Abstract
A novel PCR restriction analysis method using the RNA polymerase β-subunit- coding gene (rpoB) was employed to both detect and identify Helicobacter pylori in biopsy specimens and culture isolates. The rpoB DNAs (458 bp) were specifically amplified by PCR with the Helicobacter-specific primers (HF and HR). Based on the determined rpoB sequences of the culture isolates, an H. pylori-specific restriction site, Tru9I, was found. H. pylori can be identified by observing two discernible DNA fragments (288 and 138 bp) after Tru9I digestion and agarose gel electrophoresis. The rpoB PCR and subsequent restriction analysis (PRA) enabled the specific detection and identification of H. pylori in biopsy specimens from patients with gastroduodenal diseases. The rpoB PRA conferred a compatible or a slightly higher positive rate (53.7%) than did the Campylobacter-like organism (CLO) test (50.4%) and glmM PCR (48.8%), suggesting that it is useful for diagnosing an H. pylori infection without culture in the clinical laboratory.
Helicobacter pylori is one of the most prevalent microorganisms and is a major cause of gastrointestinal disease in humans. H. pylori is found in the stomach of patients with gastroduodenal diseases, such as duodenal and gastric ulcers (9). In addition, H. pylori is thought to be one of the major causes of stomach cancer (10, 11). It can be cultivated from biopsy specimens in 2 to 8% O2 and 10% CO2 after 3 to 5 days and is identified through means such as Gram staining, a catalase test, and a urease test. Although the culture method is a “gold standard” for diagnosing many infectious diseases, it is not easy in the case of an H. pylori infection. The sensitivity of the H. pylori isolation method shows a marked variation, because two or three biopsy specimens from different locations are needed for the culture, the histopathological examination, and the rapid urease test (CLO test; Delta West, Perth, Australia). Positive results of the CLO test and/or histological examination are not always a guarantee of a positive culture (19). Usually under normal atmospheric conditions, it vanishes easily (13); nor is it easy to cultivate H. pylori by traditional methods. Therefore, in addition to the noninvasive methods, many different kinds of methods, such as CLO testing (14), special staining (19), and PCR (2, 24), have been used to detect H. pylori in biopsy specimens.
PCR is one of the most widely used molecular methods for detecting specific pathogens. Several genes have been used to detect and identify H. pylori (2, 17, 24). However, H. pylori is a microorganism with marked genetic diversity. Although it is described as a quasispecies (25) because isolates show genetic diversity and because genetically different isolates have been cultured in the same patients, it appears that genetic identification by using housekeeping H. pylori genes is needed to accurately identify H. pylori and evaluate the causes of gastroduodenal diseases resulting from an H. pylori infection. For the genetic identification of H. pylori, several PCR methods that employ the 16S rRNA gene (ribosomal DNA [rDNA]), rpoD, ureA, ureB, and ureC have been used (2, 24). Most are protein-encoding genes except for 16S rDNA. Among them, ureC (glmM) PCR is known to be specific to H. pylori and is frequently used (17).
In this study, the RNA polymerase β-subunit-coding gene (rpoB) (1) was used for the detection and identification of H. pylori by specific PCR restriction analysis (PRA). rpoB is an important transcription apparatus in all microorganisms. It is a kind of housekeeping gene and is as stable as 16S rDNA. Recently, partial rpoB DNA sequences containing the Rifr region, which is related to rifampin resistance of Escherichia coli and Mycobacterium tuberculosis, were used to either differentiate or identify closely related species in Enterobacteriaceae (19), Mycobacterium (12), and Borrelia (15). Because this region is highly conserved, a novel PCR amplifying 458-bp DNA and a subsequent restriction analysis method were developed. This was applied to culture isolates as well as gastric biopsy specimens from patients with gastroduodenal diseases and was compared to those of glmM PCR and CLO testing.
H. pylori.
isolates (157 strains) were provided by M.-J. Cho at Gyeong-Sang University, M.-W. Chang at Kosin University Hospital, and S.-Y. Kim at Chungbuk University, all in Korea. All strains were isolated from the gastric biopsy specimens from patients with gastric disorders or from normal adults. The biopsy specimens were smeared directly on Mueller-Hinton agar containing 10% bovine serum, 10 μg of vancomycin ml−1, 25 μg of nalidixic acid ml−1, and 1 μg of amphotericin B ml−1. The inoculated contents of plates were incubated at 37°C for 4 or 5 days in 10% CO2. All isolates were identified as being H. pylori based on the culture characteristics, such as small translucent colonies under a dissecting microscope, gram-negative spiral organisms, and urease production. D. E. Berg at Washington University Medical School kindly provided the five H. pylori DNAs, which had been extracted from five Spanish strains. The Helicobacter hepaticus (ATCC 51448) strains were provided by Y.-G. Choi at the Korea Research Institute of Bioscience and Biotechnology, and the Helicobacter felis (ATCC 49179) strains were provided by J.-S. Yum at Mogam Biotechnology Research Institute. The Helicobacter cinaedi (ATCC 35683) and Helicobacter mustelae (ATCC 43772) strains were purchased from the American Type Culture Collection.
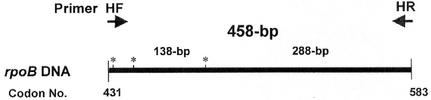
The H. pylori DNAs were prepared by using the previously described bead beater-phenol extraction method (12). A loopful culture of each isolate was suspended in 200 μl of Tris-EDTA-NaCl buffer (10 mM Tris-HCl, 1 mM EDTA, and 100 mM NaCl [pH 8.0]). A bacterial suspension was placed in a 2.0-ml screw-cap microcentrifuge tube filled with 100 μl (packed volume) of glass beads (diameter, 0.1 mm; Biospec Products, Bartlesville, Okla.) and 100 μl of phenol-chloroform-isoamyl alcohol (25:24:1) (P-2069; Sigma Chemical Co.). The tube was oscillated on a Mini-Bead Beater (Biospec Products) for 1 min and was centrifuged (12,000 × g, 5 min) to separate the phases. The aqueous phase was subsequently transferred into another clean tube; 10 μl of 3 M sodium acetate and 250 μl of ice-cold absolute ethanol were added. To precipitate the DNA, the mixture was kept at −20°C for 10 min. The harvested DNA pellets were dissolved in 60 μl of Tris-EDTA buffer (10 mM Tris-HCl and 1 mM EDTA [pH 8.0]) and were used as a template DNA for PCR. A set of primers (HF, 5′-ACTTTAAA CGCATGAAGATAT-3′; and HR, 5′-ATATTTTGACCTTCTGGGGT-3′) was used to amplify the 458-bp DNA encompassing the Rifr region (Fig. 1). The primers were selected on the basis of the known rpoB sequences from Bacillus subtilis (1), H. pylori J99, and H. pylori 26695 strains (GenBank accession nos. AE001540 and AE000625, respectively). The template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer, Daejeon, Korea) containing 1 U of Taq DNA polymerase, 250 μM each deoxynucleoside triphosphate, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and the gel loading dye. The volume was adjusted with distilled water to 20 μl. The reaction mixture was subjected to 30 amplification cycles (5 min at 95°C, 30 s at 94°C, 30 s at 52°C, 45 s at 72°C, and 5 min at 75°C) followed by a 5-min extension at 72°C (model 9600 Thermocycler; Perkin-Elmer Cetus). The PCR products were electrophoresed on a 1.2% (wt/vol) agarose gel and were purified by using the QIAEX II gel extraction kit (QIAGEN, Hilden, Germany). PCR was also performed with other bacterial DNAs, i.e., those from E. coli, B. subtilis, Moraxella catarrhalis, Corynebacterium diphtheriae, Haemophilus influenzae, Mycobacterium fortuitum, Neisseria sicca, Staphylococcus aureus, and Enterococcus faecalis, in order to evaluate the specificity of the Helicobacter-specific primers (HF and HR).
FIG. 1.
Helicobacter-specific primers used for the amplification of rpoB DNAs (458 bp). They were selected from the rpoB sequences of B. subtilis and E. coli and H. pylori 26695 (GenBank accession no. AE000625) and H. pylori J99 (accession no. AE001540). *, H. pylori-specific Tru9I restriction sites on the amplified rpoB DNA corresponding to codons 432, 441, and 487.
Another set of primers (forward primer, 5′-AAGCTTTTAGGGGTGTTAGGGGTTT-3′; and reverse primer, 5′-AAGCTTACTTTCTAACACTAACGC-3′) was used to amplify the glmM DNA (294 bp) (17). The PCR mixture was subjected to 30 amplification cycles (1 min at 93°C, 1 min at 55°C, and 1 min at 72°C).
The nucleotide sequences (363 bp) of the purified rpoB PCR products (458 bp) were determined directly by using an Applied Biosystems model 373A automatic sequencer and a BigDye Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom). For the sequencing reaction, 60 ng of PCR-amplified DNA, 3.2 pmol of either the forward or the reverse primer, and 4 μl of BigDye Terminator RR mix (part no. 4303153; Perkin-Elmer Applied Biosystems) were mixed. The final volume was adjusted to 10 μl by the addition of distilled water. The reaction was run with 5% (vol/vol) dimethyl sulfoxide for 30 cycles of 15 s at 95°C, 10 s at 50°C, and 4 min at 60°C.
The sequences were aligned by using the multiple-alignment algorithm in the MegAlign package (Windows version 3.12e; DNASTAR, Madison, Wis.). The enzyme restriction sites for sizing the DNA fragment on each rpoB sequence of H. pylori and five Helicobacter species were generated by using the MapDraw program (MapDraw version 3.14; DNASTAR).
Ten microliters of the rpoB PCR products was transferred to a fresh microcentrifuge tube and was digested with restriction enzyme according to the supplier's instruction. One microliter of Tru9I (no. R7011, 10 U/μl; Promega), 1 μl of the enzyme buffer, and 4 μl of distilled water were added to the PCR products and were placed in a 65°C water bath for 1 h. The mixture was electrophoresed (100 V for 25 min) on a 1.2% (wt/vol) agarose gel, and the DNA bands were visualized by using ethidium bromide staining and were then photographed.
The gastric biopsy specimens were obtained from 123 patients who were diagnosed with gastroduodenal diseases by gastroscopy at the Department of Internal Medicine, Dankook University Hospital. Six pieces were obtained from the biopsy specimens from the antrum of stomach. Two pieces were used for the rapid urease test, two pieces for the histological examination, and two pieces for the rpoB PRA and glmM PCR. The patients were diagnosed by using histological examinations with gastritis (41 patients), a duodenal ulcer (42 patients), a benign gastric ulcer (21 patients), and a gastric cancer (19 patients). The DNAs were extracted from the biopsy specimens as described above and were used for rpoB PCR and glmM PCR.
The rapid urease test kit (CLO test; Delta West, Bentley, Australia) was used. It was stored in the refrigerator (2 to 8°C) and was used after warming. The gastric biopsy specimens were immediately embedded into the gel in the kit. The kit was placed at room temperature for 24 h and was observed for any color change. If a color change from yellow to red was observed, it was interpreted as a positive reaction.
Two pieces of the biopsy specimens were fixed in 10% (vol/vol) buffered formalin, embedded in paraffin, and then sectioned. Hematoxylin-and-eosin staining and modified Giemsa staining were used for routine histology and to detect H. pylori, respectively.
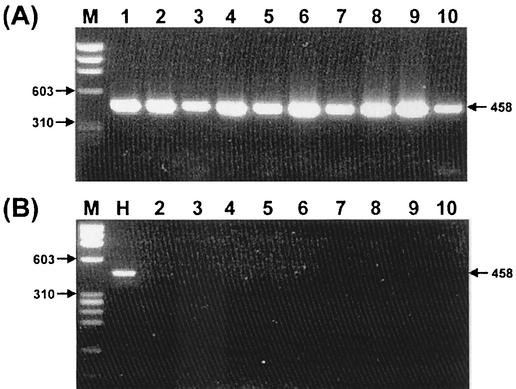
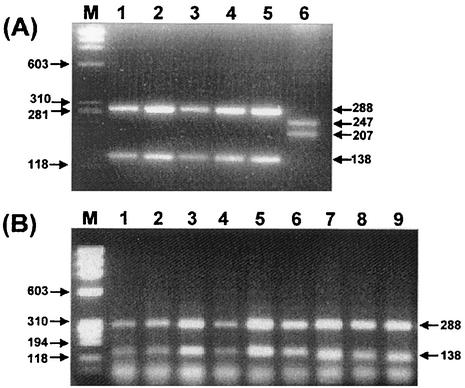
rpoB PCR for the 5 Helicobacter species and 162 H. pylori isolates including Spanish strains was performed. The PCR products (458 bp) were observed only from the Helicobacter species. There was no amplification from other bacteria, suggesting that the primers (HF and HR) are Helicobacter specific (Fig. 2). The nucleotide sequences of the amplified rpoB DNAs were determined, and portions of them (363 bp) were aligned. The percentage similarities among the rpoB sequences of 162 H. pylori strains, H. pylori 26695 (AE000625), and H. pylori J99 (AE001540) were more than 91.5%. The nucleotide sequences of the rpoB DNA among the H. pylori strains and H. cinaedi showed a 68.3% similarity. There was no significant clustering observed among the culture isolates, which had been isolated from three different areas in Korea. Through the computer-aided analysis of the rpoB DNA sequences from 164 H. pylori strains, including H. pylori 26695 and H. pylori J99, Tru9I was found to distinguish H. pylori from the other Helicobacter species (Fig. 3). Digestion with Tru9I yielded four bands (288, 138, 28, and 4 bp) from H. pylori rpoB DNA. Because of the fairly different sizes (247, 207, and 4 bp) of the DNA fragments from other species, such as H. mustelae, H. pylori can be easily identified by using the two major DNA fragments (288 and 138 bp) (Fig. 3A). Tru9I PRA for the gastric biopsy samples showed identical results, suggesting that the amplified DNAs were amplified from H. pylori (Fig. 3B). The results of rpoB PRA and glmM PCR results were compared with that of the CLO test, which was performed on the same gastric biopsy specimens. The concordance rate of rpoB PRA with the CLO test was 83.0%, while that of glmM PCR was 81.3% (Table 1). Considering the CLO test as the gold standard, the sensitivity and specificity of rpoB PRA were 85.7 and 80.0%, while those of glmM PCR were 79.3 and 83.0%, respectively. In general, the detection rate of rpoB PRA was slightly higher than those of the CLO test and glmM PCR, especially in the gastric cancer (Table 2). However, it was not statistically significant (P > 0.05). Nevertheless, this may be due to the small sample numbers. Among the 123 gastric biopsy specimens, 53 samples were histologically examined. The concordance rate between rpoB PCR and histological examination was 69.8%, while those of the CLO test and glmM PCR were 71.7 and 69.8%, respectively (Table 3).
FIG. 2.
Amplification of rpoB DNAs (458 bp) from Helicobacter species (A) and other bacteria (B). PCR products were electrophoresed on a 1.2% agarose gel. Lane M in both panels, φ X174 RF DNA/HaeIII digest. (A) Lanes: 1 to 6, H. pylori isolates; 7, H. felis; 8, H. cinaedi; 9, H. mustelae; and 10, H. hepaticus. (B) Lanes: H, H. pylori isolate; 2, E. coli, 3, C. diphtheriae; 4, S. aureus; 5, B. subtilis; 6, M. fortuitum; 7, B. catarrhalis; 8, H. influenzae; 9, N. sicca; and 10, E. faecalis.
FIG. 3.
Identification of H. pylori culture isolates (A) and H. pylori in the gastric biopsy specimens (B) by PRA using Tru9I. PCR products were digested and electrophoresed on a 1.2% agarose gel. All strains showed two major bands (288 and 138 bp), which are specific for H. pylori. Lane M in both panels, DNA marker φ X174 RF DNA/HaeIII digest. (A) Lanes 1 to 5, H. pylori culture isolates; lane 6, H. mustelae. (B) Lanes 1 to 9, gastric biopsy specimens.
TABLE 1.
Comparison of rpoB PCR and glmM PCR with CLO test in detecting H. pylori in gastric biopsy specimens (n = 123)
| Test | Result | No. of samples with CLO test result
|
|
|---|---|---|---|
| + | − | ||
| rpoB | + | 54 | 12 |
| − | 9 | 48 | |
| glmM | + | 50 | 10 |
| − | 13 | 50 | |
TABLE 2.
Comparison of methods of diagnosing H. pylori according to the patterns of gastroduodenal diseases (n = 123)
| Test | No. of patients with illness/no. of patients (%)
|
||||
|---|---|---|---|---|---|
| Gastritis | DUa | BGUb | Gastric cancer | All illnesses | |
| CLO | 16/41 (39.0) | 29/42 (69.0) | 13/21 (61.9) | 4/19 (21.1) | 62/123 (50.4) |
| rpoB | 18/41 (43.9) | 27/42 (64.3) | 14/21 (66.7) | 7/19 (36.8) | 66/123 (53.7) |
| glmM | 16/41 (39.0) | 27/42 (64.3) | 13/21 (61.9) | 4/19 (21.1) | 60/123 (48.8) |
DU, duodenal ulcer.
BGU, benign gastric ulcer.
TABLE 3.
Concordance of results from rpoB PCR, glmM PCR, and CLO test with pathology (n = 53)
| Test | Result | No. of specimens with pathology result
|
|
|---|---|---|---|
| + | − | ||
| CLO test | + | 25 | 4 |
| − | 11 | 13 | |
| rpoB PCR | + | 21 | 6 |
| − | 10 | 16 | |
| glmM PCR | + | 22 | 2 |
| − | 14 | 15 | |
There are many invasive and noninvasive methods used to diagnose an H. pylori infection. Although they may be convenient to use and safe for patients, noninvasive methods such as the urea breath test, which detects the metabolic end product, are considered to be nonspecific, as the results may be affected by other enteric bacteria (14, 19). However, gastrofiberscopy is commonly used to directly observe a pathological lesion and obtain biopsy specimens that are subsequently used for CLO test, histological examination, culture, and PCR. There is no doubt that a bacteriological culture is the best method for diagnosing a bacterial infection. However, it is not easy to cultivate H. pylori because the specimens are usually obtained from several different locations by gastrofiberscopy. In addition, the sensitivity of the culture-isolation method is low (8). Therefore, a culture is not considered to be the most practical diagnostic method. As a result, the CLO test and staining methods are preferred in many clinical laboratories. Nonetheless, they also have problems such as accuracy of species-specific identification (19). PCR (or PCR-linked methods), which is a specific and sensitive molecular method for detecting H. pylori DNA, can supplement the above methods.
PCR was applied to amplify and detect H. pylori DNAs from the ureA, ureB, ureC, cagA, and 16S rRNA genes (2, 6, 13, 17, 24). However, some of those genes have limitations as a target in detecting H. pylori. Although PCR is a good method for detecting H. pylori genes, the result will be affected by the nature of the target genes. The genetic diversity of H. pylori is well known. Even the eight repeat families varying in length from 0.47 to 3.8 kb are found in the chromosome of H. pylori (25). ureA and ureB encode urease, which is also produced from other enteric bacteria isolated from the stomach of animals. The structure of the urease-encoding gene was not the same among those bacteria, but some homologous subunits were found. Because H. pylori shows marked genetic diversity and because the sequence variations of those genes among the H. pylori strains have not been reported, the possibility of false-negative results cannot be discounted. This is more obvious in cagA, because it is not in all H. pylori strains. cagA is cytotoxin-associated gene A. Despite the relative smallness (259 to 785 bp), the targeted regions of these genes are not highly conserved for genetic identification. The 16S rRNA gene PCR with the primer set Hp1 and Hp2 was reported to be nonspecific and cannot be used to detect H. pylori in clinical specimens (2). However, in general, the sensitivity and specificity of presently available PCR methods ranged from 92 to 100% and 69 to 100%, respectively (8, 17, 18, 26, 27). Among them, PCR amplifying glmM, which was formerly called ureC and encodes phosphoglucosamine mutase (3), was reported to be the most sensitive method (17, 21).
Therefore, it is clear that PCR methods targeting a stable gene such as 16S rRNA (6) or rpoB (12) would give more reliable results. The rpoB, a kind of housekeeping gene, was newly used to detect identify H. pylori in this study. rpoB encodes the RNA polymerase β subunit and is related to rifampin resistance. The targeted region of rpoB is highly conserved in gram-positive and -negative bacteria (20). According to the sequence analysis of 162 Korean strains, the sequence dissimilarity among the H. pylori isolates was less than 8.5%. If H. pylori is a homogeneous population, the intraspecies variation of the rpoB DNA sequence would be much higher than those observed in Mycobacterium (12) and Borrelia (15). It is not surprising that H. pylori rpoB shows such high variation, because its genetic diversity is well known. However, the rpoB sequences of five Helicobacter species differed markedly from each other. The similarity of the nucleotide sequences of H. pylori and of H. cinaedi was only 68.3%, which is the lowest. Therefore, specific primers for rpoB PCR and the restriction site for H. pylori could be selected.
The results of PRA for the 162 H. pylori strains and 123 biopsy specimens suggest that it can be an H. pylori-specific method. PRA has frequently been used for molecular typing of H. pylori strains in several studies (4, 5, 7, 16, 22, 23). It could specifically amplify the DNA fragments of the urease-encoding genes. However, although restriction fragment length polymorphism analysis was efficiently used to type the H. pylori strains, it could not be applied to identify H. pylori, due to the marked complexity. The complexity, which may have originated from the size and sequence diversity of the target DNA, may be helpful for distinguishing the genotype of the H. pylori strains identified but not for the actual identification. That is why rpoB PRA is useful. Compared to other methods, rpoB PRA uses small DNA fragments, which are highly conserved. There are two or three Tru9I restriction sites on the amplified rpoB DNA of Helicobacter species. According to these results, all of the H. pylori isolates tested in this study had three identical restriction sites. Therefore, they had a simple and unique restriction pattern, which has the advantage of being able to be used for the specific identification of H. pylori. However, though only one type strain was analyzed, H. felis, which is a cat-associated species and rarely infects humans, showed the same restriction pattern.
A histological examination is known to have high sensitivity and specificity for detecting H. pylori (19). In this study, small numbers (n = 53) of gastric biopsy tissues were included. The concordance rate between the histological examination and the CLO test was similar to the concordance rate between the rpoB PRA and the glmM PCR. The concordance rate between the CLO test and the rpoB PRA was 83.0%. It is interesting that the positive rate of the rpoB PRA was higher in the gastritis and gastric cancer samples than in the duodenal and benign gastric ulcer samples. The rpoB PRA had an H. pylori detection rate similar to those of the other tests. Therefore, rpoB PRA may be useful in detecting and identifying H. pylori from gastric biopsy specimens without a culture in a clinical laboratory.
PRA may be considered an expensive, laborious, and thus impractical procedure for many samples in clinical laboratory settings. Thus, the rpoB PCR can be used alone without subsequent restriction analysis, because it is Helicobacter specific and because Helicobacter species other than H. pylori can be hardly detected in human samples. In addition, the cost, which is higher than those of other methods, including culture, will be reduced.
Acknowledgments
C.-Y. Lim and K.-H. Lee made equal contributions.
We thank D. E. Berg, Y.-G. Choi, and J.-S. Yum for providing Helicobacter strains.
This study was supported by Grants for Joint Research Projects of Korea Research Foundation (F00033, 1999) and in part by the BK21 project for Medicine, Dentistry, and Pharmacy.
REFERENCES
- 1.Boor, K. J., M. L. Duncan, and C. W. Price. 1995. Genetic and transcriptional organization of the region encoding the β subunit of Bacillus subtilis RNA polymerase. J. Biol. Chem. 270:20329-20336. [DOI] [PubMed] [Google Scholar]
- 2.Chong, S. K. F., Q. Lou, J. F. Fitzgerald, and C.-H. Lee. 1996. Evaluation of 16S rRNA gene PCR with primers Hp1 and Hp2 for detection of Helicobacter pylori. J. Clin. Microbiol. 34:2728-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Reuse, H., A. Labigne, and D. Mengin-Lecreulx. 1997. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J. Bacteriol. 179:3488-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donati, M., E. Storni, L. D'Apote, S. Moreno, A. Tucci, L. Poli, and R. Cevenini. 1999. PCR-based restriction pattern typing of the vacA gene provides evidence for a homogeneous group among Helicobacter pylori strains associated with peptic ulcer disease. J. Clin. Microbiol. 37:912-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimoto, S., B. Marshall, and M. J. Blaser. 1994. PCR-based restriction fragment length polymorphism typing of Helicobacter pylori. J. Clin. Microbiol. 32:331-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Germani, Y., C. Dauga, P. Duval, M. Huerre, M. Levy, G. Pialoux, P. Sansonetti, and P. A. Grimont. 1997. Strategy for the detection of Helicobacter species by amplification of 16S rRNA genes and identification of H. felis in a human gastric biopsy. Res. Microbiol. 148:315-326. [DOI] [PubMed] [Google Scholar]
- 7.Gibson, J. R., E. Slater, J. Xerry, D. S. Tompkins, and R. J. Owen. 1998. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J. Clin. Microbiol. 36:2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammar, M., T. Tyszkiewicz, T. Wadstrom, and P. W. O'Toole. 1992. Rapid detection of Helicobacter pylori in gastric biopsy material by polymerase chain reaction. J. Clin. Microbiol. 30:54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansson, L. E., O. Nyren, A. W. Hsing, R. Bergstrom, S. Josefsson, W. H. Chow, J. F. Fraumeni, Jr., and H. O. Adami. 1996. The risk of stomach cancer in patients with gastric or duodenal ulcer disease. N. Engl. J. Med. 335:242-249. [DOI] [PubMed] [Google Scholar]
- 10.Ho, S. A., J. A. Hoyle, F. A. Lewis, A. D. Secker, D. Cross, N. P. Mapstone, M. F. Dixon, J. I. Wyatt, D. S. Tompkins, G. R. Taylor, and P. Quirke. 1991. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J. Clin. Microbiol. 29:2543-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda, S., T. Fujioka, M. Tokieda, R. Satoh, A. Nishizono, and M. Nasu. 1998. Development of Helicobacter pylori-induced gastric carcinoma in Mongolian gerbils. Cancer Res. 58:4255-4259. [PubMed] [Google Scholar]
- 12.Kim, B.-J., S.-H. Lee, M.-A. Lyu, S.-J. Kim, G.-H. Bai, G.-T. Chae, E.-C. Kim, C.-Y. Cha, and Y.-H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 37:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lage, A. P., E. Godfroid, A. Fauconnier, A. Burette, J.-P. Butzler, A. Bollen, and Y. Glupczynski. 1995. Diagnosis of Helicobacter pylori infection by PCR: comparison with other invasive techniques and detection of cagA gene in gastric biopsy specimens. J. Clin. Microbiol. 33:2752-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laine, L., D. Lewin, W. Naritoku, R. Estrada, and H. Cohen. 1996. Prospective comparison of commercially available rapid urease tests for the diagnosis of Helicobacter pylori. Gastrointest. Endosc. 44:523-526. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S.-H., B.-J. Kim, J.-H. Kim, K.-H. Park, S.-J. Kim, and Y.-H. Kook. 2000. Differentiation of Borrelia burgdorferi sensu lato on the basis of RNA polymerase gene (rpoB) sequences. J. Clin. Microbiol. 38:2557-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, C., T. Ha, D. S. Chi, Jr., D. A. Ferguson, C. Jiang, J. J. Laffan, and E. Thomas. 1997. Differentiation of Helicobacter pylori strains directly from gastric biopsy specimens by PCR-based restriction fragment length polymorphism analysis without culture. J. Clin. Microbiol. 35:3021-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, J. J., C. L. Perng, R. Y. Shyu, C. H. Chen, Q. Lou, S. K. F. Chong, and C. H. Lee. 1999. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. J. Clin. Microbiol. 37:772-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mapstone, N. P., D. A. Lynch, F. A. Lewis, A. T. R. Axon, D. S. Tompkins, M. F. Dixon, and P. Quirke. 1993. Identification of Helicobacter pylori DNA in the mouths and stomachs of patients with gastritis using PCR. Clin. Pathol. 46:540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Megraud, F. 1997. How should Helicobacter pylori infection be diagnosed? Gastroenterology 113:S93-S98. [DOI] [PubMed] [Google Scholar]
- 20.Mollet, C., M. Drancourt, and D. Raoult. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005-1011. [DOI] [PubMed] [Google Scholar]
- 21.Moore, R. A., A. Kureishi, S. Wong, and L. E. Bryan. 1993. Categorization of clinical isolates of Helicobacter pylori on the basis of restriction digest analyses of polymerase chain reaction-amplified ureC genes. J. Clin. Microbiol. 31:1334-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen, R. J., J. Bickley, A. Hurtado, A. Fraser, and R. E. Pounder. 1994. Comparison of PCR-based restriction length polymorphism analysis of urease genes with rRNA gene profiling for monitoring Helicobacter pylori infections in patients on triple therapy. J. Clin. Microbiol. 32:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shortridge, V. D., G. G. Stone, R. K. Flamm, J. Beyer, J. Versalovic, D. W. Graham, and S. K. Tanaka. 1997. Molecular typing of Helicobacter pylori isolates from a multicenter U.S. clinical trial by ureC restriction fragment length polymorphism. J. Clin. Microbiol. 35:471-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strobel, S., S. Bereswill, P. Balig, P. Allgaier, H. G. Sonntag, and M. Kist. 1998. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J. Clin. Microbiol. 36:1285-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomb, J. F., O. White, A. R. Kerlavage, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 26.Wang, J. T., J. T. Lin, J. C. Sheu, J. C. Yang, D. S. Chen, and T. H. Wang. 1993. Detection of Helicobacter pylori in gastric biopsy tissue by polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 12:367-371. [DOI] [PubMed] [Google Scholar]
- 27.Westblom, T. U., S. Phadnis, P. Yang, and S. J. Czinn. 1993. Diagnosis of Helicobacter pylori infection by means of a polymerase chain reaction assay for gastric juice aspirates. Clin. Infect. Dis. 16:367-371. [DOI] [PubMed] [Google Scholar]