Abstract
The prevalence of the Mycobacterium bovis subsp. caprae and M. bovis subsp. bovis among German tuberculosis cases caused by the bovine tubercle bacillus from 1999 to 2001 was determined. Isolates from 166 patients living in Germany and 10 animals were analyzed by conventional laboratory procedures, spoligotyping, and partly by PCR-restriction fragment length polymorphism analysis of the gyrB gene. By spoligotyping, 55 of 176 isolates (31%) could be identified as M. bovis subsp. caprae, and 121 (69%) were confirmed as M. bovis subsp. bovis. In general, a low variability of spoligotypes with 59 distinct patterns and a cluster rate of 77% (136 isolates/19 clusters) was determined. About half of all isolates were grouped in the three main clusters with 29, 30, and 35 isolates, respectively. Differences in age and gender between the patient groups infected with M. bovis subsp. bovis and M. bovis subsp. caprae did not reach statistical significance. However, marked differences in the geographical prevalence of M. bovis subsp. caprae were observed, ranging from fewer than 10% of all M. bovis isolates in the north up to more than 80% of isolates in the south of Germany. In conclusion, M. bovis subsp. caprae accounts for a high ratio of human M. bovis-associated tuberculosis cases in Germany and was more frequently found in the southern part.
Mycobacterium bovis is a member of the M. tuberculosis complex (MTBC) that comprises the closely related species M. tuberculosis, M. bovis, M. africanum, M. microti, and M. canetii (30, 31). These species are the causative agents of tuberculosis (TB) in humans and animals. The near relatedness has been confirmed by DNA-DNA hybridization, by multilocus enzyme electrophoresis, and by sequencing of the 16S ribosomal DNA gene and the 16S-to-23S ribosomal DNA internal transcribed spacer (12, 13, 15, 18, 20, 31).
Despite this close relationship, the species of the MTBC show a large variability in their phenotypic properties, epidemiology, and importance for human TB. In contrast to M. tuberculosis and M. africanum with their natural habitat in humans, M. bovis has a broad host range and can cause TB in a wide range of domestic and wild animals and also in humans (24, 31). Cases have been observed in a great variety of mammalian species, e.g., in cattle, goat, horse, cat, dog, deer, ferret, antelope, or llama. The epidemiology of M. bovis TB is very complex and includes transmission within and between domestic and wildlife animals, as well as from animals to humans or vice versa (24). Although the clinical picture of pulmonary TB caused by M. bovis is identical to TB due to M. tuberculosis with respect to clinical, radiological, and pathological findings, direct human-to-human transmission was confirmed only in rare cases mainly among immunocompromised patients (4, 24). Contaminated food (especially milk) or direct contact with animals is considered to be the primary way M. bovis causes infection in humans (11).
The importance of M. bovis for human TB disease has dramatically decreased after the introduction of effective control measures around the mid of the last century in many developed countries (24). After the Second World War, up to 90% of cattle herds in Germany were TB positive and M. bovis accounted for 10 to 30% of human TB cases (10, 11, 19). After implementation of an efficient bovine TB control program, including tuberculin skin testing of cattle, bovine TB has been brought under control, and in 1997 Germany was officially approved as being free from bovine TB, meaning that 99.99% of cattle herds had been free of TB for the preceding 10 years (10). Accordingly, the prevalence of M. bovis among human TB cases declined to ca. 1%, a figure more likely representing the reactivation of old infections than recently acquired infections (8, 10).
Nevertheless, bovine TB continues to present a serious problem in some countries of the European Union with herd prevalence rates of up to 8.2%, and M. bovis also plays a significant role for bovine and human TB in other regions of the world, e.g., South America (2, 11, 33). The prevalence of M. bovis in developing countries is mainly unknown due to difficulties in differentiation, and thus its impact on the resurgence of the worldwide TB epidemic is unclear (11). The influence of high rates of human immunodeficiency virus and AIDS in several developing countries on M. bovis TB in humans in these regions is also an issue of considerable interest that has not yet been addressed. Since the control and prevention steps, as well as the treatment regimes, are different for M. bovis TB, these facts underline the importance of future studies analyzing the prevalence and spread of M. bovis in different regions of the world.
Routine identification of MTBC isolates can easily be performed by commercially available gene probes (ACCUProbe; GenProbe, San Diego, Calif.), whereas differentiation of M. tuberculosis and M. bovis is generally carried out by a number of biochemical tests (31). For example, M. bovis shows dysgonic growth and is negative for nitrate reduction and niacin accumulation, whereas M. tuberculosis can be identified, e.g., by its special colony morphology (eugonic), by positive test results for nitrate reduction, and by niacin accumulation (31). As a further criterion for the differentiation of M. bovis, intrinsic resistance to pyrazinamide (PZA) has been described (29, 31). However, PZA-susceptible strains of M. bovis that were characterized by specific molecular features were more recently found in Spain and Germany (3, 14, 20, 21). As a consequence, M. bovis was split into two subspecies: M. bovis subsp. bovis, which is resistant to PZA, and M. bovis subsp. caprae, which is sensitive to PZA (22). For a long time, M. bovis subsp. bovis with resistance to PZA has been considered to be the main cause of bovine TB, and PZA-susceptible strains were regarded as exceptional isolates (5, 32). However, there are no recent studies that focused on the actual contribution of both subspecies to human or bovine TB, which might have implications for both epidemiological and public health purposes.
We therefore performed this molecular epidemiological investigation of M. bovis isolates obtained from humans and some animals during a 3-year study period to gain precise data on the recent importance of M. bovis subsp. bovis and M. bovis subsp. caprae for TB due to M. bovis in Germany. Additional patient and demographic data were analyzed to define characters that are associated with TB infection with M. bovis subsp. bovis or M. bovis subsp. caprae.
MATERIALS AND METHODS
M. bovis isolates.
All available M. bovis isolates sent to the National Reference Center for Mycobacteria between 1 January 1999 and 31 December 2001 were included. Of these 176 M. bovis isolates, 166 were obtained from patients living in Germany at the time of strain isolation, and 10 were obtained from animals, including cattle (n = 8), antelope (n = 1), and pig (n = 1).
Primary isolation and culturing of the M. bovis isolates were performed as described by Kent and Kubica (17). For identification as MTBC strains, gene probes (ACCUProbe) were used. A subset of the isolates was part of our previous study (22).
Biochemical procedures and drug susceptibility testing.
All isolates were identified as M. bovis by their biochemical properties (31). Biochemical tests included colony morphology, nitrate reduction on modified Dubos broth, niacin accumulation test, growth in presence of thiophen-2-carboxylic acid hydrazide (2 μg/ml), and growth characteristics on Lebek medium and on bromcresol purple medium as described previously (1, 20, 31). Drug susceptibility testing (DST) was performed by using the proportion method on Löwenstein-Jensen medium (1) and/or the modified proportion method in BACTEC 460TB (Becton Dickinson Microbiology Systems, Cockeysville, Md.) according to the manufacturer's instructions.
Spoligotyping.
Spoligotyping was performed as described previously by Kamerbeek et al. (16). With the BioNumerics software (Windows NT, version 2.5; Applied Maths, Kortrijk, Belgium) a cluster analysis of the spoligotype patterns was performed (22). The categorical coefficient was used to calculate the similarity of spoligotype patterns, and the UPGMA (unweighted pair-group method with arithmetic averages) method was applied to calculate a dendrogram. Clusters of isolates were defined as two or more M. bovis strains with identical spoligotypes. M. tuberculosis H37Rv (ATCC 27294) and M. bovis BCG (ATCC 27289) were included as reference strains in each spoligotype experiment.
PCR-restriction fragment length polymorphism (RFLP) analysis of the gyrB DNA polymorphisms.
The DNA polymorphisms in the 1,020-bp gyrB fragment amplified with the primer pair MTUB-f and MTUB-r were analyzed by restriction with RsaI, SacII, and TaqI according to the manufacturer's instructions (New England BioLabs, Schwalbach, Germany) and as described previously (21). The analysis was performed for all strains classified as M. bovis subsp. caprae by spoligotyping and for 10 randomly chosen M. bovis subsp. bovis strains. M. tuberculosis H37Rv (ATCC 27294) and M. bovis BCG (ATCC 27289) were included as reference strains in each experiment.
Statistical analysis.
Statistical analyses were performed by using the StatView software (SAS Institute, Inc.). Differences in the means of continuous variables were calculated by using the Mann-Whitney test. Associations between proportions were assessed by using the χ2 test. Logistic regression analyses were performed to determine differences between groups; odds ratios with 95% confidence intervals were calculated. P values of <0.05 were considered statistically significant.
A geographical classification of the patient's inner-German origin in 10 regions was based on the first digit of the five-number postal code of either the patient's or the hospital's address.
RESULTS
Spoligotyping and gyrB PCR-RFLP.
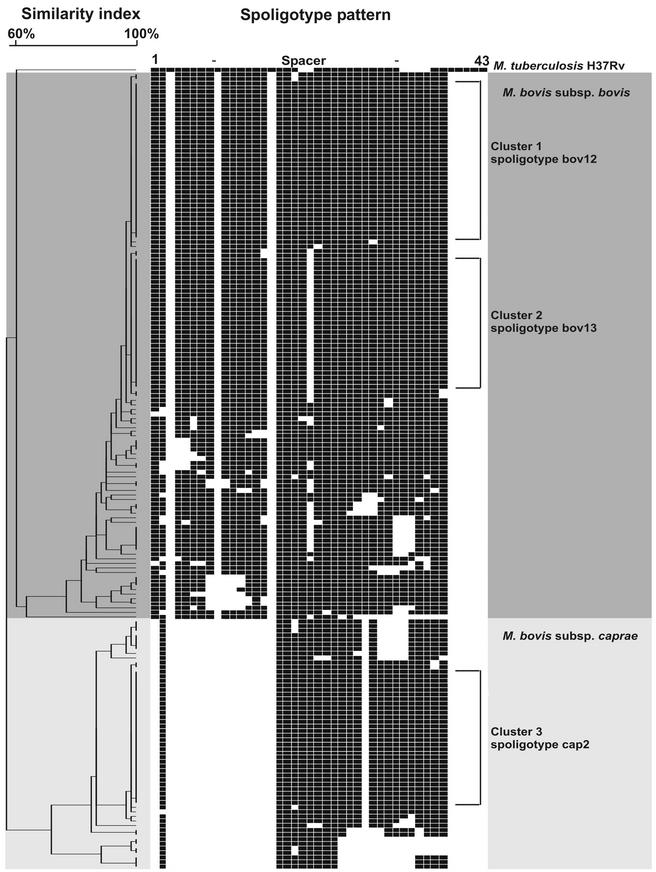
In order to differentiate between the subspecies bovis and caprae and to analyze the genetic relatedness of the M. bovis strains, spoligotyping was performed. All strains showed spoligotype patterns with the lack of spacers 39 to 43 that have been reported to be typical for M. bovis (16). A total of 55 strains (31%) were identified as M. bovis subsp. caprae by the additional absence of spacers 1 and 3 to 16, based on the description of Niemann et al. (22), and 121 strains (69%) were identified as M. bovis subsp. bovis. All M. bovis subsp. bovis strains showed no hybridization to spacers 3, 9, and 16 (Fig. 1 and 2). The 10 animal isolates were split in five M. bovis subsp. bovis isolates (obtained from cattle, antelope, and pig) and five M. bovis subsp. caprae isolates (all obtained from cattle).
FIG. 1.
Dendrogram showing the relationship of the spoligotype patterns of 121 M. bovis subsp. bovis (darker shade of gray) and 55 M. bovis subsp. caprae (lighter shade of gray) strains from Germany. The type strain M. tuberculosis H37Rv (ATCC 27294) is included as reference strain. The three main clusters—bov12, bov13, and cap2—are indicated. Banding patterns are ordered by similarity in the dendrogram, and the scale depicts the degree of similarity calculated with the categorical coefficient and the UPGMA method.
FIG. 2.
Dendrogram showing the relationship of the 59 representative spoligotype patterns (M. bovis subsp. bovis [darker shade of gray]; M. bovis subsp. caprae [lighter shade of gray]) obtained for 176 M. bovis strains from Germany. Banding patterns are ordered by similarity in the dendrogram, and the scale depicts the degree of similarity calculated with the categorical coefficient and the UPGMA method.
The similarity analysis of the 176 spoligotype patterns resulted in 59 distinct patterns (Fig. 1). The 59 representative patterns were shown in detail in Fig. 2. Among the strains analyzed, 40 showed a unique pattern, whereas 136 (77%) were grouped in 19 clusters, varying in size from 2 to 35 isolates. The low variability of spoligotype patterns and the presence of predominant spoligotypes in the study population were further underlined by the fact that 52% (n = 92) of all the strains were grouped into three large clusters (Fig. 1). Considering M. bovis subsp. caprae, 48 (87%) of the 55 isolates were grouped in eight clusters and the largest cluster (spoligotype cap2) included 30 isolates (54%). Among the 121 M. bovis subsp. bovis strains, 88 (73%) were clustered, and the two largest clusters (spoligotypes bov12, and bov13) represent 64 (52%) of all M. bovis subsp. bovis isolates (bov12, 35 isolates; bov13, 29 isolates). Spoligotype bov12 is identical to M. bovis BCG, and bov13 differed by the absence of just one spacer from this pattern.
If the two major M. bovis subsp. bovis clusters were excluded from the similarity analysis, spoligotyping showed a higher discriminatory power since only 24 (42%) of the remaining 57 isolates were clustered. Exclusion of the largest M. bovis subsp. caprae cluster does not result in a comparable improvement of strain differentiation, since 18 (72%) of 25 isolates were still in clusters. The 10 animal isolates were not separated from the human isolates in the dendrogram and were found both, within the clusters (inclusive the three large ones) and among the distinct isolates.
The gyrB PCR-RFLP analysis was used as an additional method to confirm the strain classification based on the spoligotypes. All 10 randomly chosen M. bovis subsp. bovis isolates showed the gyrB PCR-RFLP banding pattern typical for M. bovis subsp. bovis. For 52 of the M. bovis subsp. caprae isolates (95%), the additional mutation in the gyrB gene could be confirmed by restriction analysis, but 3 isolates showed a gyrB PCR-RFLP banding pattern identical to that of M. bovis subsp. bovis isolates (data not shown).
Phenotypic characteristics.
The biochemical properties tested (as listed in Materials and Methods) did not differ for subspecies bovis and caprae isolates (data not shown). The only variable phenotypic characteristic identified was resistance to PZA. In accordance with previous observations (3, 20), all strains classified as subsp. caprae were susceptible to PZA, whereas subsp. bovis strains mostly showed resistance to PZA. However, three M. bovis subsp. bovis isolates proved to be sensitive to PZA in repeated DST, and for two M. bovis subsp. bovis isolates DST was not available. All strains were susceptible to all other first-line drugs.
Statistical analysis of epidemiological and demographic data, and clinical presentation.
For further classification of the 166 patients infected with M. bovis strains, a statistical analysis of age, gender, localization of the disease, and inner German origin was performed. The male/female ratio in the study population was 1.2:1. The age of the patients ranged from 1 to 94 years, with a mean age of 64 years (data were not available for 16 patients, Table 1). The majority of patients were 60 years and older, but a marked number of cases were observed in patients younger than 40 years (n = 13, 9%). The age distribution differed significantly between men and women (mean age 63 years versus 67 years; P = 0.03). The disease was classified as respiratory in 51% (85 of 166), nonrespiratory in 34% (57 of 166), and unknown in 15% (24 of 166) of the patients. In detail, the nonrespiratory manifestations included lymph nodes in 9 of 166 cases, bones or joints in 6 of 166 cases, the genitourinary tract in 20 of 166 cases, and the central nervous system in 4 of 166 cases. Furthermore, abdominal and cutaneous manifestations, as well as abscesses and fistulas, were found. The animals had either pulmonary (n = 2) or lymph node TB (n = 6); only the antelope had abdominal TB isolated from the liver. For one cow the localization was unknown.
TABLE 1.
Age distribution of patients infected with M. bovis subsp. caprae and M. bovis subsp. bovis strainsa
| Patient age (yr) | Total (n = 150)
|
M. bovis subsp. bovis (n = 104)
|
M. bovis subsp. caprae (n = 46)
|
|||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| 0-10 | 2 | 1.3 | 1 | 1 | 1 | 2.2 |
| 11-20 | 0 | 0 | 0 | |||
| 21-30 | 7 | 4.7 | 6 | 5.8 | 1 | 2.2 |
| 31-40 | 4 | 2.7 | 3 | 2.9 | 1 | 2.2 |
| 41-50 | 11 | 7.3 | 11 | 10.6 | 0 | |
| 51-60 | 15 | 10 | 12 | 11.5 | 3 | 6.6 |
| 61-70 | 49 | 32.7 | 30 | 28.9 | 19 | 41.3 |
| 71-80 | 38 | 25.3 | 22 | 21.2 | 16 | 34.8 |
| 81-90 | 21 | 14 | 16 | 15.6 | 5 | 10.9 |
| 91-100 | 3 | 2 | 3 | 2.9 | 0 | |
Data were presented for 150 M. bovis strains. For 16 of the 166 patients, data were not available. The overall mean ages for each group were as follows: total, 64.5 years; M. bovis subsp. bovis, 63.8 years; and M. bovis subsp. caprae, 66.1 years.
In order to identify significant differences between patients infected with M. bovis subsp. bovis and M. bovis subsp. caprae strains, univariate analyses were performed. The difference in age between the patients infected with M. bovis subsp. bovis strains and M. bovis subsp. caprae strains (mean age of 63 years versus 66 years), as well as male/female ratios of 1.05:1 in the M. bovis subsp. bovis subgroup and of 1.6:1 in the M. bovis subsp. caprae subgroup, did not reach statistical significance. There was also no significant difference in the localization of TB between the two patient groups, except for the fact that lymph node TB was not present in patients infected with M. bovis subsp. caprae.
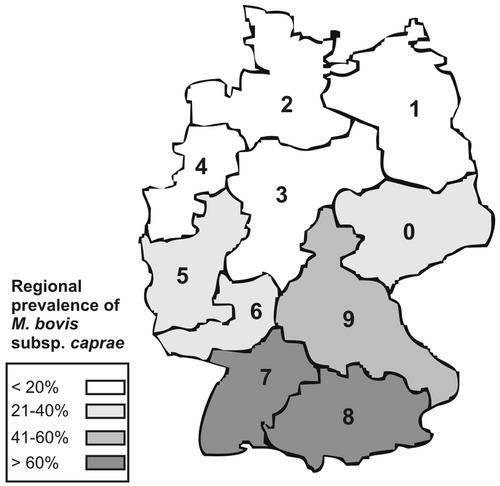
The analysis of the inner-German geographical distribution could be performed for 137 of the 166 patients (83%). The animals were regarded separately. Ten regions were distinguished based on the first digit of the five-digit postal code. The total number of patients belonging to each geographical region and the prevalence of the two subspecies are shown in Table 2. The regional percentage of the M. bovis subsp. caprae strains is shown in detail in Fig. 3. The spatial analysis revealed a marked difference in the regional prevalence of M. bovis subsp. caprae-subsp. bovis strains. In the northern regions 1, 2, 3, and 4, M. bovis subsp. caprae accounts for <20% of all M. bovis cases, whereas in the southern regions 7 and 8 >60% of all cases were M. bovis subsp. caprae. This difference is statistically significant (P < 0.001). Accordingly, ca. 45% of all the M. bovis subsp. caprae isolates were from the southern part of Germany (Table 2). Considering the animal isolates, all five M. bovis subsp. caprae strains were from the southern part. Of the M. bovis subsp. bovis strains, three were isolated from the southern parts of Germany and two were isolated from the northern parts of Germany.
TABLE 2.
Spatial analysis of the inner-German distribution for all human M. bovis isolates and for M. bovis subsp. bovis and subsp. capraea
| Region | No. (% of all isolates) | No. of isolates of M. bovis subsp.:
|
Regional proportion (%) of all isolates of M. bovis subsp.:
|
Proportion (%) within each region of M. bovis subsp.:
|
|||
|---|---|---|---|---|---|---|---|
| caprae | bovis | caprae | bovis | caprae | bovis | ||
| 0 | 11 (8) | 4 | 7 | 9.8 | 7.3 | 36 | 64 |
| 1 | 13 (9.5) | 1 | 12 | 2.4 | 12.5 | 8 | 92 |
| 2 | 15 (11) | 2 | 13 | 4.9 | 13.5 | 13 | 87 |
| 3 | 26 (19) | 3 | 23 | 7.3 | 24 | 12 | 88 |
| 4 | 18 (13) | 1 | 17 | 2.4 | 17.7 | 6 | 94 |
| 5 | 14 (10.2) | 5 | 9 | 12.2 | 9.4 | 36 | 64 |
| 6 | 10 (7.3) | 3 | 7 | 7.3 | 7.3 | 30 | 70 |
| 7 | 12 (8.8) | 10 | 2 | 24.4 | 2.1 | 83 | 17 |
| 8 | 12 (8.8) | 9 | 3 | 22 | 3.1 | 75 | 25 |
| 9 | 6 (4.4) | 3 | 3 | 7.3 | 3.1 | 50 | 50 |
| Total | 137 (100) | 41 | 96 | 100 | 100 | 30 | 70 |
The regional classification is based on the first digit of the postal code of the patient's origin. Of the 166 M. bovis isolates, 137 were analyzed, and for 29 isolates data were not available.
FIG. 3.
Spatial analysis of the inner-German origin of the M. bovis subsp. caprae strains isolated from humans. The different shades of gray indicate the regional prevalence of M. bovis subsp. caprae strains of all M. bovis isolates from humans.
DISCUSSION
In recent years, a variety of studies have focused on the molecular relationship of MTBC strains; however, epidemiological studies addressing human M. bovis-associated TB are still rare. In the present study, we present a systematic analysis of the population structure of mostly human-origin M. bovis strains isolated between 1999 and 2001 in Germany.
The recently described M. bovis subsp. caprae, being susceptible to PZA (22), was found as causative agent of one-third (31%) of the human M. bovis-associated TB cases analyzed. This proportion is surprisingly high, especially if the prevalence of M. bovis subsp. caprae strains in human or animal isolates in other countries is considered: a study on M. bovis TB in France revealed no M. bovis subsp. caprae strains among more than 1,000 animal isolates (9). M. bovis subsp. caprae strains were also not reported from animal and human isolates in the United Kingdom (27) or from animal isolates from Northern Ireland (26) and Ireland (6), South America (33), and Cameroon (23). Outside Germany, only comparably small numbers of M. bovis subsp. caprae strains have been identified in Spain (3.6% of M. bovis isolates from humans and 12% of isolates from goats and sheep [14]) and in Austria (12 cases in humans and animals in 7 years [25]). It might be assumed that M. bovis subsp. caprae represents a newly emerging genotype in Germany and is now spreading to other European countries. However, the comparably high mean age of the patients in the present study irrespective of the subspecies clearly indicates that the majority of M. bovis cases probably are due to reactivation rather than recently acquired infection. It is therefore likely that the patients have been infected before the effective control measures for bovine TB were introduced in the 1950s. As a consequence, M. bovis subsp. caprae must have been present in Germany at that time and is therefore not only recently emerging. Prior to the introduction of molecular tools for the identification and differentiation of M. bovis strains, M. bovis subsp. caprae isolates might have been misclassified due to their susceptibility to PZA, resulting in false low notification rates.
Susceptibility to PZA, however, was also observed in three M. bovis subsp. bovis strains that were obtained from two patients and one cow. These isolates showed no particular spoligotype patterns and were not related in the similarity analysis. From a phylogenetic point of view, these strains may represent ancestral M. bovis strains, from which both subspecies might have diverged. Further investigations of these strains are in progress.
Considering the characteristics of patients infected with M. bovis subsp. bovis and caprae strains, no significant differences in age, gender, and localization of the disease could be found. The absence of M. bovis subsp. caprae strains in human lymph node tissue is most likely coincidental, since M. bovis subsp. caprae strains were isolated from lymph node tissues of the animals.
The only marked difference between the two patient groups was revealed in the spatial analysis of the patients inner-German origin: the regional proportion of M. bovis subsp. caprae showed a large difference between southern and northern parts of the country, as high proportions of up to more than 80% were found in the southern parts of Germany compared to less than 10% in the northern parts of the country. This observed geographic shift in the regional proportion of both subspecies might have resulted from a similar shift in the animal population, as indicated by our finding that animals infected with M. bovis subsp. caprae strains were mainly from southern Germany. This is further supported by the presence of M. bovis subsp. caprae strains in wild and livestock animals in western Austria in a region located at the southern German border (25).
In general, the spoligotypes displayed by M. bovis isolates from Germany showed a low variability, since only 23% of all isolates had unique spoligotype patterns, whereas 77% of all isolates were clustered. However, the discriminatory power of spoligotyping is highly influenced by the presence of prominent genotypes in the study population since the three major spoligotypes identified represent ca. 50% of all isolates. Exclusion of these isolates leads to a clear reduction of the cluster rate.
The presence of prominent genotypes within M. bovis populations was also observed in other studies: the most predominant spoligotype in Germany, bov12 (20%), with the BCG-like pattern was also found frequently in France and Spain, where this spoligotype was either the most frequent (BCG-like, 26%) or the second most frequent (spb-3, 7.7%) type observed. In Italy, this spoligotype was determined in 60 and 73% of all M. bovis isolates from wild boars and cattle, respectively (28). Our second most frequent spoligotype among the M. bovis subsp. bovis strains, bov13, was also the most frequent type isolated in Spain (spb-7, 46%), and the second most frequent type isolated in France (GB54, 12%).
However, the variety of genotypes obtained differs widely in other regions of the world. The BCG-like spoligotype was found neither in the United Kingdom nor in Ireland or Northern Ireland (6, 26, 27). Furthermore, it was not present in Cameroon (23), Australia and Canada (7), and only scarcely in South America (33). The most prominent spoligotype from Australia (Sp01, 56%) is also the most prominent one in Ireland (A1, 52%), and in Northern Ireland (ST1 + ST2, 80.7%), whereas these strains are not regularly found in the other European countries. Accordingly, the spoligotype ST2 is found only once in our analysis (bov40) in a patient originally being born in Great Britain. A special clonal population of M. bovis is also described for Cameroon, where all isolates showed the absence of spacer 30 (23) that is not regularly found in other countries. Our analysis revealed only one spoligotype pattern (bov14) matching the most prominent Cameroon spoligotype C1, although no epidemiological linkage between the patient infected with this strain and Africa could be found. These findings indicate that western European countries such as Germany, France, and Spain seem to have a similar M. bovis population structure, whereas different genotypes were found in other parts of the world, e.g., Great Britain and Australia. The knowledge of a prominent genotype in a certain region is therefore of outstanding importance for the interpretation of molecular typing results of M. bovis TB.
The detection of single nucleotide polymorphisms in the gyrB gene by PCR-RFLP has been described as a rapid and accurate tool for differentiation of the MTBC (21). However, in the present study 3 of 55 (5.5%) M. bovis subsp. caprae isolates did not show the characteristic T-to-G mutation at position 1311 in the gyrB gene. Nonetheless, all M. bovis subsp. caprae and M. bovis subsp. bovis isolates showed the M. bovis specific mutation at position 756, and correct species identification can easily be achieved. In the case of discrepant results between drug susceptibility to PZA and gyrB polymorphism at position 1311, additional molecular techniques, such as spoligotyping, might be applied.
In conclusion, M. bovis subsp. caprae strains were confirmed to cause a high ratio of human M. bovis-associated cases in Germany. Thus, susceptibility to PZA among M. bovis isolates cannot longer be regarded as an exceptional finding and cannot be taken as characteristic for identification of M. bovis. Both subspecies did not differ in other biochemical characteristics and clinical presentation, but M. bovis subsp. caprae was more frequently found in the southern parts of Germany.
Acknowledgments
We thank K. Ott, B. Schlüter, I. Radzio, T. Ubben, and P. Vock (Borstel, Germany) for excellent technical assistance.
Parts of this work were supported by the Robert Koch Institute, Berlin, Germany, and the EU Concerted Action project “New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis” (QLK2-CT-2000-00630).
REFERENCES
- 1.Anonymous. 1986. Medical microbiology: diagnosis of tuberculosis. Part 9: minimum requirements for the identification of the tubercle bacilli. DIN 58943-9. Beuth Verlag, Berlin, Germany
- 2.Anonymous. 2000. Trends and sources of zoonotic agents in animals, feedingsstuffs, food, and man in the European Union and Norway in 2000. European Commission, European Union, Brussels, Belgium. [Online.] http://europa.eu.int/comm/food/fs/sfp/mr/mr_zoo_en.html
- 3.Aranaz, A., E. Liébana, A. Mateos, L. Dominguez, D. Vidal, M. Domingo, O. Gonzolez, E. F. Rodriguez-Ferri, A. E. Bunschoten, J. D. A. van Embden, and D. Cousins. 1996. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J. Clin. Microbiol. 34:2734-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blazquez, J., L. E. Espinosa de Los Monteros, S. Samper, C. Martin, A. Guerrero, J. Cobo, J. D. A. van Embden, F. Baquero, and E. Gomez-Mampaso. 1997. Genetic characterization of multidrug-resistant Mycobacterium bovis strains from a hospital outbreak involving human immunodeficiency virus-positive patients. J. Clin. Microbiol. 35:1390-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, C. H., and M. D. Yates. 1981. A study of bovine strains of Mycobacterium tuberculosis isolated from humans in southeast England, 1977-1979. Tubercle 62:113-116. [DOI] [PubMed] [Google Scholar]
- 6.Costello, E., D. O'Grady, O. Flynn, R. O'Brien, M. Rogers, F. Quigley, J. Egan, and J. Griffin. 1999. Study of restriction fragment length polymorphism analysis and spoligotyping for epidemiological investigation of Mycobacterium bovis. J. Clin. Microbiol. 37:3217-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cousins, D., S. Williams, E. Liebana, A. Aranaz, A. Bunschoten, J. van Embden, and T. Ellis. 1998. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J. Clin. Microbiol. 36:168-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutsches Zentralkomitee zur Bekämpfung der Tuberkulose. 2002. Informationsbericht. Pmi-Verlag, Frankfurt am Main, Germany.
- 9.Haddad, N., A. Ostyn, C. Karoui, M. Masselot, M. F. Thorel, S. L. Hughes, J. Inwald, R. G. Hewinson, and B. Durand. 2000. Spoligotype diversity of Mycobacterium bovis strains isolated in France from 1979 to 2000. J. Clin. Microbiol. 39:3623-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartung, M. 2001. Bericht über die epidemiologische Situation der Zoonosen in Deutschland für 2000-Übersicht über die Meldungen der Bundesländer. RKI-Hausdruckerei, Berlin, Germany.
- 11.Fanning, E. A. 1994. Mycobacterium bovis infection in animals and humans, p. 351-364. In P. D. O. Davies (ed.), Clinical tuberculosis. Chapman & Hall Medical, London, England.
- 12.Feizabadi, M. M., I. D. Robertson, D. V. Cousins, and D. J. Hampson. 1996. Genomic analysis of Mycobacterium bovis and other members of the Mycobacterium tuberculosis complex by isoenzyme analysis and pulsed-field gel electrophoresis. J. Clin. Microbiol. 34:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frothingham, R., H. G. Hills, and K. H. Wilson. 1994. Extensive DNA sequence conservation throughout the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 32:1639-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutiérrez, M., S. Samper, M. S. Jiménez, J. D. A. van Embden, J. F. Marín, and C. Martín. 1997. Identification by spoligotyping of a caprine genotype in Mycobacterium bovis strains causing human tuberculosis. J. Clin. Microbiol. 35:3328-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imaeda, T. 1985. Deoxyribonucleotide acid relatedness among selected strains of Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium bovis BCG, Mycobacterium microti, and Mycobacterium africanum. Int. J. Syst. Bacteriol. 35:147-150. [Google Scholar]
- 16.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strains differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health and Human Services, Centers for Disease Control, Atlanta, Ga.
- 18.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Böttger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meissner, G. 1974. Bovine tuberculosis in man before and after the eradication of tuberculosis in cattle. Prax. Pneumol. 28:123-128. [PubMed] [Google Scholar]
- 20.Niemann, S., E. Richter, and S. Rüsch-Gerdes. 2000. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis. J. Clin. Microbiol. 38:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niemann, S., D. Harmsen, S. Rüsch-Gerdes, and E. Richter. 2000. Differentiation of clinical Mycobacterium tuberculosis complex strains by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemann, S., E. Richter, and S. Rüsch-Gerdes. 2002. Biochemical and genetic evidence for the transfer of Mycobacterium tuberculosis subsp. caprae Aranaz et al. 1999 to the species Mycobacterium bovis Karlson and Lessel 1970 (approved lists 1980) as Mycobacterium bovis subsp. caprae comb. nov. Int. J. Syst. Evol. Microbiol. 52:433-436. [DOI] [PubMed] [Google Scholar]
- 23.Njanpop-Lafourcade, B. M., J. Inwald, A. Ostyn, B. Durand, S. Hughes, M. F. Thorel, G. Hewinson, and N. Haddad. 2001. Molecular typing of Mycobacterium bovis isolates from Cameroon. J. Clin. Microbiol. 39:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Reilly, L. M., and C. J. Daborn. 1995. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuberc. Lung Dis. 76(Suppl. 1):1-46. [DOI] [PubMed] [Google Scholar]
- 25.Prodinger, W. M., A. Eigentler, F. Allerberger, M. Schonbauer, and W. Glawischnig. 2002. Infection of red deer, cattle, and humans with Mycobacterium bovis subsp. caprae in western Austria. J. Clin. Microbiol. 40:2270-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roring, S., M. S. Hughes, L. A. Beck, R. A. Skuce, and S. D. Neill. 1998. Rapid diagnosis and strain differentiation of Mycobacterium bovis in radiometric culture by spoligotyping. Vet. Microbiol. 61:71-80. [DOI] [PubMed] [Google Scholar]
- 27.Sales, M. P., G. M. Taylor, S. Hughes, M. Yates, G. Hewinson, D. B. Young, and R. J. Shaw. 2001. Genetic diversity among Mycobacterium bovis isolates: a preliminary study of strains from animal and human sources. J. Clin. Microbiol. 39:4558-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serraino, A., G. Marchetti, V. Sanguinetti, M. C. Rossi, R. G. Zanoni, L. Catozzi, A. Bandera, W. Dini, W. Mignone, F. Franzetti, and A. Gori. 1999. Monitoring of transmission of tuberculosis between wild boars and cattle: genotypical analysis of strains by molecular epidemiology techniques. J. Clin. Microbiol. 37:2766-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662-667. [DOI] [PubMed] [Google Scholar]
- 30.Van Soolingen, D., T. Hoogenboezem, P. E. de Haas, P. W. Hermans, M. A. Koedam, K. S. Teppema, P. J. Brennan, G. S. Besra, F. Portaels, J. Top, L. M. Schouls, and J. D. A. van Embden. 1997. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol. 47:1236-1245. [DOI] [PubMed] [Google Scholar]
- 31.Wayne, L. G., and G. P. Kubica. 1986. The mycobacteria, p. 1435-1457. In P. H. A. Sneath and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams and Wilkins Co., Baltimore, Md.
- 32.Wayne, L. G., R. C. Good, M. I. Krichevsky, Z. Blacklock, H. L. David, D. Dawson, W. Gross, J. Hawkins, V. V. Levy-Frebault, C. McManus, F. Portaels, S. Rüsch-Gerdes, K. H. Schröder, V. A. Silcox, M. Tsukamura, L. van den Breen, and M. A. Yakrus. 1991. Fourth report of the cooperative, open-ended study of slowly growing mycobacteria by the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 41:463-472. [DOI] [PubMed] [Google Scholar]
- 33.Zumarraga, M. J., C. Martin, S. Samper, A. Alito, O. Latini, F. Bigi, E. Roxo, M. E. Cicuta, F. Errico, M. C. Ramos, A. Cataldi, D. van Soolingen, and M. I. Romano. 1999. Usefulness of spoligotyping in molecular epidemiology of Mycobacterium bovis-related infections in South America. J. Clin. Microbiol. 37:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]