Abstract
A duplex PCR (D-PCR) amplifying portions of the Brachyspira hyodysenteriae NADH oxidase gene and the B. pilosicoli 16S rRNA gene was developed and then tested on DNA extracted from 178 porcine fecal samples. The feces also underwent anaerobic culture and species-specific PCRs. Fecal extraction-D-PCR detected seven additional samples containing B. hyodysenteriae and five more containing B. pilosicoli.
Pigs are colonized by two pathogenic species of anaerobic intestinal spirochetes, Brachyspira hyodysenteriae and B. pilosicoli. B. hyodysenteriae causes swine dysentery, a severe mucohemorrhagic diarrheal disease (10), while B. pilosicoli causes porcine intestinal spirochetosis, a milder colitis that also occurs in other host species (9). Because of the economic impact of these diseases on pig production, rapid diagnostic methods are needed to detect and distinguish between them. Historically, a positive diagnosis of swine dysentery or porcine intestinal spirochetosis has required isolation of the associated spirochete and confirmation of its identity by performance of phenotypic tests (11). However, the fastidious and slow-growing nature of intestinal spirochetes, together with the limited range of phenotypic differences between them, has hampered this approach. To overcome such problems, a variety of PCR assays for B. hyodysenteriae and B. pilosicoli have been developed (1-3, 7, 8, 14, 15). These PCRs target conserved genes such as the 16S rRNA gene (rDNA), the 23S rDNA, the NADH oxidase (nox) gene, or the hemolysin (tly) gene. The assays typically are conducted on intestinal spirochete isolates or on spirochetal growth harvested from primary isolation plates. Difficulties have been encountered when trying to extract spirochetal DNA directly from feces for use as a PCR template, and there appears to be only one report of the use of PCR (plus hybridization) for direct detection of B. hyodysenteriae from feces (5) and one recent report of its use for B. pilosicoli (4).
A multiplex PCR for the simultaneous detection of B. hyodysenteriae, Lawsonia intracellularis (the agent of porcine proliferative enteropathy), and Salmonella spp. has been described previously (6), but to date, no PCRs amplifying and distinguishing between B. hyodysenteriae and B. pilosicoli in one reaction mixture have been reported. The purpose of the present study was to develop a reliable and robust duplex PCR (D-PCR) system that can be used to detect these two species by using DNA extracted directly from pig feces. To evaluate the test as a diagnostic tool, it was compared with selective anaerobic culture, followed by individual species-specific PCRs conducted on growth harvested from the primary isolation plates.
Control spirochete strains and culture conditions.
Control spirochete strains were obtained from the culture collection held at the Reference Centre for Intestinal Spirochetes, Murdoch University, Western Australia, Australia. These included the type strains of B. hyodysenteriae (B78T) and B. pilosicoli (P43/6/78T), which were used to seed feces, as well as another 48 strains of B. hyodysenteriae, 18 strains of B. pilosicoli, 12 strains of B. intermedia, 8 strains of B. innocens, 8 strains of B. murdochii, 2 strains of “B. canis,” 1 strain of B. alvinipulli, and 1 strain of B. aalborgi, all of which were used to confirm the specificity of the D-PCR. The strains originated from Australia, Europe, Scandinavia, and North America, and their identities had previously been established by phenotypic testing, PCR assays, and/or multilocus enzyme electrophoresis (13, 16). The strains were propagated at 37°C in Kunkle's prereduced anaerobic broth containing 2% (vol/vol) fetal bovine serum and a 1% (vol/vol) ethanolic cholesterol solution (12). Cells were harvested from a mid-log-phase culture and counted with a hemocytometer.
Fecal samples and culture conditions.
A total of 178 fecal samples from grower pigs on seven Australian farms were collected. Fecal samples were kept at 4°C and transported to Murdoch University, where they were processed within 48 h of collection. Farm 1 was a high-health status farm with no history of diarrhea, while the other farms all had a history of diarrhea in growers. Bacteriological swabs were inserted into the feces and streaked onto selective Trypticase soy agar (BBL, Becton Dickinson Microbiology Systems, Cockeysville, Md.) plates containing 5% (vol/vol) defibrinated ovine blood, 400 μg of spectinomycin per ml, and 25 μg each of colistin and vancomycin (Sigma-Aldrich Pty. Ltd., Castle Hill, Australia) per ml. The plates were incubated for 5 to 7 days at 37°C in an anaerobic environment of 94% H2 and 6% CO2 generated with anaerobic Gaspak plus sachets (BBL). The presence of a low, flat, spreading growth of spirochetes on the plate was recorded, as was any hemolysis around the growth. Areas of suspected spirochetal growth were picked off, resuspending in phosphate-buffered saline, and examined under a phase-contrast microscope.
DNA preparation from isolation plates.
For the field samples, PCR assays for the two spirochete species were applied to growth harvested from the primary isolation plates. This standard detection method was designated culture-PCR. The tip of a sterile wooden toothpick was used to stab areas of spirochete growth. Where both hemolytic and nonhemolytic growth areas were observed on a plate, both areas were stabbed. The adherent material was resuspended in 50 μl of ultrapure water and boiled for 30 s. A 2.5-μl volume was added to each of the separate B. hyodysenteriae and B. pilosicoli PCR mixtures, which were designed to amplify portions of the NADH oxidase gene and the 16S rDNA, respectively. These PCRs and their optimized conditions have been described previously (2, 15).
DNA extraction from feces.
DNA was extracted from fecal samples with the QIAamp DNA Stool Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. Briefly, 200 mg of feces was resuspended in 2 ml of ASL buffer by vortexing for 1 min, and then 1.6 ml of the lysate was transferred into a new tube. The suspension was boiled for 5 min and centrifuged at 20,000 × g for 1 min, and 1.2 ml of supernatant was transferred to a new tube containing an InhibitEX tablet. The tube was vortexed for 1 min and incubated at room temperature for 1 min. The tube was centrifuged at 20,000 × g for 3 min, and 200 μl of supernatant was transferred to a new tube containing 15 μl of proteinase K. Two hundred microliters of AL buffer was added, and the tube was vortexed before incubation at 70°C for 10 min. Two hundred microliters of absolute ethanol was added to the lysate, and the tube was vortexed. The contents were applied to a spin column and centrifuged at 10,000 × g for 1 min. The column was washed once with 500 μl of AW1 buffer at 10,000 × g for 1 min and once with 500 μl of AW2 buffer at 10,000 × g for 3 min. DNA was eluted from the column at 10,000 × g with 100 μl of AE buffer heated to 70°C. The DNA extracted from all of the field samples was subjected to the D-PCR assay. In addition, DNA extracted from each of the 51 samples collected at farm 4 was also subjected to the two separate species-specific PCR assays, as used on the bacterial growth harvested from the isolation plates.
Primer design for D-PCR.
The D-PCR used the same basic targets as the individual PCRs for B. hyodysenteriae and B. pilosicoli. Two primer pairs were used. H1 (5′-ACTAAAGATCCTGATGTATTTG-3′) and H2 (5′-CTAATAAACGTCTGCTGC-3′) targeted a 354-bp region of the NADH oxidase (nox) gene of B. hyodysenteriae, while P1 (5′-AGAGGAAAGTTTTTTCGCTTC-3′) and P2 (5′-GCACCTATGTTAAACGTCCTTG-3′) targeted an 823-bp region of the 16S rDNA of B. pilosicoli. Primer H2 (formerly NOX1-R) and P1 (formerly Acoli1) have been described previously (2, 15). Primers H1 and P2 were selected from gene sequences available from the GenBank database and were designed to give specific PCR products of easily distinguishable sizes.
D-PCR.
The fecal extraction-D-PCR protocol was undertaken by an operator with no knowledge of the results obtained by culture-PCR. Purified fecal DNA was amplified by a hot-start PCR in a 25-μl total volume with HotStarTaq DNA polymerase (QIAGEN GmbH). Briefly, amplification mixtures consisted of 1× PCR buffer (containing 1.5 mM MgCl2), 0.5 U of HotStarTaq DNA polymerase, 0.2 mM each deoxynucleoside triphosphate (Amersham Pharmacia Biotech AB, Uppsala, Sweden), the first primer pair (H1 and H2) at 0.5 μM, the second primer pair (P1 and P2) at 0.17 μM, and 2.5 μl of chromosomal template DNA. The cycling conditions used involved an initial 15-min HotStarTaq DNA polymerase activation step at 95°C, followed by 31 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and primer extension at 72°C for 1 min. The PCR products were subjected to electrophoresis in 1.5% (wt/vol) agarose gels in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA), stained with ethidium bromide, and viewed under UV light.
Detection limits of the D-PCR test.
The detection limits of the fecal extraction-D-PCR test were determined by seeding fecal samples with different concentrations of spirochetal cells, extracting the DNA from the feces, and then subjecting it to the D-PCR assay. Sterile tubes containing 200 mg of feces from two healthy pigs, which were culture-PCR negative for intestinal spirochetes, were individually resuspended with 200-μl serial 10-fold dilutions of spirochete cells in TE (10 mM Tris-HCl-1 mM EDTA [pH 8.0]) buffer ranging from 1 to 108 cells/ml. The samples tested contained B. hyodysenteriae B78T, B. pilosicoli P43/6/78T, or equal numbers of cells of both strains. A tube consisting of 100 μl of TE buffer was included as a negative control. The fecal suspensions were used for column extraction of chromosomal DNA and D-PCR.
For both series of seeded samples and for both components of the D-PCR assay, a PCR product from samples containing 103 cells/g was faintly visible and a PCR product from samples containing 104 cells/g was clearly visible. The practical limits of detection therefore were between 103 and 104 cells/g of feces for both species. The same limits of detection applied when both species were present individually, as well as when they were present together in equal numbers. These detection limits were comparable to those routinely achieved by culture-PCR (1), and these, in turn, are better than those achieved by culture alone (1).
Specificity of the D-PCR assay.
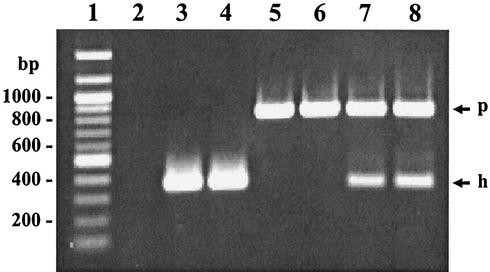
The specificity of the D-PCR assay for the two species was assessed by applying it to chromosomal DNAs extracted from 100 different intestinal spirochete strains representing seven species and one proposed species (“B. canis”) of the genus Brachyspira. All 49 strains of B. hyodysenteriae and 19 strains of B. pilosicoli gave specific amplification of the correct predicted product size, while no products were generated from the other spirochete strains. Examples of the products generated are shown in Fig. 1.
FIG. 1.
Example of the D-PCR products of B. hyodysenteriae and B. pilosicoli from column-extracted feces. Lanes: 1, 100-bp molecular size ladder; 2, no-template PCR control; 3, B. hyodysenteriae B78T-spiked sample; 4, farm 2 sample containing B. hyodysenteriae; 5, B. pilosicoli P43/6/78T-spiked sample; 6, farm 3 sample containing B. pilosicoli; 7 and 8, farm 2 samples containing mixed cultures of B. hyodysenteriae and B. pilosicoli. Arrows: p, B. pilosicoli D-PCR product; h, B. hyodysenteriae D-PCR product.
Sequencing of PCR products.
To check the specificity of the reactions further, the D-PCR amplicons from 18 fecal samples from the farms were chosen for direct sequencing with an ABI 373A DNA sequencer (PE Applied Biosystems, Foster City, Calif.). The fecal DNA was amplified in a 25-μl total volume with Pfu DNA polymerase (Promega, Madison, Wis.). Briefly, amplification mixtures consisted of 1× PCR buffer (containing 2 mM Mg2SO4), 0.6 U of Pfu DNA polymerase, 0.2 mM each deoxynucleoside triphosphate (Amersham Pharmacia Biotech AB), the appropriate primer pair (H1 and H2 or P1 and P2) at 0.5 μM, and 2.5 μl of chromosomal template DNA. Cycling conditions involved an initial 2-min denaturation step at 95°C, followed by 31 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 30 s, and primer extension at 72°C for 2 min. The PCR products were purified with an UltraClean PCR clean-up kit (Mo Bio Laboratories, Solana Beach, Calif.). Sequencing of the B. hyodysenteriae product was performed with the H1 and H2 primers, and that of the B. pilosicoli product was performed with the P1 and P2 primers. Each sequencing reaction was performed in duplicate in a 10-μl volume consisting of approximately 300 ng of PCR product, 5 pmol of primer, and 4 μl of the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Mix (PE Applied Biosystems, GmbH, Weiterstadt, Germany). Cycling conditions were a 2-min denaturing step at 96°C, followed by 25 cycles of denaturation at 96°C for 10 s, annealing at 52°C for 5 s, and primer extension at 60°C for 4 min. Residual rhodamine dye terminators were removed from the sequencing products by precipitation with 95% (vol/vol) ethanol containing 120 mM sodium acetate (pH 4.6), and the sequencing products were vacuum dried. The sequencing products were analyzed with an ABI 373A DNA sequencer (PE Applied Biosystems). Type strain controls for B. hyodysenteriae (B78T) and B. pilosicoli (P43/6/78T) were also included for sequencing.
Field samples.
The results of a comparison of culture-PCR and fecal extraction-D-PCR for the 178 field samples are shown in Table 1. Both methods classified all of the samples from three of the farms as negative for the pathogenic intestinal spirochetes, samples from two farms as containing both B. hyodysenteriae and B. pilosicoli, and samples from two farms as just containing B. hyodysenteriae. These complementary results provide further evidence that the D-PCR assay has a specificity similar to that of culture-PCR.
TABLE 1.
Comparison of results for detection of B. hyodysenteriae and B. pilosicoli in 178 porcine fecal samples from seven farms by either anaerobic culture with PCR on the primary growth or D-PCR on DNA extracted from feces
| Farm no. | No. of samples | Culture-PCR from isolation plate
|
D-PCR on DNA extracted from feces
|
|||||
|---|---|---|---|---|---|---|---|---|
| B. hyodysenteriae positive | B. pilosicoli positive | Negative | B. hyodysenteriae positive | B. pilosicoli positive | B. hyodysenteriae and B. pilosicoli positive | Negative | ||
| 1a | 26 | 0 | 0 | 26 | 0 | 0 | 0 | 26 |
| 2 | 40 | 10 | 2 | 28 | 11 | 2 | 3 | 24 |
| 3 | 18 | 2 | 3 | 13 | 2 | 5 | 0 | 11 |
| 4 | 51 | 17 | 0 | 34 | 20 | 0 | 0 | 31 |
| 5 | 20 | 0 | 0 | 20 | 0 | 0 | 0 | 20 |
| 6 | 13 | 0 | 0 | 13 | 0 | 0 | 0 | 13 |
| 7 | 10 | 10 | 0 | 0 | 10 | 0 | 0 | 0 |
| Total | 178 | 39 | 5 | 134 | 43 | 7 | 3 | 125 |
High-health status farm.
In addition, compared to culture-PCR, the fecal extraction-D-PCR method detected seven extra samples containing B. hyodysenteriae DNA and five extra samples containing B. pilosicoli DNA. These were made up of four additional samples containing B. hyodysenteriae and three containing B. pilosicoli on farm 2, two extra samples containing B. pilosicoli on farm 3, and three extra samples containing B. hyodysenteriae on farm 4. Furthermore, three samples from farm 2 were found to contain both spirochete species by fecal extraction-D-PCR, whereas only B. hyodysenteriae was detected by culture-PCR. This overall increase in the detection rate for both spirochete species represents a clear advantage for the fecal extraction-D-PCR method. The increased detection rate is probably attributable to the step of DNA extraction from feces rather than to the D-PCR itself, since the results of the individual PCR assays of the DNAs extracted from the 51 fecal samples from farm 4 agreed completely with those of the D-PCR assay, including detection of the presence of B. hyodysenteriae in 3 samples that was not detected by culture-PCR.
Sequencing of the D-PCR amplicons generated from 18 of the field samples further supported the specificity of the D-PCR. All 14 B. hyodysenteriae products had the same sequence over 354 bp of the nox gene as did type strain B78T. The seven B. pilosicoli products had between 99.3% (703 bp) and 99.7% (706 bp) sequence homology with the corresponding 708 bp of the 16S rDNA sequence of type strain P43/6/78T. These 21 sequences included those from three fecal samples that simultaneously generated products for both B. pilosicoli and B. hyodysenteriae, two fecal samples that produced a B. pilosicoli product in D-PCR but were negative by culture-PCR, and one fecal sample that produced a product for B. hyodysenteriae in the D-PCR assay but was negative by culture-PCR.
Small numbers of spirochete-like bacteria were observed by phase-contrast microscopy in the primary growth from nine fecal samples, i.e., seven from farm 2 and two from farm 3. These could not be isolated in pure culture, and the samples were negative in the PCRs used on the growth. The D-PCR assays of the corresponding fecal samples were also negative. It is assumed that these organisms were nonpathogenic intestinal spirochete species such as B. innocens.
The overall objective of this study was achieved. The D-PCR method was specific when tested with a large number of intestinal spirochete strains, and the sequences of the products generated from field samples were identical to (B. hyodysenteriae) or very similar to (B. pilosicoli) the sequences of the respective type strains of the species, again confirming the specificity of the assay. Compared to culture-PCR, the fecal extraction-D-PCR method detected additional samples that contained B. hyodysenteriae and/or B. pilosicoli on farms where these organisms were known to be present. The fecal extraction-D-PCR method also had the major advantage that results could be obtained for both spirochete species within 5 h of specimen receipt. If there was a need to obtain isolates for strain typing or testing of antimicrobial sensitivities, positive samples could then be cultured. In comparison, culture-PCR takes 3 to 5 days for the culture component and then several hours to perform both PCR tests. The costs of laboratory consumables for the two techniques are comparable, so the major financial saving of the fecal extraction-D-PCR method results from the reduced handling times and labor components.
Acknowledgments
We thank Francis Brigg for assistance with sequencing and Robyn Smith, Ross Buddle, Samantha Wright, and Kellie Smith for organizing the collection of pig feces.
REFERENCES
- 1.Atyeo, R. F., S. L. Oxberry, B. G. Combs, and D. J. Hampson. 1998. Development and evaluation of polymerase chain reaction tests as an aid to diagnosis of swine dysentery and intestinal spirochaetosis. Lett. Appl. Microbiol. 26:126-130. [DOI] [PubMed] [Google Scholar]
- 2.Atyeo, R. F., T. B. Stanton, N. S. Jensen, D. S. Suriyaarachichi, and D. J. Hampson. 1999. Differentiation of Serpulina species by NADH oxidase gene (nox) sequence comparisons and nox-based polymerase chain reaction tests. Vet. Microbiol. 67:47-60. [DOI] [PubMed] [Google Scholar]
- 3.Barcellos, D. E., M. R. Mathiesen, M. de Uzeda, I. I. Kader, and G. E. Duhamel. 2000. Prevalence of Brachyspira species isolated from diarrhoeic pigs in Brazil. Vet. Rec. 146:398-403. [DOI] [PubMed] [Google Scholar]
- 4.Choi, C., D. U. Han, J. Kim, W.-S. Cho, H.-K. Chung, T. Jung, B. S. Yoon, and C. Chae. 2002. Prevalence of Brachyspira pilosicoli in Korean pigs, determined using a nested PCR. Vet. Rec. 150:217-218. [DOI] [PubMed] [Google Scholar]
- 5.Elder, R. O., G. E. Duhamel, R. W. Schafer, M. R. Mathiesen, and M. Ramanathan. 1994. Rapid detection of Serpulina hyodysenteriae in diagnostic specimens by PCR. J. Clin. Microbiol. 32:1497-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elder, R. O., G. E. Duhamel, M. R. Mathiesen, E. D. Erickson, C. J. Gebhart, and R. D. Oberst. 1997. Multiplex polymerase chain reaction for simultaneous detection of Lawsonia intracellularis, Serpulina hyodysenteriae, and salmonellae in porcine intestinal specimens. J. Vet. Diagn. Investig. 9:281-286. [DOI] [PubMed] [Google Scholar]
- 7.Fellstrom, C., B. Pettersson, J. Thomson, A. Gunnarsson, M. Persson, and K. E. Johansson. 1997. Identification of Serpulina species associated with porcine colitis by biochemical analysis and PCR. J. Clin. Microbiol. 35:462-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fellstrom, C., U. Zimmerman, A. Aspan, and A. Gunnarsson. 2001. The use of culture, pooled samples and PCR for identification of herds infected with Brachyspira hyodysenteriae. Anim. Health Res. Rev. 2:37-43. [PubMed] [Google Scholar]
- 9.Hampson, D. J., and D. J. Trott. 1999. Spirochetal diarrhea/porcine intestinal spirochetosis, p. 553-562. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa State University, Ames.
- 10.Harris, D. L., D. J. Hampson, and R. D. Glock. 1999. Swine dysentery, p. 579-600. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa State University, Ames.
- 11.Jensen, N. S. 1997. Detection, identification and subspecific differentiation of intestinal spirochaetes, p. 323-341. In D. J. Hampson, and T. B. Stanton (ed.), Intestinal spirochaetes in domestic animals and humans. CAB International, Wallingford, England.
- 12.Kunkle, R. A., D. L. Harris, and J. M. Kinyon. 1986. Autoclaved liquid medium for propagation of Treponema hyodysenteriae. J. Clin. Microbiol. 24:669-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, J. I., D. J. Hampson, A. J. Lymbery, and S. J. Harders. 1993. The porcine intestinal spirochaetes: identification of new genetic groups. Vet. Microbiol. 34:273-285. [DOI] [PubMed] [Google Scholar]
- 14.Leser, T. D., K. Moller, T. K. Jensen, and S. E. Jorsal. 1997. Specific detection of Serpulina hyodysenteriae and potentially pathogenic weakly beta-haemolytic porcine intestinal spirochetes by polymerase chain reaction targeting 23S rDNA. Mol. Cell. Probes 11:363-372. [DOI] [PubMed] [Google Scholar]
- 15.Park, N. Y., C. Y. Chung, A. J. McLaren, R. F. Atyeo, and D. J. Hampson. 1995. Polymerase chain reaction for identification of human and porcine spirochaetes recovered from cases of intestinal spirochaetosis. FEMS Microbiol. Lett. 125:225-229. [DOI] [PubMed] [Google Scholar]
- 16.Trott, D. J., S. L. Oxberry, and D. J. Hampson. 1997. Evidence for Serpulina hyodysenteriae being recombinant, with an epidemic population structure. Microbiology 143:3357-3365. [DOI] [PubMed] [Google Scholar]