Abstract
Resistance to fluoroquinolones among clinical isolates of Staphylococcus aureus has become a clinical problem. Therefore, a rapid method to identify S. aureus and its susceptibility to fluoroquinolones could provide clinicians with a useful tool for the appropriate use of these antimicrobial agents in the health care settings. In this study, we developed a rapid real-time PCR assay for the detection of S. aureus and mutations at codons Ser-80 and Glu-84 of the grlA gene encoding the DNA topoisomerase IV, which are associated with decreased susceptibility to fluoroquinolones. The detection limit of the assay was 10 genome copies per reaction. The PCR assay was negative with DNA from all 26 non-S. aureus bacterial species tested. A total of 85 S. aureus isolates with various levels of fluoroquinolone resistance was tested with the PCR assay. The PCR assay correctly identified 100% of the S. aureus isolates tested compared to conventional culture methods. The correlation between the MICs of ciprofloxacin, levofloxacin, and gatifloxacin and the PCR results was 98.8%. The total time required for the identification of S. aureus and determination of its susceptibility to fluoroquinolones was about 45 min, including DNA extraction. This new rapid PCR assay represents a powerful method for the detection of S. aureus and its susceptibility to fluoroquinolones.
In recent years, the clinical efficacy of fluoroquinolones against infections with Staphylococcus aureus has been undermined by the widespread emergence of decreased susceptibility to these compounds (1, 8). Resistance to fluoroquinolones, and in particular to ciprofloxacin, is considerably higher among methicillin-resistant S. aureus (MRSA) strains than among methicillin-susceptible S. aureus strains (8). Indeed, 60 to 90% of the MRSA organisms isolated worldwide are now resistant to ciprofloxacin (3). Recently, considerable efforts have been made to improve the spectra of activity of quinolones against S. aureus, including MRSA. New fluoroquinolones, such as levofloxacin, gatifloxacin, and moxifloxacin, have been developed and are presently on the market. However, despite their improved activities, newer fluoroquinolones have been shown to be less active against ciprofloxacin-resistant bacteria, and an increased resistance to these new compounds has been observed, especially with MRSA strains (8).
Fluoroquinolone resistance in S. aureus has been well studied and is associated, in most isolates, with single mutations in DNA topoisomerase IV and DNA gyrase (8, 25). Resistance mutations most often occur within the quinolone resistance-determining regions located in the grlA and gyrA genes encoding the A subunits of topoisomerase IV and DNA gyrase, respectively. In S. aureus, DNA topoisomerase IV is the primary target of fluoroquinolones, and first-step resistance to most of these antimicrobial agents has been associated with mutations in grlA (8, 25). Indeed, data from various studies have shown that most S. aureus isolates having decreased susceptibility to fluoroquinolones harbor mutations at codons 80 (Ser-80 to Phe or Tyr) and/or 84 (Glu-84 to Lys or Val or Gly) of grlA (23, 26, 27, 30). Single mutations in grlA appear to be sufficient to reach MICs of ciprofloxacin that exceed the laboratory breakpoints for susceptibility (8). On the other hand, for new fluoroquinolones, single grlA mutations are associated with a reduced susceptibility to these compounds but are not sufficient for clinical resistance. Additional mutations in gyrA and, less commonly, in grlB and gyrB, encoding the B subunits of the DNA topoisomerase IV and DNA gyrase, respectively, will usually generate full resistance to new fluoroquinolones and an increased level of ciprofloxacin resistance (8). In addition to mutations in these loci, altered expression of norA, a gene encoding a multidrug efflux pump, can also confer low-level resistance to fluoroquinolones (8, 25). It has been shown that S. aureus isolates with existing first-step grlA mutations are more likely to acquire subsequent mutations that result in clinical resistance to the new fluoroquinolones (5). Therefore, the development of novel methods that can rapidly identify mutations in the S. aureus grlA gene would provide a useful tool for the appropriate use of new fluoroquinolones in clinical settings.
In this study, we describe a rapid real-time PCR assay for the detection of S. aureus and of mutations in the grlA gene associated with fluoroquinolone resistance. We compare this PCR assay with conventional culture methods for identification of S. aureus isolates and determination of their susceptibility to fluoroquinolones.
(This study was partially presented at the 101st General Meeting of the American Society for Microbiology, Orlando, Fla., 20 to 24 May 2001.)
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study were obtained from the American Type Culture Collection (ATCC) (Manassas, Va.) (n = 2), the Montreal General Hospital (Montréal, Québec, Canada) (n = 28), Pavillon Centre Hospitalier de l'Université Laval (Sainte-Foy, Québec, Canada) (n = 1), the Mount Sinai Hospital (Toronto, Ontario, Canada) (n = 5), the Huashan Hospital (Shanghai, China) (n = 19), Universidad de Buenos Aires (Buenos Aires, Argentina) (n = 10), Statens Serum Institut (Copenhagen, Denmark) (n = 10), Pfizer (Groton, Conn.) (n = 1), Institut Pasteur (Paris, France) (n = 2), the Centers for Disease Control and Prevention (Atlanta, Ga.) (n = 5), and the Biological Research Laboratory, Sankyo Co., Ltd. (Tokyo, Japan) (n = 4). Confirmation of the identification of the S. aureus clinical isolates was performed by using the MicroScan WalkAway panel type positive breakpoint combo 13 (Dade Behring Canada Inc., Mississauga, Ontario, Canada).
MIC determination.
The MICs of ciprofloxacin, levofloxacin, gatifloxacin, and oxacillin were determined by the Etest method (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. The results were interpreted according to the standards of the NCCLS (20). S. aureus ATCC 29213 was used as a quality control for antimicrobial susceptibility testing.
Primers and probes.
Alignment of publicly available sequences of grlA genes from several staphylococcal species was performed with the program Pileup from the GCG package (version 10; Accelrys). Two primers (grlA205 and grlA396) specific to the S. aureus grlA gene were designed with the help of the Oligo primer analysis software, version 6.65 (Molecular Biology Insights, Cascade, Colo.). Three molecular beacon probes (MBPs) were also designed with a DNA fold program (M. Zuker, http://www.bioinfo.rpi.edu/applications/mfold/old/dna/). The stem of each molecular beacon was formed by adding 5-nucleotide complementary sequences at either end of the probe sequence. To avoid nonspecific fluorescence emission, the stem sequence was designed to ensure that the molecular beacons adopt a hairpin structure at the optimal annealing temperature of the PCR, when no target amplicons are present. Two MBPs (S80-FAM and E84-FAM) targeting the wild-type sequence of grlA were used to detect the presence of mutations at codons 80 and 84, respectively. A third MBP (Sau-TET), targeting an invariant region of grlA, was used to identify S. aureus and as a positive amplification control. Resistance to fluoroquinolones was identified when there was a positive signal from the S. aureus probe and no signal from either or both of the wild-type alleles of the probes targeting the regions potentially containing mutations at codons 80 and/or 84 of grlA. Primers were synthesized with a DNA synthesizer (model 391, PE Applied Biosystems, Foster City, Calif.), and MBPs were obtained either from Stratagene (La Jolla, Calif.) or from Biosearch Technology (Novato, Calif.). PCR primers and probes used in this study are listed in Table 1.
TABLE 1.
Amplification primers and molecular beacon probes used in this study
| Oligonucleotide | Sequence | Positiona |
|---|---|---|
| Primers | ||
| grlA205 (Forward) | 5′-GAT GTT ATT GGT CAA TAT CAT CCA-3′ | 205-228 |
| grlA396 (Reverse) | 5′-AAG AAA CTG TCT CTT TAT TAA TAT CAC GT-3′ | 396-424 |
| Probesb | ||
| S80-FAM | 5′ (FAM)-CAG CGT ACA CTG AGG AGT CTC CGC TG-3′ (DABCYL) | 233-248 |
| E84-FAM | 5′ (FAM)-CGC GAA GTG TAC GAA GCA ATG GTC GCG-3′ (DABCYL) | 243-259 |
| Sau-TET | 5′ (TET)-CCG GGT ATC GAT AAT GAT CCG CCA GCC CGG-3′ (DABCYL) | 324-343 |
Nucleotide positions in the grlA gene (start codon, 1 to 3) from GenBank accession no. D67075.
The underlined sequences constitute the stem of each molecular beacon. FAM, 6-carboxylfluorescein; TET, tetrachloro-6-carboxylfluorescein.
PCR amplification.
For all bacterial strains, amplification was performed using either purified genomic DNAs or crude DNA extracts prepared from bacterial suspensions whose turbidity was adjusted to that of a 0.5 McFarland standard, which corresponds to approximately 1.5 × 108 bacteria/ml. Genomic DNA was purified with the G Nome kit (Qbiogene Inc., Carlsbad, Calif.) according to the manufacturer's instructions. Crude DNA extracts were prepared for PCR by using a rapid DNA extraction kit (Infectio Diagnostic [I.D.I.] Inc., Sainte-Foy, Canada) (13). Either 0.1 ng of purified genomic DNA or 1 μl of crude DNA extract was transferred directly to a 24-μl PCR mixture containing 0.4 μM concentrations of the primers grlA205 and grlA396, a 0.2 μM concentration of each of the MBPs, a 200 μM concentration of each of the four deoxyribonucleotide triphosphates (Pharmacia Biotech, Baie d'Urfé, Québec, Canada), 10 mM Tris-HCl (pH 9.1), 50 mM KCl, 0.1% Triton X-100, 3.5 mM MgCl2, 3.3 mg of bovine serum albumin/ml (Sigma-Aldrich Canada, Oakville, Ontario, Canada), and 0.5 U of Taq DNA polymerase (Promega, Madison, Wis.) combined with the TaqStart antibody (BD Biosciences Clontech, Palo Alto, Calif.). For each assay, two separate PCRs containing either probes S80-FAM and Sau-TET or probes E84-FAM and Sau-TET were performed. The thermal cycling protocol was 3 min at 96°C for initial denaturation followed by 45 cycles of 3 steps consisting of 5 s at 95°C for the denaturation step, 15 s at 60°C for the annealing step, and 10 s at 72°C for the extension step. Real-time detection of the PCR products was performed on a Smart Cycler (Cepheid, Sunnyvale, Calif.) by measuring the fluorescent signal emitted by the MBPs hybridized to their genetic target at the end of each annealing step. The specificity of the PCR assay was verified by using a panel of 27 bacterial species (31 strains) genetically close to S. aureus (Table 2). For determination of the analytical sensitivity of the PCR assay, serial twofold dilutions of purified genomic DNA were used to determine the minimal number of genomes that can be detected.
TABLE 2.
Evaluation of the real-time PCR assay for the detection of fluoroquinolone-resistant Staphylococcus aureus using a variety of bacteria
| Bacterial species (strain number) | Fluorescent signal with the indicated test and probea
|
|||
|---|---|---|---|---|
| PCR no. 1
|
PCR no. 2
|
|||
| Sau-TET | S80-FAM | Sau-TET | E84-FAM | |
| Abiotrophia defectiva (ATCC 49176) | − | − | − | − |
| Bacillus cereus (ATCC 13472) | − | − | − | − |
| Bacillus mycoides (ATCC 10206) | − | − | − | − |
| Enterococcus faecalis (ATCC 29212) | − | − | − | − |
| Enterococcus faecalis (ATCC 19433) | − | − | − | − |
| Enterococcus flavescens (ATCC 49996) | − | − | − | − |
| Granulicatella adiacens (ATCC 49175) | − | − | − | − |
| Lactococcus lactis (ATCC 11454) | − | − | − | − |
| Listeria innocua (ATCC 33090) | − | − | − | − |
| Listeria monocytogenes (ATCC 15313) | − | − | − | − |
| Staphylococcus auricularis (ATCC 33753) | − | − | − | − |
| Staphylococcus capitis subsp. urealyticus (ATCC 49326) | − | − | − | − |
| Staphylococcus carnosus (ATCC 51365) | − | − | − | − |
| Staphylococcus chromogenes (ATCC 43764) | − | − | − | − |
| Staphylococcus epidermidis (ATCC 35983) | − | − | − | − |
| Staphylococcus epidermidis (ATCC 35984) | − | − | − | − |
| Staphylococcus gallinarum (ATCC 35539) | − | − | − | − |
| Staphylococcus haemolyticus (ATCC 29970) | − | − | − | − |
| Staphylococcus haemolyticus (LSPQ 2514) | − | − | − | − |
| Staphylococcus hominis subsp. hominis (ATCC 27844) | − | − | − | − |
| Staphylococcus lentus (ATCC 29070) | − | − | − | − |
| Staphylococcus lugdunensis (ATCC 43809) | − | − | − | − |
| Staphylococcus saccharolyticus (ATCC 14953) | − | − | − | − |
| Staphylococcus saprophyticus (ATCC 15305) | − | − | − | − |
| Staphylococcus saprophyticus (ATCC 35552) | − | − | − | − |
| Staphylococcus saprophyticus (ATCC 43867) | − | − | − | − |
| Staphylococcus simulans (ATCC 27848) | − | − | − | − |
| Staphylococcus warneri (ATCC 27836) | − | − | − | − |
| Staphylococcus xylosus (ATCC 29971) | − | − | − | − |
| Streptococcus agalactiae (ATCC 12400) | − | − | − | − |
| Streptococcus pneumoniae (ATCC 49619) | − | − | − | − |
| S. aureus (ATCC 25923)b | + | + | + | + |
| S. aureus (ATCC 29213)b | + | + | + | + |
| S. aureus (CCRI-2024)b | + | − | + | + |
| S. aureus (9191)b | + | − | + | + |
| S. aureus (9299)b | + | − | + | + |
| S. aureus (9015)b | + | + | + | − |
| S. aureus (9283)b | + | + | + | − |
| S. aureus (CCRI-12876)b | + | − | + | − |
All PCR tests were performed using 0.1 ng of purified genomic DNA.
These strains were chosen to represent different mutations at codons 80 and 84 of grlA. The quinolone resistance-determining regions of grlA from these strains were sequenced in this study. S. aureus strains ATCC 25923 and ATCC 29213 have no mutation at codons 80 and 84; strains CCRI-2024, 9191, and 9299 have a Ser-80-to-Phe (TCC→TTC) mutation; strain 9015 has a Glu-84-to-Gly (GAA→GGA) mutation; strain 9283 has a Glu-84-to-Lys (GAA→AAA) mutation; and strain CCRI-12876 has both Ser-80-to-Tyr (TCC→TAC) and Glu-84 to Gly (GAA→GGA) mutations.
DNA sequencing.
A 220-bp portion of the grlA gene encompassing the nucleotide mutations at codons Ser-80 and Glu-84 of this gene was sequenced for eight S. aureus strains that were used as genotype control strains (ATCC 25923, ATCC 29213, CCRI-2024, CCRI-12876, 9015, 9191, 9283, and 9299). Amplification was performed as previously described (14) with primers grlA205 and grlA396 (Table 1). The PCR products were purified and sequenced as previously described (14).
Nucleotide sequence accession number.
GenBank accession numbers for the 220-bp partial sequence of the S. aureus grlA gene are as follows: AY234038 for strain ATCC 25923, AY214125 for strain ATCC 29213, AY214123 for strain CCRI-2024, AY214124 for strain CCRI-12876, AY214128 for strain 9015, AY214127 for strain 9191, AY214126 for strain 9283, and AY214129 for strain 9299.
RESULTS
Specificity and sensitivity of the PCR assay.
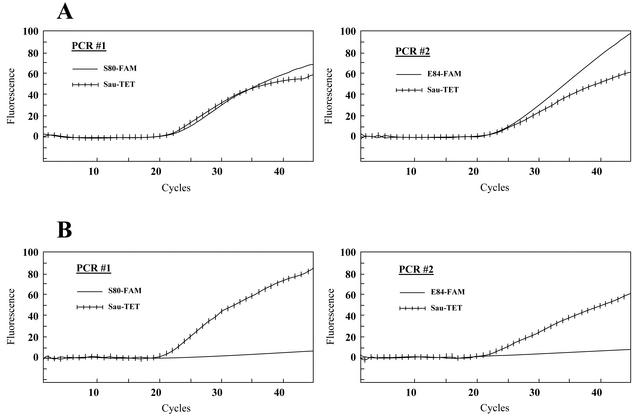
In this study, we have developed a PCR primer pair (grlA205 and grlA396) specific to the S. aureus grlA gene and three fluorescent MBPs that allow the detection of the amplified grlA gene product (Sau-TET) together with mutations at codons 80 (S80-FAM) and 84 (E84-FAM) of this gene. Two PCRs were performed for each assay. In the first reaction, the S. aureus grlA-specific primer pair was combined with the Sau-TET and S80-FAM probes, whereas in the second reaction, this primer pair was combined with the Sau-TET and E84-FAM probes. The specificity of the real-time PCR assay for S. aureus was first demonstrated with a panel of 27 bacterial species (31 strains) genetically close to S. aureus (Table 2). DNA from all bacterial species other than S. aureus were PCR negative, showing that this assay is specific to S. aureus. The specificity of the PCR assay for the detection of mutations at codons 80 and 84 of the S. aureus grlA gene was then tested against eight S. aureus strains with or without mutations in grlA (Table 2). Figure 1 shows an example of the detection of fluoroquinolone-sensitive and -resistant S. aureus strains on a Smart Cycler. A positive fluorescent signal was detected with the three probes for the fluoroquinolone-sensitive strain with no mutation at codons 80 and 84 of grlA, whereas a positive signal was detected with the Sau-TET probe only for the fluoroquinolone-resistant strain with mutations at both codons. The assay correctly identified the six S. aureus strains with mutations at codons 80 and/or 84 of grlA (Table 2). It also correctly identified the two S. aureus strains with no mutation at these codons. The analytical sensitivity of the assay was determined by using genomic DNA purified from three S. aureus strains with or without mutations at codons 80 and/or 84 of grlA. The detection limit was about 10 genomic copies per PCR (data not shown).
FIG. 1.
Example showing the real-time detection on a Smart Cycler of a fluoroquinolone-sensitive S. aureus strain (ATCC 25923) containing no mutation in the grlA gene (A) and a fluoroquinolone-resistant S. aureus strain (CCRI-12876) containing mutations at both codons 80 and 84 (B). For every sample tested, two separate PCRs, one containing probes S80-FAM and Sau-TET and one containing probes E84-FAM and Sau-TET, were performed. The left graphs show the fluorescence curves for the Sau-TET probe (hatched curve) and the S80-FAM probe (plain curve). The right graphs show the fluorescence curves for the Sau-TET probe (hatched curve) and the E84-FAM probe (plain curve). PCR assays were performed with crude DNA extracts prepared from bacterial suspensions whose turbidities were adjusted to that of a 0.5 McFarland standard.
Correlation between PCR and conventional culture methods.
We have compared the results of conventional culture methods for identification of S. aureus and the susceptibility to ciprofloxacin, levofloxacin, and gatifloxacin determined by the Etest method for 85 S. aureus clinical isolates from various geographic areas with those obtained by the PCR assay. For these 85 S. aureus strains, MICs ranged from 0.094 to >32 μg/ml for ciprofloxacin and levofloxacin and from 0.047 to >32 μg/ml for gatifloxacin. All S. aureus strains tested were correctly identified by PCR. Correlation between fluoroquinolone susceptibility and the presence of mutations in grlA is summarized in Table 3. Of the 85 S. aureus strains tested, 35 (41.2%) were susceptible to ciprofloxacin (MIC, ≤1 μg/ml). All these strains were negative for mutation at codons 80 and 84 based on PCR. Of the 50 ciprofloxacin-resistant strains (58.8%) (ciprofloxacin MIC, ≥2 μg/ml), 49 had mutations at codons 80 and/or 84 detected by PCR. No mutation was detected in grlA in one ciprofloxacin-resistant strain. Thirty-seven S. aureus strains (43.5%) were susceptible to levofloxacin (MIC, ≤2 μg/ml). No mutation was detected in grlA for 36 of these strains. One levofloxacin-sensitive strain had a mutation at codon 84 of grlA detected by PCR. Forty-eight strains (56.5%) were resistant to levofloxacin (MIC, ≥4 μg/ml), and all had mutations at codons 80 and/or 84 detected by PCR. Of the 37 S. aureus strains (43.5%) susceptible to gatifloxacin (MIC, ≤2 μg/ml), 36 were negative for mutation at codons 80 and/or 84 by PCR. A mutation was detected at codon 84 of grlA by PCR in one gatifloxacin-sensitive strain. Forty-eight S. aureus strains (56.5%) were resistant to gatifloxacin (MIC, ≥4 μg/ml), and all had mutations at codons 80 and/or 84 detected by PCR. Overall, among the 49 S. aureus strains with mutations in grlA, 26 (53%) had a mutation at codon 80, 2 (4.1%) had a mutation at codon 84, and 21 (42.9%) had mutations at both codons.
TABLE 3.
Correlation between fluoroquinolone susceptibility and the real-time PCR assay
| Fluoroquinolone | Etest result (no.) | PCR result
|
|
|---|---|---|---|
| Susceptible (no.) | Resistant (no.) | ||
| Ciprofloxacin | Susceptible (35) | 35 | 0 |
| Resistant (50) | 1 | 49 | |
| Levofloxacin | Susceptible (37) | 36 | 1 |
| Resistant (48) | 0 | 48 | |
| Gatifloxacin | Susceptible (37) | 36 | 1 |
| Resistant (48) | 0 | 48 | |
DISCUSSION
The emergence of fluoroquinolone resistance in S. aureus is of great concern. In the treatment of staphylococcal infections, fluoroquinolones should not be administered empirically but only after determination of the antimicrobial susceptibility of the clinical isolates. For these reasons, it has become important to develop rapid diagnostic tests to monitor the clinical utility of fluoroquinolones and also to minimize the emergence of resistance. Many molecularly-based tests have been recently developed to identify S. aureus and to determine its susceptibility to various antibiotics (10, 11, 17, 22). While all these tests detected the presence of a specific gene to establish the antibiotic susceptibility profile, mutations in the targets of fluoroquinolones must be detected to determine the susceptibility to this class of antibiotics.
In the present study, we developed a rapid real-time PCR assay that can be used to simultaneously identify S. aureus and the first-step mutations associated with decreased susceptibility to fluoroquinolones. This assay is based on the detection of the S. aureus grlA gene encoding the A subunits of DNA topoisomerase IV and of mutations at codons Ser-80 and/or Glu-84, which are associated with fluoroquinolone resistance. We have compared the PCR assay for the detection of S. aureus and mutations in grlA with classical methods for identification of S. aureus and determination of susceptibility to fluoroquinolones. Overall, we found correlations between these methods of 100% for S. aureus identification and of 98.8% for ciprofloxacin, levofloxacin, or gatifloxacin susceptibility testing. Two strains did not show any correlation between the PCR results and the MICs. One ciprofloxacin-resistant strain, for which the ciprofloxacin MIC was 2 μg/ml, had no mutation, based on PCR. The decreased susceptibility to ciprofloxacin in this strain may be associated with other resistance mechanisms, such as NorA efflux pump-mediated resistance or mutations in other regions of grlA (12, 19, 23, 24). Another strain which was resistant to ciprofloxacin, for which the ciprofloxacin MIC was 4 μg/ml, had a mutation at codon 80 but was clinically susceptible to levofloxacin and gatifloxacin (MICs, 1.5 and 0.75 μg/ml, respectively). However, the MICs of these two antibiotics were six to eight times higher than those for the fluoroquinolone-sensitive S. aureus control strain ATCC 29213, showing that the former strain exhibits reduced susceptibility to these fluoroquinolones. The low level of resistance to ciprofloxacin and reduced susceptibility to the new fluoroquinolones suggested that this strain may have a single mutation at codon 80 of grlA. Therefore, these data corroborated those from others which have shown that a single grlA mutation is not sufficient to cause clinical resistance to levofloxacin and gatifloxacin but greatly decreased the susceptibility to these antimicrobial agents (9, 30).
In the method described in this study, mutations in gyrA, gyrB, or grlB were not sought. However, detection of mutations in grlA provided an excellent correlation with susceptibility to all the fluoroquinolones tested. The good correlation between grlA mutations and resistance to gatifloxacin and levofloxacin may be explained by accumulation of mutations in both grlA and gyrA genes in the strains studied. Indeed, 96% (49 of 51) of the fluoroquinolone-resistant S. aureus isolates tested in this study exhibited high-level resistance to ciprofloxacin (MIC, >8 μg/ml). This high level of resistance is usually associated with mutations in grlA as well as in gyrA, gyrB, and/or grlB genes which generate cross-resistance to new fluoroquinolones (4, 9, 26). Among the 85 S. aureus isolates studied, 56 were resistant to oxacillin (MRSA strains). Forty-six (82.1%) of the 56 MRSA isolates exhibited resistance to both ciprofloxacin (MIC, >32 μg/ml) and new fluoroquinolones, whereas only three of the forty (7.5%) methicillin-susceptible S. aureus isolates exhibited this resistance phenotype. Therefore, these data suggest a high prevalence of resistance to the new fluoroquinolones among MRSA isolates, as also described recently by other groups (7, 23). The high prevalence of fluoroquinolone resistance among MRSA isolates has been attributed to both selection by quinolone exposure and transmission of clonal strains in health care settings (8). The increasing resistance of S. aureus isolates to new fluoroquinolones suggests that physicians should be cautious when using these antibiotics, especially against isolates with grlA mutations, as the potential to acquire subsequent mutations that lead to full resistance is higher in those strains (5). The PCR assay described in this study should therefore provide a useful tool for the appropriate use of new fluoroquinolones against S. aureus infections.
In the real-time PCR assay developed in this study, fluorescent MBPs were used to detect the S. aureus grlA gene and the mutations associated with fluoroquinolone resistance. MBPs are fluorescently labeled single-stranded nucleic acid probes that possess a stem-loop structure (28). It has been shown that the structure of MBPs allows a more effective detection of single-nucleotide polymorphisms over a wide range of temperatures than do the corresponding linear probes, such as TaqMan probes (2). MBPs have been used in several applications, including detection of mutations associated with antibiotic resistance, such as mutations in the rpoB gene of Mycobacterium tuberculosis that confer resistance to the antibiotic rifampin (21). Other groups have used adjacent probes or TaqMan probes for the detection of gyrA mutations associated with ciprofloxacin resistance in gram-negative bacteria (15, 16, 29, 31). The present study shows that our MBPs were highly specific for detection of single-nucleotide polymorphisms in the S. aureus grlA gene associated with fluoroquinolone resistance.
Several methodological approaches for the detection of mutations in the S. aureus targets of fluoroquinolones, such as sequencing, single-strand conformation polymorphism, restriction fragment length polymorphism, and denaturing high-performance liquid chromatography, have been developed (6, 18, 27, 30). Because of the complexity and the time required to perform the experiments, these methods are not appropriate for routine use in clinical microbiology laboratories. The use of the real-time PCR assay described here greatly decreased the time required to identify S. aureus isolates and to detect the grlA mutations associated with reduced susceptibility to fluoroquinolones compared to conventional methods. The procedure, including DNA extraction, can be completed in less than 1 h. Furthermore, this PCR assay was sensitive, as it detected as few as 10 genome copies per PCR, showing that its potential for detection of S. aureus directly from clinical specimens.
With a correlation of 98.8% between the MICs and the PCR assay for all three fluoroquinolones tested, this real-time PCR assay represents a promising diagnostic tool that can replace conventional culture methods for rapid identification of S. aureus along with susceptibility testing against fluoroquinolones.
Acknowledgments
This study was supported by Infectio Diagnostic (I.D.I.) Inc. (Sainte-Foy, Québec, Canada) and by grant PA-15586 from the Canadian Institutes of Health Research (CIHR). M.O. is a CIHR investigator.
We thank Richard Giroux for his excellent technical assistance. We also thank D. Centron (Universidad de Buenos Aires, Buenos Aires, Argentina), D. E. Low (Mount Sinai Hospital, Toronto, Ontario, Canada), F. C. Tenover (Centers for Disease Control and Prevention, Atlanta, Ga.), W. Fu (Huashan Hospital, Shanghai, China), L. V. Pallesen (Statens Serum Institut, Copenhagen, Denmark), J. Sutcliffe (Pfizer, Groton, Conn.), P. Courvalin (Institut Pasteur, Paris, France), and T. Takenouchi (Sankyo Co., Ltd., Tokyo, Japan) for providing clinical isolates of S. aureus.
REFERENCES
- 1.Acar, J. F., and F. W. Goldstein. 1997. Trends in bacterial resistance to fluoroquinolones. Clin. Infect. Dis. 24(Suppl. 1):S67-S73. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet, G., S. Tyagi, A. Libchaber, and F. R. Kramer. 1999. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA 96:6171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S114-S132. [DOI] [PubMed] [Google Scholar]
- 4.Fukuda, H., S. Hori, and K. Hiramatsu. 1998. Antibacterial activity of gatifloxacin (AM-1155, CG5501, BMS-206584), a newly developed fluoroquinolone, against sequentially acquired quinolone-resistant mutants and the norA transformant of Staphylococcus aureus. Antimicrob. Agents Chemother. 42:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gootz, T. D., R. P. Zaniewski, S. L. Haskell, F. S. Kaczmarek, and A. E. Maurice. 1999. Activities of trovafloxacin compared with those of other fluoroquinolones against purified topoisomerases and gyrA and grlA mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1845-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannachi-M'Zali, F., J. E. Ambler, C. F. Taylor, and P. M. Hawkey. 2002. Examination of single and multiple mutations involved in resistance to quinolones in Staphylococcus aureus by a combination of PCR and denaturing high-performance liquid chromatography (DHPLC). J. Antimicrob. Chemother. 50:649-655. [DOI] [PubMed] [Google Scholar]
- 7.Hoogkamp-Korstanje, J. A. 1997. In-vitro activities of ciprofloxacin, levofloxacin, lomefloxacin, ofloxacin, pefloxacin, sparfloxacin and trovafloxacin against gram-positive and gram-negative pathogens from respiratory tract infections. J. Antimicrob. Chemother. 40:427-431. [DOI] [PubMed] [Google Scholar]
- 8.Hooper, D. C. 2002. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 9.Ince, D., and D. C. Hooper. 2001. Mechanisms and frequency of resistance to gatifloxacin in comparison to AM-1121 and ciprofloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaffe, R. I., J. D. Lane, S. V. Albury, and D. M. Niemeyer. 2000. Rapid extraction from and direct identification in clinical samples of methicillin-resistant staphylococci using the PCR. J. Clin. Microbiol. 38:3407-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonas, D., H. Grundmann, D. Hartung, F. D. Daschner, and K. J. Towner. 1999. Evaluation of the mecA femB duplex polymerase chain reaction for detection of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 18:643-647. [DOI] [PubMed] [Google Scholar]
- 12.Kaatz, G. W., S. M. Seo, and C. A. Ruble. 1993. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1086-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ke, D., C. Ménard, F. J. Picard, M. Boissinot, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin. Chem. 46:324-331. [PubMed] [Google Scholar]
- 14.Ke, D., F. J. Picard, F. Martineau, C. Ménard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1999. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 37:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Z., S. Yokoi, Y. Kawamura, S. Maeda, T. Ezaki, and T. Deguchi. 2002. Rapid detection of quinolone resistance-associated gyrA mutations in Neisseria gonorrhoeae with a LightCycler. J. Infect. Chemother. 8:145-150. [DOI] [PubMed] [Google Scholar]
- 16.Lindler, L. E., W. Fan, and N. Jahan. 2001. Detection of ciprofloxacin-resistant Yersinia pestis by fluorogenic PCR using the LightCycler. J. Clin. Microbiol. 39:3649-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martineau, F., F. J. Picard, N. Lansac, C. Ménard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2000. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messina, C., V. Cafiso, F. Campanile, M. Santagati, and S. Stefani. 2001. Rapid method for detection of gyrA and grlA mutations in unrelated strains of staphylococci susceptible and resistant to levofloxacin. New Microbiol. 24:347-353. [PubMed] [Google Scholar]
- 19.Muñoz-Bellido, J. L., M. Alonzo Manzanares, J. A. Martínez Andrés, M. N. Guttiérrez Zufiaurre, G. Ortiz, M. Segovia Hernández, and J. A. García-Rodríguez. 1999. Efflux pump-mediated quinolone resistance in Staphylococcus aureus strains wild type for gyrA, gyrB, grlA, and norA. Antimicrob. Agents Chemother. 43:354-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial susceptibility testing. Eleventh informational supplement. Approved standard M100-S11. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 22.Reischl, U., H. J. Linde, M. Metz, B. Leppmeier, and N. Lehn. 2000. Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence PCR. J. Clin. Microbiol. 38:2429-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz, F. J., A. C. Fluit, S. Brisse, J. Verhoef, K. Kohrer, and D. Milatovic. 1999. Molecular epidemiology of quinolone resistance and comparative in vitro activities of new quinolones against European Staphylococcus aureus isolates. FEMS Immunol. Med. Microbiol. 26:281-287. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz, F. J., A. C. Fluit, M. Luckefahr, B. Engler, B. Hofmann, J. Verhoef, H. P. Heinz, U. Hadding, and M. E. Jones. 1998. The effect of reserpine, an inhibitor of multidrug efflux pumps, on the in-vitro activities of ciprofloxacin, sparfloxacin and moxifloxacin against clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 42:807-810. [DOI] [PubMed] [Google Scholar]
- 25.Schmitz, F. J., P. G. Higgins, S. Mayer, A. C. Fluit, and A. Dalhoff. 2002. Activity of quinolones against gram-positive cocci: mechanisms of drug action and bacterial resistance. Eur. J. Clin. Microbiol. Infect. Dis. 21:647-659. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz, F. J., B. Hofmann, B. Hansen, S. Scheuring, M. Luckefahr, M. Klootwijk, J. Verhoef, A. Fluit, H. P. Heinz, K. Kohrer, and M. E. Jones. 1998. Relationship between ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin (BAY 12-8039) MICs and mutations in grlA, grlB, gyrA and gyrB in 116 unrelated clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 41:481-484. [DOI] [PubMed] [Google Scholar]
- 27.Schmitz, F. J., M. E. Jones, B. Hofmann, B. Hansen, S. Scheuring, M. Luckefahr, A. Fluit, J. Verhoef, U. Hadding, H. P. Heinz, and K. Kohrer. 1998. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob. Agents Chemother. 42:1249-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 29.Walker, R. A., N. Saunders, A. J. Lawson, E. A. Lindsay, M. Dassama, L. R. Ward, M. J. Woodward, R. H. Davies, E. Liebana, and E. J. Threlfall. 2001. Use of a LightCycler gyrA mutation assay for rapid identification of mutations conferring decreased susceptibility to ciprofloxacin in multiresistant Salmonella enterica serotype Typhimurium DT104 isolates. J. Clin. Microbiol. 39:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, T., M. Tanaka, and K. Sato. 1998. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob. Agents Chemother. 42:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson, D. L., S. R. Abner, T. C. Newman, L. S. Mansfield, and J. E. Linz. 2000. Identification of ciprofloxacin-resistant Campylobacter jejuni by use of a fluorogenic PCR assay. J. Clin. Microbiol. 38:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]