0.2 respectively. A third construct pGL2-IRP1P0.3 was generated by insertion of a 19 bp fragment of the mouse IRP1 promoter containing the GAS sequence upstream of the −532 to +73 bp fragment in pGL2-IRP1P0.2. Sequences corresponding to the putative Sp1-, MTF1- and STAT-binding sites are shown. (C) Basal and NO-dependent transcriptional activity of mouse IRP1 promoter fragments. HuH7 cells were transfected with luciferase reporter gene constructs containing 194 bp (pGL2-IRP1P0.1), 605 bp (pGL2-IRP1P0.2) and 624 bp (pGL2-IRP1P0.3) IRP1 promoter fragments as described in the Experimental section. After 5 h, the cells were treated with 250 μM DETA/NO (solid bars) or left untreated (open bars). After 48 h, the cells were harvested, lysed and assayed for luciferase activity. β-Galactosidase activity produced by the co-transfected plasmid pSV40 β-gal was used to correct for transfection efficiencies and to normalize the luciferase activity of each sample. Results represent the means±S.D. for three separate experiments. Asterisks indicate a statistically significant decrease in promoter activity in NO-treated cells compared with non-NO-treated cells (*P<0.05).
Transfections and reporter gene assays
Twenty-four hours before transfection, HuH7 cells were plated in six-well culture plates (Nunc) at 50–70% confluence. The cells were transiently transfected with 1 μg of plasmid DNA using Lipofectamine™ reagent according to the manufacturer's recommendations. Cells were incubated, 6 h after transfection, with 250 μM DETA/NO for 48 h. After incubation the cells were harvested, total cellular extracts were prepared and luciferase activity was assessed using the Luciferase Assay System kit according to the manufacturer's instructions (Promega). As an internal control to assess transfection efficiency, cells were co-transfected with a reporter vector (pSV β-gal) containing the β-galactosidase gene driven by the SV40 (simian virus 40) virus promoter (Promega). Luciferase activities were normalized relative to co-expressed β-galactosidase activity values measured in a standard assay in which ONPG hydrolysis was quantified by absorbance at 420 nm using a Hitachi U-2000 spectrophotometer. As negative and positive controls for transient transfection experiments, the pGL2-basic vector without any promoter sequence and a pGL3 control vector containing the SV40 promoter were used respectively.
EMSA
Nuclear protein extracts were prepared from cells as previously described [33]. Three double-stranded DNA fragments, created by annealing oligonucleotides, were used as probes: 5′-CCAGGCTCCGCCCCTCGGGG-3′, 5′-CCCCGAGGGGCGGAGCCTGG-3′; 5′-GAAGGGAGGGCGCACCCGGACT-3′, 5′-AGTCCGGGTGCGCCCTCCCTTC-3′ and 5′-AAGGCATTTCTAG-GAAAATAAGAT-3′, 5′-ATCTTATTTTCCTAGAAATGCCTT-3′. These sequences were identical with the following regions of the mouse IRP1 promoter: −70 to −50 bp containing an Sp1-binding site (underlined), −141 to −119 bp containing an MRE sequence (underlined) and −1363 to −1339 bp containing a GAS sequence (underlined). EMSAs were performed as described previously [33]. Briefly, all fragments were labelled with [32P]ATP (NEN Life Science Products–DuPont, Bad Homburg, Germany) using T4 polynucleotide kinase (Fermentas). The radiolabelled probes (0.1–0.5 ng) were incubated with nuclear protein extracts (2.5 μg) for 20 min at room temperature in 20 μl reaction mixtures containing 10 mM Hepes (pH 7.9), 100 mM KCl, 1.5 mM MgCl2, 1 mM DTT (dithiothreitol), 1 mM EGTA and 10% glycerol. The specificity of DNA binding was determined by competition assays performed using a 25- or 50-fold excess of the unlabelled fragments. Reactions were examined by electrophoresis on nondenaturing 6% polyacrylamide gels prepared in a buffer composed of 45 mM Tris (pH 8.0), 45 mM borate and 1 mM EDTA. After running for 2 h, the gels were dried and the radioactive signal visualized using a Bio-Rad Molecular Imager FX and quantified using Quantity One software (Bio-Rad).
Western blot analysis
A total of 50 μg of nuclear or cytoplasmic proteins, prepared as described previously [7,34], were separated by electrophoresis on SDS/8% polyacrylamide gels and transferred to Hybond nitrocellulose membranes (Amersham Biosciences) using Novex® Western Transfer apparatus (Invitrogen). STAT5a and STAT5b proteins were detected by incubation of the membranes with anti-STAT5a- and anti-STAT5b-specific rabbit antibodies kindly provided by Dr Frederic Verdier (INSERM U363, Paris, France) [35]. Nuclear extracts from UT7 cells (kindly provided by Dr Frederic Verdier) overexpressing STAT5a and STAT5b proteins were used as a positive control. Immunoreactive bands were revealed following incubation with a goat anti-rabbit horse-radish peroxidase-conjugated secondary IgG (Sigma) using the chemiluminescence ECL®-plus kit (Amersham Life Sciences). Quantification was performed relative to β-actin detected using a specific antibody obtained from Sigma.
RT (reverse transcriptase)–PCR analysis of STAT5a mRNA level
For RT–PCR analysis, total cellular RNA was extracted from RAW 264.7 macrophages (107 cells) using TRIzol® reagent (Invitrogen, Carlsbad, CA, U.S.A.) according to the manufacturer's protocol. Total RNA (2 μg) was used for reverse transcription with superscript II RNAse H RT (Invitrogen) in a 30 μl reaction mixture incubated at 45 °C for 1 h. Of this reaction mixture, 2 μl were then used for PCR amplification with Platinum Pfx DNA Polymerase (Invitrogen), using oligonucleotide primers specific for STAT5a: forward: 5′-CCGTGGGATGCTATTGACTT-3′, reverse: 5′-ACGTGTTCTGGAGCTGTGTG-3′. PCR products were analysed on 2% agarose gels run with TBE (Tris/borate/EDTA; 45 mM Tris/borate and 1 mM EDTA) buffer and stained with ethidium bromide. The specific band of 169 bp was quantified with a Molecular Imager using Quantity One software (Bio-Rad) and normalized to a 489 bp fragment of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) housekeeping gene amplified from duplicate samples using the forward primer 5′-GACCACAGTCCATGCCATCAC-3′ and the reverse primer 5′-TCCACCACCCTGTTGCTGTAG-3′.
Real-time quantitative RT–PCR
STAT5a mRNA levels in RAW 264.7 cells were measured by realtime quantitative RT–PCR using a Light Cycler (Roche Diagnostics, Mannheim, Germany). The same cDNA templates and primers as those described above were used. Detection of amplification products was carried out using SYBR Green (Roche) as described previously [36]. To confirm amplification specificity, the PCR products from each primer pair were subjected to melting- curve analysis and subsequent agarose-gel electrophoresis. For data analysis, Light Cycler 3.5 Software was used [36]. Quantification was performed relative to GAPDH control reactions.
Statistical analysis
For statistical evaluation, one-way ANOVA was applied. For estimation of the significance of difference between arithmetic means, the Scheffe test was used. All calculations were done using Statgraphics Plus 6.0 software.
RESULTS
Identification of putative binding sites for TFs regulated by NO and iron in the IRP1 promoter
The −2790 to +90 bp fragment of the mouse IRP1 gene (GenBank® accession no. AJ427344) was searched for potential binding sites for TFs regulated by NO and/or changes in intracellular iron level using the TESS (Transcription Element Search System) software (http://www.cbil.upenn.edu/tess/) and TRANSFAC database (http://www.gene-regulation.com/pub/databases.html#transfac). We found the following conserved motifs: one Sp1 sequence, 55 bp away from the transcription start site; one MRE sequence, 128 bp away from the transcription start site; and one GAS motif, a potential binding site for STAT family proteins, 1345 bp upstream of the transcription start site (Figure 1A). All identified sequences are also present in the human IRP1 promoter (GenBank® accession no. AL161783) at approximately the same distances from the transcription start site (Figure 1A).
Predominant role of the GAS sequence in the inhibition of mouse IRP1 promoter activity by NO in transfected HuH7 cells
To perform functional studies of the IRP1 promoter and analyse its regulation by NO, three luciferase-based reporter constructs containing sub-fragments of the IRP1 promoter sequence, with sizes ranging from 195 to 624 bp (Figure 1B), were transiently transfected into HuH7 cells and the cells were grown for 48 h with or without DETA/NO as an NO donor. The pGL3 control vector expressing the luciferase under the control of the SV40 promoter was highly expressed in these cells and there was a 60% reduction in luciferase activity following DETA/NO treatment (results not shown), as previously reported [37]. Transient transfections of HuH7 cells with the pGL2-IRP1P0.1 construct containing the minimal promoter fragment showed that the basal luciferase activity produced by this construct was very low and treatment of the cells for 48 h with a chemical NO donor (DETA/NO, 250 μM), induced a statistically significant 2-fold reduction (P<0.05) in luciferase activity (Figure 1C). The longer 605 bp fragment from pGL2-IRP1P0.2 construct containing both Sp1 and MRE showed a higher transcriptional activity, with a 3–4-fold increase in luciferase activity as compared with HuH7 pGL2-IRP1P0.1-transfected cells. However, the NO treatment only induced a moderate decrease in luciferase activity. Another construct was made by adding an oligonucleotide with the GAS sequence upstream of pGL2-IRP1P0.2 (Figure 1B). The basal activity produced by the pGL2-IRP1P0.3 plasmid was the highest among all the analysed IRP1 promoter-luciferase constructs. Treatment with DETA/NO of pGL2-IRP1P0.3 vector-transfected HuH7 cells revealed a substantial (90%) inhibition of luciferase activity compared with basal conditions (Figure 1C). Considering that the pGL2-IRP1P0.3 construct was generated by the addition, upstream of the 605 bp fragment in pGL2-IRP1P0.2, of a short 19 bp fragment comprising the upstream GAS sequence from the IRP1 promoter, this result suggests the effective and predominant role of the STAT-binding site in both the basal transcriptional activity and the NO-dependent regulation of the mouse IRP1 promoter.
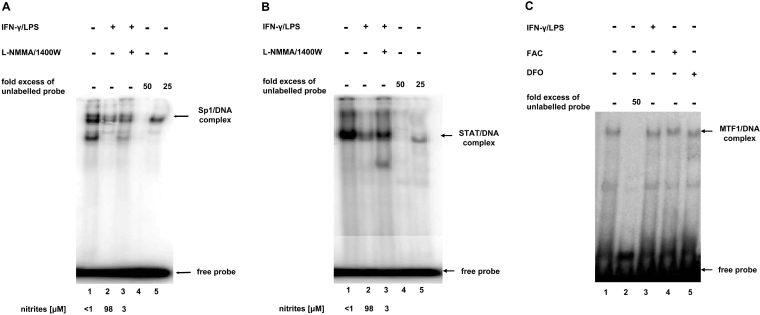
NO inhibits Sp1– and STAT–DNA binding, but has no effect on MTF1 DNA binding
Since the Sp1- and the STAT-binding sites appear to be functional and play a role in NO-mediated down-regulation of the IRP1 promoter we decided to study the effect of NO on the DNA binding activity of Sp1 and STATs in RAW 264.7 macrophage nuclear extracts. We performed EMSAs using short DNA probes comprising the Sp1-binding site or the GAS sequence of the mouse IRP1 promoter. The specificity of the constitutive DNA-binding reactions (Figures 2A and 2B, lanes 1) was confirmed by competition with a 50-fold (Figures 2A and 2B, lanes 4) and a 25-fold (Figures 2A and 2B, lanes 5) molar excess of the unlabelled probes. NO production was stimulated by treating the cells with IFN-γ and LPS and was followed by measuring nitrite (a stable oxidation product of NO) released in the culture medium (as indicated below Figures 2A and 2B). The formation of Sp1–DNA (Figure 2A, lane 2) and STAT–DNA (Figure 2B, lane 2) complexes in reactions including nuclear extracts from NO-producing RAW 264.7 cells was markedly reduced as compared with those using extracts from control cells (Figures 2A and 2B, lanes 1). This decrease was 65 and 47% respectively as assessed by densitometric analysis. The effect of NO was largely reversed when EMSAs were performed using nuclear extracts from stimulated macrophages treated with 1400W and L-NMMA, two NOS2 inhibitors (Figures 2A and 2B, lanes 3). EMSA was also used to visualize nuclear protein complexes with the MRE sequence overlapping the −128 to −135 region of the IRP1 promoter in NO-producing macrophages (Figure 2C). The specificity of the basal MTF1 binding to MRE in control RAW 264.7 macrophages (Figure 2C, lane 1) was confirmed by competition with a 50-fold excess (Figure 2C, lane 2) of the unlabelled MRE probe. NO has been shown to affect LIP levels in various cell types [53,54], and thus may indirectly influence the DNA-binding activity of MTF1, a TF known to bind MRE and to be regulated by heavy metals [38]. However, we detected no changes in MTF1 binding to MRE in RAW macrophages using nuclear extracts from cells stimulated for NO synthesis with IFN-γ/LPS (Figure 2C, lane 3). Similarly, in macrophages incubated either with FAC (Figure 2C, lane 4) or DFO, an iron chelator (Figure 2C, lane 5), the level of MTF1–DNA complexes was unaffected compared with control cells, clearly indicating that fluctuations in intracellular iron level had no effect on MTF1–DNA binding.
Figure 2. Effect of endogenous NO on Sp1–, STAT– and MTF1–DNA binding activity in mouse 264.7 macrophages.
Nuclear protein (2 μg) and a molar excess of 32P-labelled probes containing the (A) Sp1, (B) GAS or (C) MRE sequences from the mouse IRP1 promoter were used in EMSA analysis as described in the Experimental section. Cultured RAW 264.7 macrophages were stimulated for 16 h with 20 units/ml IFN-γ plus 50 ng/ml LPS in the presence or absence of 1 mM L-NMMA and 25 μM 1400W (A, B). NO production was measured by assaying nitrites using the Griess method in culture supernatants of stimulated and unstimulated cells. RAW 264.7 macrophages were incubated with 250 μM DFO and/or 100 μM FAC for 24 h (C). To check the specificity of the DNA binding, unlabelled competitor fragments were added to reaction mixtures at 25- and 50-fold molar excess over the labelled probes. Reaction products were separated by electrophoresis on non-denaturing 6% polyacrylamide gels. The experiments were performed at least three times, and representative gels are shown. The positions of the DNA–protein complexes and the free probes are indicated.
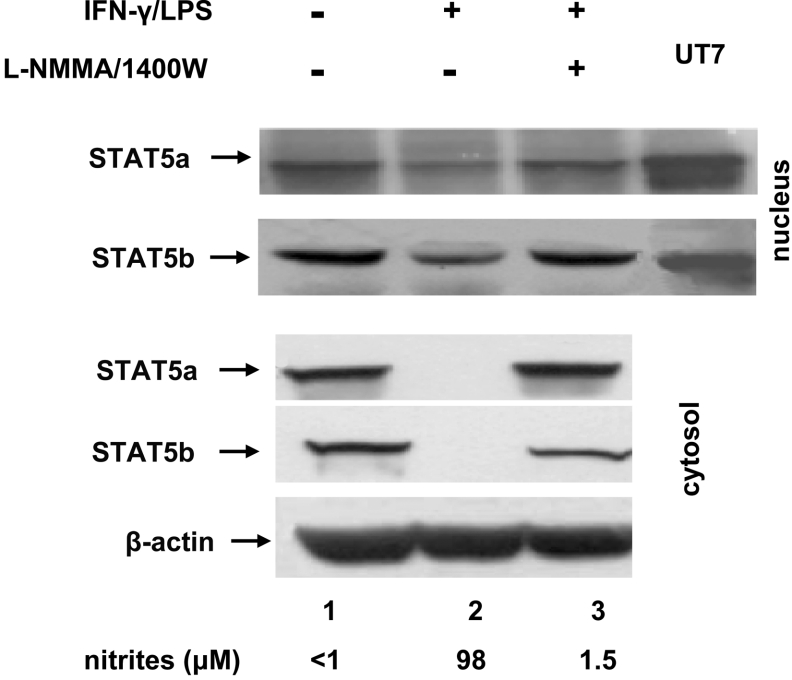
NO decreases STAT5a and STAT5b protein levels in the cytosol and nuclei of NO-producing macrophages
The mechanism underlying the NO-mediated decrease in STAT–DNA complex formation and inhibition of transcriptional activity of the mouse IRP1 promoter, could involve impaired nuclear translocation of STAT proteins in response to NO. Since the STAT protein family comprises seven members showing apparently similar binding affinity for the GAS sequence [39], we focused our attention on the STAT5 proteins whose involvement in signalling pathways inhibited by NO is well recognized [32,40]. We measured STAT proteins in cytosolic and nuclear extracts of RAW 264.7 macrophages, using nuclear extracts from UT7 cells overexpressing STAT5a and STAT5b as positive controls. The results showed a marked reduction in STAT5b protein levels in nuclei of NO-producing RAW 264.7 macrophages as compared with control macrophages (Figure 3, upper panel, lane 2 versus lane 1). However, an even more drastic reduction in both STAT5a and STAT5b proteins, i.e. an almost total absence (Figure 3, lower panel, lane 2), was observed in the cytosol, which indicates that impaired translocation is not the reason for the reduction in STAT–DNA complex formation. This characteristic pattern of changes in STAT protein levels in these two cellular compartments was specific for NO-producing cells and it was largely reversed when NO synthesis had been abolished by inhibitors of NOS2 (Figure 3, lane 3).
Figure 3. Reduced STAT5a and STAT5b protein levels in NO-producing RAW 274.7 macrophages.
RAW 264.7 macrophages were stimulated for 16 h with 20 units/ml IFN-γ plus 50 ng/ml LPS in the presence or absence of 1 mM L-NMMA and 25 μM 1400W. NO production was measured by assaying nitrite in the culture medium. STAT5a and STAT5b levels were assessed in nuclear (upper panel) and cytosolic (lower panel) extracts by Western blotting as described in the Experimental section. To assist identification of STAT5a and STAT5b bands, nuclear extracts from UT7 cells overexpressing STAT5a and STAT5b proteins (a gift from Dr Frederic Verdier) were electrophoresed on the same gels. Each blot is representative of three blots showing similar results.
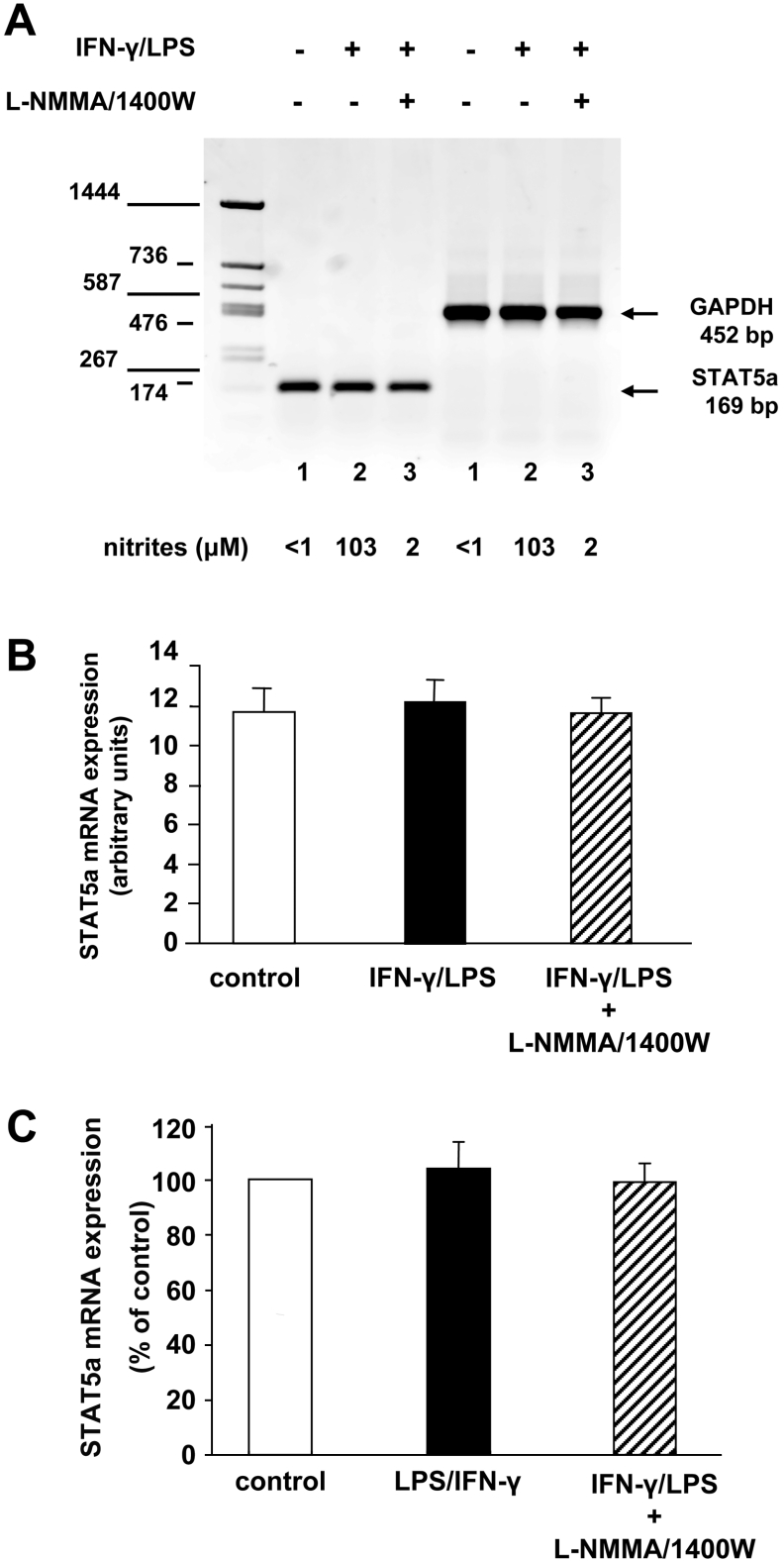
LPS/IFN-γ-induced reduction of STAT5a expression is not due to mRNA down-regulation
We next examined whether this reduction in the amount of STAT5a protein resulted from a decrease in the level of the corresponding mRNA transcript. Both semi-quantitative RT–PCR (Figures 4A and 4B) and real-time quantitative RT–PCR (Figure 4C) consistently showed that STAT5a mRNA expression in NO-producing RAW 264.7 macrophages was unchanged relative to control cells or in comparison with macrophages in which NO synthesis was severely reduced. These analyses suggest that the substantial decrease in STAT5a protein content in response to NO is not due to less efficient transcription of the STAT5a gene or decreased stability of STAT5a mRNA.
Figure 4. Analysis of STAT5a mRNA abundance in NO-producing RAW 264.7 macrophages by RT–PCR.
RAW 264.7 macrophages were stimulated for 16 h with 20 units/ml IFN-γ plus 50 ng/ml LPS in the presence or absence of 1 mM L-NMMA and 25 μM 1400W. NO production was measured by assaying nitrite in the culture medium. RNA was isolated from harvested cells and used for semi-quantitative RT–PCR as described in the Experimental section. The GAPDH amplicon was used as a control for the amount of cDNA used in the PCR reactions. The experiments were performed three times, and a representative result is shown (A). PCR product bands were quantified using a Molecular Imager and the means±S.D. are plotted in arbitrary units (n=3) (B). STAT5a mRNA abundance in RAW 264.7 macrophages was measured by real-time RT–PCR (C). Each column represents the mean (±S.D.) for three amplification reactions performed on a single cDNA sample reverse transcribed from RNA prepared from cells from three independent experiments. The level of STAT5b mRNA in control macrophages was assigned the 100% value.
DISCUSSION
Most studies on the regulation of IRP1 activities have been focused on post-translational mechanisms underlying the insertion/extrusion of a [4Fe-4S] cluster into/from the IRP1 molecule [5,13]. However, we have recently shown that changes in intracellular protein IRP1 levels could account for the observed modifications in its IRE-binding affinity [16,41,42]. It has long been known that in mammals, IRP1 displays highly variable expression patterns in different tissues [43]. The biological significance of regulation of IRP1 expression at the protein level are indicated by studies on NO-producing macrophages, in which NO was shown not only to induce IRP1 RNA binding activity but also to reduce IRP1 protein level [16]. The aim of the present study was to investigate the transcriptional regulation of IRP1 in response to NO and to identify regulatory elements within the IRP1 promoter responsible for NO-dependent inhibition of IRP1 expression. Although previous studies have reported no changes in the total amount of immunologically detectable IRP1 following treatment of various cell types with iron sources and iron chelators [44–46], recent data from closely related mouse lymphoblastic cell lines show a close quantitative correlation between the amount of IRP1 and LIP levels [41]. Furthermore, it has been demonstrated that hepatic IRP1 protein levels in haemochromatosis patients are altered [47]. With this in mind, we also attempted to test the influence of iron on transcriptional regulation of the IRP1 gene. Using bioinformatics software, the mouse IRP1 promoter sequence was screened for putative binding sites for NO- and iron-dependent transcriptional regulators. We identified potential binding sites for Sp1 and STAT proteins, two TFs known to be molecular targets for NO [30,32,40]. We also found one potential MRE sequence similar to those present in the promoter region of genes encoding metallothionein-IIA [48], a heavy-metal binding protein, or bivalent metal transporter 1 [49], a mediator of apical iron uptake in duodenal enterocytes and iron transfer from the transferrin receptor endosomal cycle into the cytosol.
To gain further insights into the role of the GAS and Sp1 sequences in NO-mediated repression of the mouse IRP1 gene, we transiently transfected human hepatoma HuH7 cells with constructs containing fragments of the mouse IRP1 promoter fused to the luciferase reporter gene. The minimal 55 bp promoter had a very low basal transcriptional activity, which was reduced by NO production. The mouse IRP1 promoter lacks typical TATA or CCAAT boxes. In many genes lacking a TATA box, a proximally positioned Sp1 site may be critical for basal transcription and can determine the start site [50]. Our results indicate that the proximal Sp1 binding site is of limited importance for basal transcription of IRP1. However, NO treatment of the transfected cells induced a significant reduction in basal transcriptional activity, and accordingly, nuclear extracts from activated macrophages reduced the DNA binding activity of Sp1. This result is in line with those of previous studies demonstrating inhibition or a lack of DNA binding by native Sp1 derived from different cell types treated with chemical NO donors [30,51,52]. It is important to note that the NO-mediated loss of Sp1 DNA binding activity may result in either a decrease [30,51] or an increase [52] in gene expression, depending on whether this factor is acting as an activator or a repressor of transcription. Thus the results of our EMSA experiments are not conclusive concerning the role of Sp1 in NO-dependent down-regulation of IRP1 gene transcription.
Transfection of HuH7 cells with the second construct, pGL2-IRP1P0.2, containing both Sp1 and MRE sequences, resulted in substantially higher basal transcriptional activity, but no decrease in luciferase activity in response to NO. Accordingly, there was no change in the DNA-binding activity to the MRE sequence in response to fluctuations in cellular iron content or to NO, recently shown to alter LIP level [53,54]. This result agrees with those of previous studies showing that interaction of MTF1 with multiple MREs present in 5′-regulatory regions of several genes is induced by heavy metals other than iron [48]. It is thus possible that the effect of MTF1-mediated modulation of gene transcription may only involve genes containing several copies of the MRE sequence in their promoters [48]. The insensitivity of the binding of MTF-1, a zinc sensor protein [55], to DNA in response to NO may result from the opposite effects of NO on zinc finger-dependent transcription. Zinc finger disruption by NO may inhibit or activate transcription, depending on the structure of the promoter [56]. NO-mediated inhibition of zinc finger-dependent gene transcription [51] may be counteracted by an indirect effect resulting from NO-mediated release of zinc bound to cytosolic low-molecular mass chelators. The activation of MTF1–DNA binding by such a mechanism has been reported in response to Cd (cadmium) and oxidative stress upon Cd binding or oxidation of metal-co-ordinating thiolates respectively [55].
Functional analysis of the IRP1 promoter using the third construct, pGL2-IRP1P0.3, clearly showed that the GAS sequence plays a major role in both the basal expression level and NO-dependent regulation of the IRP1 gene. It should be stressed that only a 19 bp fragment comprising the GAS sequence from the IRP1 promoter was added to pGL2-IRP1P0.2 in order to generate pGL2-IRP1P0.3. The demonstration that NO produced a 70% decrease in binding of STAT proteins to the GAS sequence located in the IRP1 promoter strongly suggests the involvement of this family of TFs in the regulation of IRP1 gene expression by NO. In agreement with this finding, negative gene regulation in response to NO by STAT5 [32,40], and also by STAT1 [57], has been reported for several cellular functions in various cell types. Taken together, the results of the EMSAs and those from the functional activity assays indicate that the GAS sequence is an important regulatory element responsible for down-regulation of IRP1 expression by NO.
The GAS sequence was originally identified as an element required for the rapid transcriptional induction of genes in response to IFN-γ [58]. The protein complex binding to GAS sequences in IFN-γ-treated cells is a dimer of STAT1, the prototype of the STAT protein family. To date, seven different STATs have been recognized, six of which bind to a GAS element [59]. Because STAT5 proteins are those most frequently shown to take part in nuclear signalling in response to NO [32,40], we investigated the roles of STAT5a and STAT5b in the mechanism underlying NO-dependent down-regulation of IRP1. Although these two STAT5 proteins share a high degree of amino acid sequence identity and show considerable functional overlap they are not functionally redundant [59]. According to a general model for receptor-mediated activation of STAT, there are several cytokine receptor-associated Janus kinases that induce dimerization, nuclear translocation and DNA binding of STAT through tyrosine phosphorylation [39]. To identify the mechanism through which NO exerts its repressive effect on IRP1 expression via the GAS sequence, we initially investigated whether it is associated with impaired nuclear translocation of STAT5. Endogenous NO was found to markedly reduce nuclear levels of both STAT5a and STAT5b in RAW 264.7 macrophages. A similar effect was observed when RAW 264.7 cells were incubated for 18 h with DETA/NO (results not shown). Surprisingly, an identical analysis of cytosolic extracts from RAW 264.7 macrophages, activated for NO synthesis, showed an even greater decrease in STAT5a and STAT5b content than that observed in nuclei. The results of these Western blot analyses indicated an overall decline in STAT5 protein levels rather than one restricted to nuclei. The reduction in the amount of STAT5a at the protein level was not associated with a decrease in the expression or stability of the transcript, since STAT5a mRNA levels were not influenced by NO. Taken together, our results on STAT5a mRNA and protein levels in NO-producing macrophages strongly suggest that NO sensitizes STAT5 proteins for accelerated degradation. This conclusion is supported by the description [60] of a proteosome proteolytic pathway for STAT5 and the proposal that this plays a role in down-regulation of STAT5 signalling. A hypothetical mechanism for the down-regulation of STAT5 activity by NO may involve nitration of key tyrosine residue(s) in STAT5 molecules and subsequent proteolytic degradation by the ubiquitin–proteosome pathway as has been described for other proteins [61].
The findings of the present study permit us to outline a plausible regulatory loop between STAT family proteins, NO and IRP1. TFs from the STAT family participate in signal transduction initiated by IFN-γ, leading to the activation of NOS2 expression and subsequent high output of NO [62]. The action of NO rapidly generates the RNA binding form of IRP1 [13], which down-regulates ferritin and up-regulates transferrin receptor via a post-transcriptional mechanism, thus contributing to the expansion of the catalytically active free iron pool. On the other hand, NO would promote degradation of STAT5 proteins, which inhibits expression of the IRP1 gene at the transcriptional level. The biological importance of this regulation may be in minimizing the NO-dependent reaction that boosts RNA binding by IRP1 and, consequently, attenuates the NO-induced rise in the LIP, a major factor in increasing the likelihood of oxidative injury.
Finally, a question arises as to the possible role of STAT5-driven regulation of IRP1 described here, in the cross-talk between NO and iron in cells. It is conceivable to propose that iron released from iron-containing proteins upon NO enters LIP and transits through the cytosolic compartment before being transported across the cellular membrane as nitrosyl–iron complexes by multidrug resistance-associated protein 1, as suggested recently [22]. This so-called transient iron may be easily taken up by ferritin, a main regulator of LIP levels [63]. By decreasing the amount of IRP1 via STAT5, NO minimizes its potential to inhibit ferritin expression at the post-transcriptional level, and may thus maintain and safeguard the capacity of this protein to sequester potentially harmful free iron.
Acknowledgments
This work was supported by a NATO (North Atlantic Treaty Organisation) Collaborative Linkage grant (PST.CGL 979519). We thank John Gittins for a critical reading of this paper.
References
- 1.Hentze M. W., Muckenthaler M. U., Andrews N. C. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 2.Meyron-Holtz E. G., Ghosh M. C., Iwai K., LaVaute T., Brazzolotto X., Berger U. V., Land W., Olivierre-Wilson H., Grinberg A., Love P., et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooperman S. S., Meyron-Holtz E. G., Olivierre-Wilson H., Ghosh M. C., McConnell J. P., Rouault T. A. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106:1084–1091. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galy B., Ferring D., Minana B., Bell O., Janser H. G., Muckenthaler M., Schumann K., Hentze M. W. Generation of conditional alleles of the murine Iron Regulatory Protein (IRP)-1 and -2 genes. Blood. 2005;106:2580–2589. [Google Scholar]
- 5.Bouton C., Drapier J. C. Iron regulatory proteins as NO signal transducers. Science STKE 2003. 2003:182. doi: 10.1126/stke.2003.182.pe17. [DOI] [PubMed] [Google Scholar]
- 6.Meyron-Holtz E. G., Ghosh M. C., Rouault T. A. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science. 2004;306:2087–2090. doi: 10.1126/science.1103786. [DOI] [PubMed] [Google Scholar]
- 7.Drapier J. C., Hirling H., Wietzerbin J., Kaldy P., Kühn L. C. Biosynthesis of nitric oxide activates iron regulatory factor in macrophages. EMBO J. 1993;12:3643–3649. doi: 10.1002/j.1460-2075.1993.tb06038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss G., Goossen B., Doppler W., Fuchs D., Pantopoulos K., Werner-Felmayer G., Wachter H., Hentze M. W. Translational regulation via iron-responsive elements by the nitric oxide/NO-synthase pathway. EMBO J. 1993;12:3651–3657. doi: 10.1002/j.1460-2075.1993.tb06039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martins E. A., Robalinho R. L., Meneghini R. Oxidative stress induces activation of a cytosolic protein responsible for control of iron uptake. Arch. Biochem. Biophys. 1995;316:128–134. doi: 10.1006/abbi.1995.1019. [DOI] [PubMed] [Google Scholar]
- 10.Pantopoulos K., Hentze M. W. Rapid responses to oxidative stress mediated by iron regulatory protein. EMBO J. 1995;14:101–108. doi: 10.1002/j.1460-2075.1995.tb07291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schalinske K. L., Anderson S. A., Tuazon P. T., Chen O. S., Kennedy M. C., Eisenstein R. S. The iron–sulfur cluster of iron regulatory protein 1 modulates the accessibility of RNA binding and phosphorylation sites. Biochemistry. 1997;36:3950–3958. doi: 10.1021/bi9624447. [DOI] [PubMed] [Google Scholar]
- 12.Hanson E. S., Leibold E. A. Regulation of iron regulatory protein 1 during hypoxia and hypoxia/reoxygenation. J. Biol. Chem. 1998;273:7588–7593. doi: 10.1074/jbc.273.13.7588. [DOI] [PubMed] [Google Scholar]
- 13.Drapier J. C., Bouton C., Oliveira L. Redox modulation of iron regulatory proteins by nitric oxide and peroxynitrite. In: Ignarro L. J., editor. Nitric Oxide Biology and Pathobiology. San Diego: Academic Press; 2001. pp. 315–328. [Google Scholar]
- 14.Soum E., Brazzolotto X., Goussias C., Bouton C., Moulis J. M., Mattioli T. A., Drapier J. C. Peroxynitrite and nitric oxide differently target the iron–sulfur cluster and amino acid residues of human iron regulatory protein 1. Biochemistry. 2003;42:7648–7654. doi: 10.1021/bi030041i. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez D., Drapier J. C., Bouton C. Endogenous nitration of iron regulatory protein-1 (IRP-1) in nitric oxide-producing murine macrophages: further insight into the mechanism of nitration in vivo and its impact on IRP-1 functions. J. Biol. Chem. 2004;279:43345–43351. doi: 10.1074/jbc.M401889200. [DOI] [PubMed] [Google Scholar]
- 16.Oliveira L., Drapier J. C. Down-regulation of iron regulatory protein 1 gene expression by nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6550–6555. doi: 10.1073/pnas.120571797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watts R. N., Ponka P., Richardson D. R. Effects of nitrogen monoxide and carbon monoxide on molecular and cellular iron metabolism: mirror-image effector molecules that target iron. Biochem. J. 2003;369:429–440. doi: 10.1042/BJ20021302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipiński P., Drapier J. C. Interplay between ferritin metabolism, reactive oxygen species and nitric oxide. J. Biol. Inorg. Chem. 1997;2:559–566. [Google Scholar]
- 19.Recalcati S., Taramelli D., Conte D., Cairo G. Nitric oxide-mediated induction of ferritin synthesis in J774 macrophages by inflammatory cytokines: role of selective iron regulatory protein-2 downregulation. Blood. 1998;91:1059–1066. [PubMed] [Google Scholar]
- 20.Wang J., Chen G., Pantopoulos K. Nitric oxide inhibits the degradation of IRP2. Mol. Cell. Biol. 2005;25:1347–1353. doi: 10.1128/MCB.25.4.1347-1353.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hibbs J. B., Jr, Taintor R. R., Vavrin Z. Iron depletion: possible cause of tumor cell cytotoxicity induced by activated macrophages. Biochem. Biophys. Res. Commun. 1984;123:716–723. doi: 10.1016/0006-291x(84)90288-2. [DOI] [PubMed] [Google Scholar]
- 22.Watts R. N., Hawkins C., Ponka P., Richardson D. R. Nitrogen monoxide (NO)-mediated iron release from cells is linked to NO-induced glutathione efflux via multidrug resistance-associated protein 1. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7670–7675. doi: 10.1073/pnas.0602515103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry Y., Lepoivre M., Drapier J. C., Ducrocq C., Boucher J. L., Guissani A. EPR characterization of molecular targets for NO in mammalian cells and organelles. FASEB J. 1993;7:1124–1134. doi: 10.1096/fasebj.7.12.8397130. [DOI] [PubMed] [Google Scholar]
- 24.Vanin A. F. Dinitrosyl iron complexes and S-nitrosothiols are two possible forms for stabilization and transport of nitric oxide in biological systems. Biochemistry (Mosc.) 1998;63:782–793. [PubMed] [Google Scholar]
- 25.Durante W., Kroll M. H., Christodoulides V., Peyton K. J., Schafer A. I. Nitric oxide induces heme oxygenase-1 gene expression and carbon monoxide production in vascular smooth muscle cells. Circ. Res. 1997;80:557–564. doi: 10.1161/01.res.80.4.557. [DOI] [PubMed] [Google Scholar]
- 26.Bouton C., Demple B. Nitric oxide-inducible expression of heme oxygenase-1 in human cells. Translation-independent stabilization of the mRNA and evidence for direct action of nitric oxide. J. Biol. Chem. 2000;275:32688–32693. doi: 10.1074/jbc.275.42.32688. [DOI] [PubMed] [Google Scholar]
- 27.Bogdan C. Nitric oxide and the regulation of gene expression. Trends Cell Biol. 2001;11:66–75. doi: 10.1016/s0962-8924(00)01900-0. [DOI] [PubMed] [Google Scholar]
- 28.Shin W. S., Hong Y. H., Peng H. B., De Caterina R., Libby P., Liao J. K. Nitric oxide attenuates vascular smooth muscle cell activation by interferon-gamma. The role of constitutive NF-κB activity. J. Biol. Chem. 1996;271:11317–11324. doi: 10.1074/jbc.271.19.11317. [DOI] [PubMed] [Google Scholar]
- 29.Pilz R. B., Suhasini M., Idriss S., Meinkoth J. L., Boss G. R. Nitric oxide and cGMP analogs activate transcription from AP-1-responsive promoters in mammalian cells. FASEB J. 1995;9:552–558. doi: 10.1096/fasebj.9.7.7737465. [DOI] [PubMed] [Google Scholar]
- 30.Sellak H., Yang X., Cao X., Cornwell T., Soff G. A., Lincoln T. Sp1 transcription factor as a molecular target for nitric oxide- and cyclic nucleotide-mediated suppression of cGMP-dependent protein kinase-Iα expression in vascular smooth muscle cells. Circ. Res. 2002;90:405–412. doi: 10.1161/hh0402.105898. [DOI] [PubMed] [Google Scholar]
- 31.Metzen E., Zhou J., Jelkmann W., Fandrey J., Brune B. Nitric oxide impairs normoxic degradation of HIF-1α by inhibition of prolyl hydroxylases. Mol. Biol. Cell. 2003;14:3470–3481. doi: 10.1091/mbc.E02-12-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bingisser R. M., Tilbrook P. A., Holt P. G., Kees U. R. Macrophage-derived nitric oxide regulates T cell activation via reversible disruption of the Jak3/STAT5 signaling pathway. J. Immunol. 1998;160:5729–5734. [PubMed] [Google Scholar]
- 33.Andrews N. C., Faller D. V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roe A. L., Howard G., Blouin R. A. Characterization of nuclear protein binding (AP-1, GR, and STAT) in the genetically obese (fa/fa) Zucker rat. Life Sci. 1998;15:1339–1346. doi: 10.1016/s0024-3205(98)00397-x. [DOI] [PubMed] [Google Scholar]
- 35.Verdier F., Rabionet R., Gouilleux F., Beisenherz-Huss C., Varlet P., Mulle O., Mayeux P., Lacombe C., Gisselbrecht S., Chretien S. A sequence of the CIS gene promoter interacts preferentially with two associated STAT5A dimers: a distinct biochemical difference between STAT5A and STAT5B. Mol. Cell. Biol. 1998;18:5852–5860. doi: 10.1128/mcb.18.10.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson D. A. C., Feeney S., Boyle C., Sitt A. W. Retinal VEGF mRNA measured by SYBR green I fluorescence: a versatile approach to quantitative PCR. Mol. Vision. 2000;6:178–183. [PubMed] [Google Scholar]
- 37.Fan X., Roy E., Zhu L., Murphy T. C., Kozlowski M., Nanes M. S., Rubin J. Nitric oxide donors inhibit luciferase expression in a promoter-independent fashion. J. Biol. Chem. 2003;278:10232–10238. doi: 10.1074/jbc.M209911200. [DOI] [PubMed] [Google Scholar]
- 38.Radtke F., Georgiev O., Muller H. P., Brugnera E., Schaffner W. Functional domains of the heavy metal-responsive transcription regulator MTF-1. Nucleic Acids Res. 1995;23:2277–2286. doi: 10.1093/nar/23.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horvath C. M. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 40.Mazzoni A., Bronte V., Visintin A., Spitzer J. H., Apolloni E., Serafini P., Zanovello P., Segal D. M. Myeloid suppressor lines inhibit T cell responses by an NO-dependent mechanism. J. Immunol. 2002;168:689–695. doi: 10.4049/jimmunol.168.2.689. [DOI] [PubMed] [Google Scholar]
- 41.Lipiński P., Drapier J. C., Oliveira L., Retmańska H., Sochanowicz B., Kruszewski M. Intracellular iron status as a hallmark of mammalian cell susceptibility to oxidative stress: a study of L5178Y mouse lymphoma cell lines differentially sensitive to H2O2. Blood. 2000;95:2960–2966. [PubMed] [Google Scholar]
- 42.Starzyński R. R., Lipiński P., Drapier J. C., Diet A., Smuda E., Bartłomiejczyk T., Gralak M. A., Kruszewski M. Down-regulation of iron regulatory protein 1 activities and expression in superoxide dismutase 1 knock-out mice is not associated with alterations in iron metabolism. J. Biol. Chem. 2005;280:4207–4212. doi: 10.1074/jbc.M411055200. [DOI] [PubMed] [Google Scholar]
- 43.Müllner E. W., Rothenberger S., Müller A. M., Kühn L. C. In vivo and in vitro modulation of the mRNA-binding activity of iron-regulatory factor. Tissue distribution and effects of cell proliferation, iron levels and redox state. Eur. J. Biochem. 1992;208:597–605. doi: 10.1111/j.1432-1033.1992.tb17224.x. [DOI] [PubMed] [Google Scholar]
- 44.Tang C. K., Chin J., Harford J. B., Klausner R. D., Rouault T. A. Translational repressor activity is equivalent and is quantitatively predicted by in vitro RNA binding for two iron-responsive element-binding proteins, IRP1 and IRP2. J. Biol. Chem. 1992;267:24466–24470. doi: 10.1074/jbc.270.10.4983. [DOI] [PubMed] [Google Scholar]
- 45.Henderson B. R., Seiser C., Kuhn L. C. Characterization of a second RNA-binding protein in rodents with specificity for iron-responsive elements. J. Biol. Chem. 1993;268:27327–27334. [PubMed] [Google Scholar]
- 46.Guo B., Phillips J. D., Yu Y., Leibold E. A. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 1995;270:21645–21651. doi: 10.1074/jbc.270.37.21645. [DOI] [PubMed] [Google Scholar]
- 47.Neonaki M., Graham D. C., White K. N., Bomford A. Down-regulation of liver iron-regulatory protein 1 in haemochromatosis. Biochem. Soc. Trans. 2002;30:726–728. doi: 10.1042/bst0300726. [DOI] [PubMed] [Google Scholar]
- 48.Palmiter R. D. Regulation of metallothionein genes by heavy metals appears to be mediated by a zinc-sensitive inhibitor that interacts with a constitutively active transcription factor, MTF-1. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1219–1223. doi: 10.1073/pnas.91.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee P. L., Gelbart T., West C., Halloran C., Beutler E. The human Nramp2 gene: characterization of the gene structure, alternative splicing, promoter region and polymorphisms. Blood Cells Mol. Dis. 1998;2:199–215. doi: 10.1006/bcmd.1998.0186. [DOI] [PubMed] [Google Scholar]
- 50.Sauerwald A., Hoesche C., Oschwald R., Kilimann M. W. The 5′-flanking region of the synapsin I gene. A G+C-rich, TATA- and CAAT-less, phylogenetically conserved sequence with cell type-specific promoter function. J. Biol. Chem. 1990;265:14932–14937. [PubMed] [Google Scholar]
- 51.Berendji D., Kolb-Bachofen V., Zipfel P. F., Skerka C., Carlberg C., Kroncke K. D. Zinc finger transcription factors as molecular targets for nitric oxide-mediated immunosuppression: inhibition of IL-2 gene expression in murine lymphocytes. Mol. Med. 1999;5:721–730. [PMC free article] [PubMed] [Google Scholar]
- 52.Wang S., Wang W., Wesley R. A., Danner R. L. A Sp1 binding site of the tumor necrosis factor α promoter functions as a nitric oxide response element. J. Biol. Chem. 1999;274:33190–33193. doi: 10.1074/jbc.274.47.33190. [DOI] [PubMed] [Google Scholar]
- 53.Lipiński P., Starzyński R. R., Drapier J. C., Bouton C., Bartłomiejczyk T., Sochanowicz B., Smuda E., Gajkowska A., Kruszewski M. Induction of iron regulatory protein 1 RNA-binding activity by nitric oxide is associated with a concomitant increase in the labile iron pool: implications for DNA damage. Biochem. Biophys. Res. Commun. 2005;327:349–355. doi: 10.1016/j.bbrc.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 54.Chenais B., Morjani H., Drapier J. C. Impact of endogenous nitric oxide on microglial cell energy metabolism and labile iron pool. J. Neurochem. 2002;81:615–623. doi: 10.1046/j.1471-4159.2002.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giedroc D. P., Chen X., Apuy J. L. Metal response element (MRE)-binding transcription factor-1 (MTF-1): structure, function, and regulation. Antioxide Redox Signal. 2001;3:577–596. doi: 10.1089/15230860152542943. [DOI] [PubMed] [Google Scholar]
- 56.Kroncke K. D. Nitrosative stress and transcription. Biol. Chem. 2003;384:1365–1377. doi: 10.1515/BC.2003.153. [DOI] [PubMed] [Google Scholar]
- 57.Giustizieri M. L., Albanesi C., Scarponi C., De Pita O., Girolomoni G. Nitric oxide donors suppress chemokine production by keratinocytes in vitro and in vivo. Am. J. Pathol. 2002;161:1409–1418. doi: 10.1016/S0002-9440(10)64416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Decker T., Kovarik P., Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 59.Grimley P. M., Dong F., Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–157. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 60.Wang D., Moriggl R., Stravopodis D., Carpino N., Marine J. C., Teglund S., Feng J., Ihle J. N. A small amphipathic α-helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosine-phosphorylated Stat5. EMBO J. 2000;19:392–399. doi: 10.1093/emboj/19.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schopfer F. J., Baker P. R., Freeman B. A. NO-dependent protein nitration: a cell signaling event or an oxidative inflammatory response? Trends Biochem. Sci. 2003;28:646–654. doi: 10.1016/j.tibs.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 62.lanchette J., Jaramillo M., Olivier M. Signalling events involved in interferon-gamma-inducible macrophage nitric oxide generation. Immunology. 2003;108:513–522. doi: 10.1046/j.1365-2567.2003.01620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Picard V., Epsztejn S., Santambrogio P., Cabantchik Z. I., Beaumont C. Role of ferritin in the control of the labile iron pool in murine erythroleukemia cells. J. Biol. Chem. 1998;273:5382–5386. doi: 10.1074/jbc.273.25.15382. [DOI] [PubMed] [Google Scholar]