Abstract
A novel and highly sensitive multiplex nested PCR assay has been developed for the simultaneous glycoprotein B (gB) typing of four gB genotypes of human cytomegalovirus (CMV) directly from clinical specimens. Specifically, a pair of primers to conserved regions of all gB genotypes within the gB gene (gB and gpUL55) was used for primary amplification. A mixture of nested primers to specific and conserved regions of each gB genotype was used for a secondary PCR amplification, yielding amplicons of different sizes for each gB genotype that could easily be differentiated by agarose gel electrophoresis. A total of 40 of 44 serum specimens and 26 of 26 cerebrospinal fluid (CSF) samples, which had previously tested positive for CMV in 66 of 70 AIDS patients with different CMV disease conditions, were gB genotyped by this novel assay. Significant differences in glycoprotein B genotype distribution between serum and CSF specimens were found (P = 0.001). gB genotype 3 (gB3) and gB2 were the most prevalent genotypes in sera (42.5%) and CSF (38.5%), respectively. A different distribution was also observed when only patients with CMV retinitis were studied (P = 0.005), suggesting a gB2 neuron tropism. Neither CMV disease nor any particular clinical manifestation of CMV disease was associated with gB genotypes (P > 0.05). A high incidence of mixed infection with the gB1 and gB3 genotypes (27.5%) was detected in serum specimens, indicating that reinfection and reactivation are common traits in advanced AIDS patients.
In advanced AIDS patients, disseminated cytomegalovirus (CMV) disease causes significant morbidity and mortality, despite a decrease in frequency with the introduction of highly active antiretroviral therapy. In these patients, the central nervous system is a major target for CMV (17), and virus infection frequently causes encephalitis and retinitis (12). Many clinical cases are the result of reactivation of latent infection and are linked to the immune status of the patient. However, a reactivation of latent infection after CMV reinfection is not rare and may lead to an increased chance of recombination and formation of new CMV variants.
There is evidence indicating that the existence of CMV variants plays an important role in the pathogenesis of disease. These variants affecting several genes may be responsible for the different diseases related to CMV infection. Moreover, the diversity of organs and cell types infected by CMV in vivo has led to the hypothesis that disease and tissue tropism may be associated with sequence variation among strains (6, 10, 14, 15, 24).
The most widely characterized polymorphic gene is gpUL55, which encodes a glycoprotein essential for virus penetration and cell fusion (16) and represents the major target for neutralizing antibodies (8). The interaction of this glycoprotein, named glycoprotein B (gB), with its cellular binding partner has recently been revealed to be the main mechanism by which CMV alters cellular transcription early during infection (23). There are four major gB genotypes, determined based on the region surrounding the proteolytic cleavage site (5), although additional genotypes have also been described (21).
A number of studies have attempted to correlate gB genotype with specific CMV disease manifestations (1, 4, 10, 15, 22, 27, 28) in immunocompromised patients, although no such relationship has yet been clearly established. Among AIDS patients, gB genotype distribution has been widely studied in blood, urine, semen, vitreous, and saliva specimens (1, 4, 9, 12, 18). However, the frequency distribution of gB genotypes in cerebrospinal fluid (CSF) specimens has not been studied yet. There is a high incidence of neurological infections caused by CMV in advanced AIDS patients, and for this reason it would be of great interest to determine the gB genotype involved in these cases.
To date, the gB genotypes of CMV have been determined by PCR of the gB sequences, with subsequent analysis by restriction fragment length polymorphism, single-stranded conformation polymorphism, heteroduplex mobility analysis, and direct DNA sequencing. Our study represents the first approach to genotyping CMV gB directly from clinical specimens by using multiplex nested-PCR (M-nPCR) technology. This method has proven to be a valuable tool for differentiation, subgrouping, subtyping, and genotyping of viruses (19, 25, 26).
Differences among gB genotypes may influence pathogenesis, perhaps by distinct cell tropism, and therefore the frequency distributions of gB genotypes in serum and CSF were the subject of further investigation and comparison. We also investigated whether there is any effect of gB genotype variation on the clinical outcome related to CMV disease. To achieve these objectives, we carried out a retrospective study, reported here, in which clinical samples from 70 AIDS patients with different CMV disease conditions were directly gB genotyped.
MATERIALS AND METHODS
Clinical samples and patients.
A single CSF (n = 26) or serum (n = 44) specimen from each of 70 advanced AIDS patients was retrospectively studied and used to evaluate the efficacies of M-nPCR assays. Twenty-four CSF samples were obtained from our bank collection that had previously been determined to be DNA positive for CMV by using a multiplex nested-PCR assay for human herpesviruses (26). The remaining 46 specimens (2 CSF and 44 serum samples) were obtained from the Infectious Diseases Department at Ramón y Cajal Hospital. These were previously determined to be CMV pp65 antigen assay positive and/or culture positive for CMV. All clinical samples had been stored at −70°C.
We obtained 2 CSF and 14 serum samples from patients with asymptomatic CMV infection. Seven additional sera were from patients who remained unclassified due to incomplete fulfillment of CMV disease criteria. A total of 24 CSF samples and 23 sera were obtained from patients with clinical CMV disease manifestations, including 24 CSF specimens and 11 serum specimens obtained from patients diagnosed with CMV retinitis (n = 35). A patient was diagnosed with CMV disease on the basis of the agreed criteria outlined in Ljungman et al. (13). Differences in geographical origin and demographic characteristics among patients were not significant. Patients were predominantly Caucasian Spanish males from the Madrid area for whom injected drug abuse was the mode of human immunodeficiency virus (HIV) transmission and who had a similar degree of immunosuppression induced by HIV (CD4 counts of <200 × 106 cells/liter). Clinical information was obtained by review of each subject's medical record.
Primer design and preparation.
Synthetic specific DNA oligonucleotide primers were based on previously published sequences of the CMV gB gene obtained from GenBank. All available sequences were aligned by using the MACAW 1.01 program (20), and proposed primers were subsequently tested by using the HINT PCR program (TDI, Madrid, Spain) to satisfy the general conditions of primer design. The first PCR round was designed to amplify a 751-bp segment containing the major region of sequence variability within the gB gene. Consensus primers for primary PCR amplification were designed on highly conserved regions for five gB genotypes described for the gpUL55 (i.e., gB1 to gB5). A nested PCR round was carried out with five pairs of genotype-specific consensus primers that were designed to achieve optimal performance in a multiplex reaction (Table 1). All primers were prepared by using a model 394 ABI DNA synthesizer (Perkin-Elmer). The expected band sizes were 420, 613, 190, 465, and 139 bp for CMV gB1, CMV gB2, CMV gB3, CMV gB4, and CMV gB5, respectively.
TABLE 1.
Multiplex primers for CMV gBs
| Amplification reaction and primer | Polaritya | Position (nucleotides)b | Sequence (5′→3′) |
|---|---|---|---|
| Primary | |||
| CMVQ1+ | HS5GLYBG+ | 868-885 | TTTGGAGAAAACGCCGAC |
| CMVQ1- | HS5GLYBG- | 1619-1597 | CGCGCGGCAATCGGTTTGTTGTA |
| Nestedc | |||
| CMVGT1+ | HS5GLYBG+ | 1111-1130 | ATGACCGCCACTTTCTTATC |
| CMVGT2+ | HEHCMVGB+ | 1074-1096 | TTCCGACTTTGGAAGACCCAACG |
| CMVGT3+ | HS5GLYBM+ | 1341-1359 | TAGCTCCGGTGTGAACTCC |
| CMVGT4+ | HS5GLYBD+ | 1057-1082 | ACCATTCGTTCCGAAGCCGAGGAGTCA |
| CMVGT5+ | AF043721+ | 307-325 | TACCCTATCGCTGGAGAAC |
| CMVQ2- | HS5GLYBG- | 1531-1513 | GTTGATCCACRCACCAGGC |
+, forward; −, reverse.
The numbering positions for HS5GLYBG (6), HEHCMVGB (7), HS5GLYBM (6), HS5GLYBD (6), and AF043721 (21) are as described previously.
The reverse primer was shared by all CMV gBs.
M-nPCR.
DNA from CSF samples and serum samples was extracted as described by Casas et al. (3). Briefly, 50 μl of each clinical specimen was incubated for 10 min at room temperature with 200 μl of a guanidinium thiocyanate lysis buffer. DNA was extracted by isopropanol-ethanol precipitation, and the pellet was dissolved in 10 μl of sterile double-distilled water. The primary PCR amplification and the multiple secondary PCR amplification of gB DNA were optimized by testing a wide range of Mg2+ concentrations, two different PCR buffers, two different primer concentrations, a wide range of deoxynucleoside triphosphate (dNTP) concentrations and two Taq concentrations. Several annealing temperatures, starting at 5°C below the average melting point of primers, were used. Serial dilutions of each CMV gB genotype were used to test improvement in sensitivity. A 5-μl portion of the extracted DNA was added to a PCR mixture containing 60 mM Tris (pH 8.5), 15 mM (NH4)2SO4, 3 mM MgCl2, 0.4 mM concentrations of dNTPs, 10 pmol each of CMVQ1+ and CMVQ1− primers (Table 1), and 2.5 U of Taq polymerase in a final volume of 50 μl. Amplification was performed in a PTC-200 Peltier thermal cycler (MJ Research, Watertown, Mass.) programmed for 1 denaturation step at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 30 s, and ending with an extension step at 72°C for 5 min. From this reaction, a 1-μl sample of the product was amplified in the M-nPCR in a reaction mixture containing PCR buffer (Perkin-Elmer), 1 mM MgCl2, 0.2 mM concentrations of dNTPs, 2.5 U of Taq polymerase, and an equimolar mixture of 10 pmol of each inner primer (CMVGT1+, CMVGT2+, CMVGT3+, CMVGT4+, CMVGT5+, and CMVQ2−; Table 1) in a final volume of 50 μl. The second PCR round was carried out under conditions identical to those used in the first PCR round except the annealing temperature was 58°C instead of 60°C. The M-nPCR products were analyzed by electrophoresis on 2% high-resolution agarose gels (MS-8; Hispanlab, Madrid, Spain) stained with ethidium bromide. Serial dilutions of a previously quantified CMV DNA sample from CMV AD169 strain (EMBL accession no. X17403) were used as positive controls for CMV gB2. To control for contamination in the nested step, negative controls (tubes without CMV DNA) were used. In addition, all samples were retested, yielding identical results. The way to avoid false-positive and false-negative results, as well as contamination in the different steps of nested PCR, was described in detail in a previous study carried out in our laboratory (2).
DNA sequencing and analysis of sequences.
Four samples that yielded single bands, with molecular weights corresponding to each of the putative gB genotypes, were selected for DNA sequencing. PCR products amplified by primers CMVQ1+ and CMVQ1− were purified by precipitation with 2.5 M ammonium acetate and isopropyl alcohol and subsequently washed with 70% ethanol to remove the dNTPs and excess primers. The pellet was diluted in 20 μl of RNase-free sterile water, and a 5-μl aliquot was analyzed by agarose gel electrophoresis. The DNA concentration was estimated after visualization of bands and comparison with a standard DNA marker (50-bp ladder; Gibco-BRL). For direct sequencing of purified PCR products in both directions (forward and reverse), 2 μl of purified DNA (20 to 50 ng) was included in a mixture containing 4 μl of BigDye terminator reaction mix (ABI Prism BigDye terminator cycle sequencing kit; Perkin-Elmer Applied Biosystems) and 20 pmol of each sense or antisense inner primer to a final volume of 10 μml. After an initial denaturation step at 94°C for 3 min, 25 PCR cycles consisting of 96°C for 10 s and 60°C for 4 min in a PTC-200 thermal cycler were programmed. PCR products were cleaned by ethanol precipitation and loaded into an automated ABI Prism 377 model sequencer. Forward and reverse sequence data of each sample were aligned by using the MEGALIGN program (DNAStar, Inc., Madison, Wis.). The consensus sequences of each gB genotype were compared and aligned to the four sequences obtained from the samples. A second analysis of all CMV gB sequences published in GenBank and the sequences obtained in our laboratory was made by pairwise alignment with MACAW 1.01 software.
Statistics.
The relative frequencies of the gB genotypes were compared by using 5×2 contingency tables, and P values were calculated for the associated χ2 statistics by using the SPSS version 8.0 statistical package (SPSS, Inc., Chicago, Ill.).
RESULTS
Performance of experiments and evaluation and validation of M-nPCR with clinical specimens.
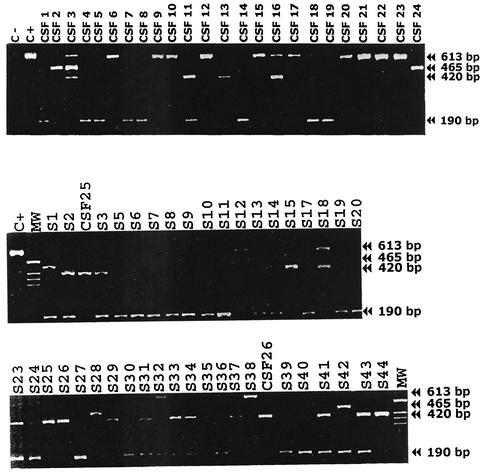
PCR product of the expected size (613 bp for gB2) was obtained by the M-nPCR assay with the reference CMV AD-169 strain that had been previously established as being gB2 genotype (6). Band sizes for gB genotypes 1 to 4 (420, 613, 190, and 465 bp for gB1, gB2, gB3, and gB4, respectively) were obtained singly in 47 samples and combined in another 19 samples (Fig. 1). No PCR product corresponding to the band size for gB5 was obtained in any sample tested. Serial dilution experiments with plasmidic copies containing cloned CMV DNA of each gB genotype showed that the M-nPCR was sufficiently sensitive to detect between 10 and 100 plasmids per PCR tube. No amplification was observed with other members of herpesviridae tested. All samples were retested for each pair of primers, and identical results were obtained when the multiplex was used. Direct sequencing of the polymorphic region amplified by primers CMVQ1+ and CMVQ1− from four samples that previously yielded single bands corresponding to each of the gB genotypes and subsequent analysis of the sequences obtained were in agreement with published consensus gB sequences, and no atypical or additional genotypes were found.
FIG. 1.
M-nPCR analysis for CMV gB typing. Lanes: C+, positive control; MW, base-pair ladder; S plus number, serum samples; CSF plus number, CSF samples. Sizes: gB1, 420 bp; gB2, 613 bp; gB3, 190 bp; gB4, 465 bp.
gB genotyping of clinical samples.
Of the clinical samples from which CMV had previously been detected, 66 of 70 (94.3%) samples were successfully gB genotyped. The remaining four serum samples with nonvisible bands were subsequently found to to be negative for CMV DNA by another highly sensitive PCR assay (26). Table 2 summarizes the frequency of gB genotypes according to specimens, nondisease versus CMV disease (including retinitis), and retinitis. In addition, the M-nPCR assay for gB genotyping was able to detect 19 samples (28.7%) containing a mixture of two or even three genotypes. Table 3 summarizes the frequency of CMV gB genotype mixed infections according to specimens.
TABLE 2.
Distribution of gB genotypes
| Specimen (n) | No. (%) of samples bearing genotype:
|
||||
|---|---|---|---|---|---|
| gB1 | gB2 | gB3 | gB4 | Mixed infection | |
| Sera (40) | 5 (12.5) | 1 (2.5) | 17 (42.5) | 1 (2.5) | 16 (40) |
| CSF (26) | 3 (11.5) | 10 (38.5) | 8 (30.8) | 2 (7.7) | 3 (11.5) |
| M-nPCR+ samplesa (66) | (12.1) | (16.7) | (37.9) | (4.5) | (28.7) |
| Outcome | |||||
| Asymptomatic patientsb (12) | 2 (16.7) | 0 (0) | 7 (58.3) | 1 (8.3) | 2 (16.7) |
| Symptomatic patientsc (47) | 3 (6.4) | 11 (23.4) | 16 (34) | 2 (4.3) | 15 (32) |
| CMV retinitis (35) | 1 (2.9) | 10 (28.6) | 11 (31.4) | 2 (5.7) | 11 (31.5) |
Of 70 tested samples, 66 were gB genotyped.
Of 16 clinical samples obtained from asymptomatic patients 4 were found to be gB M-nPCR assay negative.
A total of 47 clinical samples were obtained from symptomatic patients, including 35 patients diagnosed with retinitis.
TABLE 3.
Distribution of CMV mixed infection by different gB genotypes
| Specimen (n) | No. (%) of samples bearing genotypes:
|
||||
|---|---|---|---|---|---|
| gB1-gB2 | gB1-gB3 | gB2-gB3 | gB3-gB4 | gB1-gB2-gB4 | |
| Sera (40) | 1 (2.5) | 11 (27.5) | 2 (5) | 2 (5) | 0 (0) |
| CSF (26) | 1 (3.8) | 1 (3.8) | 0 (0) | 0 (0) | 1 (3.8) |
| Total + samples (66) | 2 (3) | 12 (18.2) | 2 (3) | 2 (3) | 1 (1.5) |
We examined whether there were any statistical differences in gB genotype frequency between specimens (serum versus CSF), between patient groups (CMV disease versus nondisease), and within the CMV disease patient group between patients with retinitis versus those without retinitis. Further differences and/or correlations could not be studied because frequencies in some of the 5×2 fields were insufficient for statistical analysis. Significant differences in gB genotype distribution between serum and CSF specimens were found (P = 0.001). gB3 was more frequently found in serum specimens than in CSF specimens. On the other hand, gB2 was more frequently found in CSF specimens but was rarely found in serum. This difference was also noted in the retinitis patient group (P = 0.005) (Table 4). However, there was no correlation between the gB genotype and CMV disease versus nondisease (P > 0.05) or between retinitis versus nonretinitis (P > 0.05).
TABLE 4.
Distribution of gB genotypes in patients with retinitis
| Specimen (n) | No. (%) of samples bearing genotype:
|
||||
|---|---|---|---|---|---|
| gB1 | gB2 | gB3 | gB4 | Mixed infection | |
| Sera (11) | 0 (0) | 0 (0) | 3 (27.3) | 0 (0) | 8 (72.7) |
| CSF(24) | 1 (4.2) | 10 (41.6) | 8 (33.3) | 2 (8.3) | 3 (12.5) |
| Total + samples (35) | 1 (2.8) | 10 (28.6) | 11 (31.4) | 2 (8.3) | 11 (31.4) |
DISCUSSION
In this retrospective study, CMV was detected and gB genotyped by M-nPCR in 94.3% of the studied samples in which CMV had previously been detected by PCR assays or by other classical assays. The remaining ungenotyped samples did not contain CMV DNA, as demonstrated by subsequent analysis of the sample by a highly sensitive PCR assay for CMV.
In the light of the essential role of gB in virus-host interaction, it is reasonable to surmise that variations in the gB genotype could be involved in cell tropism or the development of a particular end-organ disease and, accordingly, the characterization of the distribution of this genotype is clearly relevant. Of particular interest in the present study with a homogeneous patient demographic population was the finding that CMV gB genotypes are significantly differently distributed among specimen types: gB3 predominated in serum samples, regardless of CMV infection status and CMV disease status, whereas gB2 was observed to be more prevalent in CSF samples. Statistical analysis of gB genotype distribution in serum and CSF samples from patients diagnosed with retinitis reinforced this different pattern of gB distribution depending on the body site. In another study, four of nine paired samples of ocular fluid and blood showed a difference in gB genotype between these compartments (18). Comparison of genotype frequency in blood and CSF in patients with retinitis suggests that CSF CMV replication does not result exclusively from CMV reactivation in blood and, more importantly, the central nervous system could be a preferential site of infection by CMV gB2. We have demonstrated that CMV gB genotypes differ in prevalence in serum and CSF. This result clearly indicates the existence of additional neurotropic gB types. This finding is of great importance since it is the first report of a gB2 neuron tropism with clinical utility in the prognosis of CMV infection.
Infection by genotype gB3 alone (37.9%) or mixed infection with gB1 (18.2%) predominated in serum specimens, and these infection levels are higher than those reported in other gB genotype distribution studies in AIDS patients. Other studies have suggested that the distribution of CMV gB genotypes may be associated with geographic and/or demographic differences among AIDS patients (29, 30). These characteristics are mainly related to black women versus white homosexual men or sexually acquired AIDS versus injected-drug users. In our retrospective study, the overrepresentation of Spanish AIDS patients infected through the use of injected drugs precludes any precise analysis of links between the distribution of these subtypes and geographic and/or demographic differences and risk factors for HIV transmission. An increased rate of CMV gB2 type in the blood of Italian and American homosexual AIDS patients with respect to injected drug abusers has been described. Surprisingly, gB3 is more common in the blood of Italian AIDS patients belonging to the intravenous-drug-user risk group (30), and we have also observed this in the sera of our patients. This coincidence could be consistent with the patient demographic distribution of gB genotypes hypothesis. Although gB3 was the most prevalent gB genotype in serum specimens, gB2 clearly predominated in CSF specimens obtained from a population with the same demographic features.
In AIDS patients, gB2 and gB3 detected in serum samples have been associated with poor outcome and the development of retinitis (22). However, no significant correlation of the CMV gB genotype viremia with retinitis, nor with the clinical outcome of infection, was found in our study. In another study, gB2 was the most common gB genotype (43%) detected in 141 vitreous of 120 AIDS patients with CMV retinitis, and the ratio of gB subtypes was similar among the three geographically distinct patient populations (4).
gB2 and gB3 genotypes have also been associated with the expression of adhesion molecules, which may increase the spreading of these types in lymphocytes (11). In an in vitro study of CMV infection, it was demonstrated that, after extensive propagation in fibroblasts, CMV loses tropism for a number of otherwise-natural host cells and that the loss is associated with the appearance of genomic variants (24). Moreover, gB genotypes differ in their tropism for peripheral blood leukocytes in vivo: gB1 does not infect T lymphocytes, but the ability of gB2 and gB3 to infect monocytes and lymphocytes may explain the tropism and brain damage caused by these genotypes (14).
We have also found a higher prevalence of mixed infections by two different genotypes (mainly gB1 and gB3 in serum specimens) than in other studies. This may be because of reinfection by different CMV gB genotypes and/or because reactivation of a latent CMV infection is a frequent trait in advanced AIDS patients. Not surprisingly, mixed gB genotype infection was found more often in serum than in CSF, suggesting either selective virulence or specific organ or compartment susceptibility.
In conclusion, the M-nPCR assay described here provides a sensitive and specific assay for gB genotyping CMV directly from clinical specimens and allows mixed infection due to different gB genotypes to be easily detected. To our knowledge, this is the first study to show differences in CMV gB genotype distribution in the CSF and blood of AIDS patients with CMV retinitis of the same geographic origin and with the same demographic characteristics.
Although the gB gene is a candidate virulence factor for CMV strains causing infection in AIDS patients, the large coding capacity and slow cell-associated replication cycle of CMV suggest that pathogenesis involves a complex interaction of viral proteins with multiple host cell targets and that the genetic differences found between CMV strains are not limited to the gB gene but occur throughout the CMV genome. The study of gB gene polymorphism is an approach for identifying differences that influence the virulence of a CMV strain, although we do not rule out the possibility that the gB gene may be genetically linked to other unknown traits that are the true virulence genes. However, the recently discovered role of gB in the transcription regulation of infected cells has focused attention on this glycoprotein, highlighting the need for further pathogenesis studies.
Acknowledgments
We thank G. C. Fedele, F. Pozo, M. P. Sánchez-Seco, I. Casas, and M. Mosquera for technical assistance and A. Avellón and M. D. Pérez-Vázquez for performing statistical analyses.
This work was supported by the Spanish Ministry of Science and Technology grants (grant CICYT95-0393-OP).
REFERENCES
- 1.Bongarts, A., D. von Laer, C. Vogelberg, K. Ebert, J. van Lunzen, J. Garweg, P. Vaith, F. T. Hufert, O. Haller, and U. Meyer-König. 1996. Glycoprotein B genotype of human cytomegalovirus: distribution in HIV-infected patients. Scand. J. Infect. Dis. 28:447-449. [DOI] [PubMed] [Google Scholar]
- 2.Casas, I., F. Pozo, G. Trallero, J. M. Echevarria, and A. Tenorio. 1999. Viral diagnosis of neurological infection by RT multiplex PCR: a search for entero- and herpesviruses in a prospective study. J. Med. Virol. 57:145-151. [DOI] [PubMed] [Google Scholar]
- 3.Casas, I., L. Powell, P. E. Klapper, and G. M. Cleator. 1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction. J. Virol. Methods 53:25-36. [DOI] [PubMed] [Google Scholar]
- 4.Chern, K. C., D. B. Chandler, D. F. Martin, B. D. Kuppermann, R. A. Wolitz, and T. P. Margolis. 1998. Glycoprotein B subtyping of cytomegalovirus (CMV) in the vitreous of patients with AIDS and CMV retinitis. J. Infect. Dis. 178:1149-1153. [DOI] [PubMed] [Google Scholar]
- 5.Chou, S. W. 1990. Differentiation of cytomegalovirus strains by restriction analysis of DNA sequences amplified from clinical specimens. J. Infect. Dis. 162:738-742. [DOI] [PubMed] [Google Scholar]
- 6.Chou, S. W. 1992. Comparative analysis of sequence variation in gp116 and gp55 components of glycoprotein B of human cytomegalovirus. Virology 188:388-390. [DOI] [PubMed] [Google Scholar]
- 7.Chou, S. W., and K. M. Dennison. 1991. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J. Infect. Dis. 163:1229-1234. [DOI] [PubMed] [Google Scholar]
- 8.Cranage, M. P., T. Kouzarides, A. T. Bankier, S. Satchwell, K. T. Weston, B. Barrell, H. Hart, S. E. Bell, A. C. Minson, and G. L. Smith. 1986. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. 5:3057-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fidouh-Houhou, N., X. Duval, F. Bissuel, V. Bourbonneux, P. Flandre, J. L. Ecobichon, M. C. Jordan, J. L. Vilde, F. Brun-Vezinet, and C. Leport. 2001. Salivary cytomegalovirus (CMV) shedding, glycoprotein B genotype distribution, and CMV disease in human immunodeficiency virus-seropositive patients. Clin. Infect. Dis. 33:1406-1411. [DOI] [PubMed] [Google Scholar]
- 10.Fries, B. C., S. Chou, M. Boeckh, and B. Torok-Storb. 1994. Frequency distribution of cytomegalovirus envelope glycoprotein genotypes in bone marrow transplant recipients. J. Infect. Dis. 169:769-774. [DOI] [PubMed] [Google Scholar]
- 11.Grundy, J. E., and K. L. Downes. 1993. Upregulation of LFA-3 and ICAM-1 on the surface of fibroblast infected with cytomegalovirus. Immunology 78:405-412. [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy, P. G., D. A. Newsome, J. Hess, O. Narayan, D. L. Suresch, W. R. Green, and R. C. Gallo. 1986. Cytomegalovirus but not HTLV-III detected by in situ hybridization in retinal lesions in patients with AIDS. BMJ 293:162-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ljungman, P., P. Griffiths, C. Payaand, and S. A. Plotkin. 2002. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin. Infect. Dis. 34:1094-1097. [DOI] [PubMed]
- 14.Meyer-König, U., C. Vogelberg, A. Bongarrts, D. Kampa, R. Delbrück, G. Wolf-Volbeck, G. Kiste, M. Gaberland, F. T. Hufert, and D. von Laer. 1998. Glycoprotein B genotype correlates with cell tropism in vivo of human cytomegalovirus infection. J. Med. Virol. 55:75-81. [PubMed] [Google Scholar]
- 15.Meyer-König, U., M. Haberland, D. von Laer, O. Haller, and F. T. Hufert. 1998. Intragenic variability of human cytomegalovirus glycoprotein B in clinical strains. J. Infect. Dis. 177:1162-1169. [DOI] [PubMed] [Google Scholar]
- 16.Navarro, D., P. Pedro, S. Tugizov, K. Topp, J. La Vail, and L. Pereira. 1993. Glycoprotein B of human cytomegalovirus promotes virion penetration into cells, transmission of infection from cell to cell, and fusion of infected cells. Virology 197:143-158. [DOI] [PubMed] [Google Scholar]
- 17.Nelson, J. A., C. Reynolds-Kohler, M. B. A. Oldstone, and C. A. Wiley. 1988. HIV and CMV coinfect brain cells in patients with AIDS. Virology 165:286-290. [DOI] [PubMed] [Google Scholar]
- 18.Peek, R., F. Verbraak, M. Bruinenberg, A. van der Lelij, G. van der Horn, and A. Kijlstra. 1998. Cytomegalovirus glycoprotein B genotyping in ocular fluids and blood of AIDS patients with cytomegalovirus retinitis. Investig. Ophthalmol. Vis. Sci. 39:1183-1187. [PubMed] [Google Scholar]
- 19.Pring-Akerblom, P., F. E. J. Trijssenaar, T. Adrian, and H. Hoyer. 1999. Multiplex polymerase chain reaction for subgenus-specific detection of human adenovirus in clinical samples. J. Med. Virol. 58:87-92. [PubMed] [Google Scholar]
- 20.Schuler, G. D., S. F Altschul, and D. J. Lipman. 1991. A work-bench for multiple alignment construction analysis. Protein Struct. Funct. Genet. 9:180-190. [DOI] [PubMed] [Google Scholar]
- 21.Shepp, D. H., M. E. Match, S. M. Lipson, and R. G. Pergolizzi. 1998. A fifth human cytomegalovirus glycoprotein B genotype. Res. Virol. 149:109-114. [DOI] [PubMed] [Google Scholar]
- 22.Shepp, D. H., M. E. Match, A. B. Ashraf, S. M. Lipson, C. Millan, and R. Pergolizzi. 1996. Cytomegalovirus glycoprotein B groups associated with retinitis in AIDS. J. Infect. Dis. 174:184-187. [DOI] [PubMed] [Google Scholar]
- 23.Simmen, K. A., J. Singh, B. G. M. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Früh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinzger, C., K. Schmidt, J. Knapp, M. Kahl, R. Beck, J. Waldman, H. Hebart, H. Einsele, and G. Jahn. 1999. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J. Gen. Virol. 80:2867-2877. [DOI] [PubMed] [Google Scholar]
- 25.Stockton, J., J. S. Ellis, M. Saville, J. P. Clewley, and M. C. Zambon. 1998. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J. Clin. Microbiol. 36:2990-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenorio, A., J. E. Echevarría, I. Casas, J. M. Echevarría, and E. Tabarés. 1993. Detection and typing of human herpesviruses by multiplex polymerase chain reaction. J. Virol. Methods 44:261-269. [DOI] [PubMed] [Google Scholar]
- 27.Torok-Storb, B., M. Boeckh, C. Hoy, W. Leisenring, D. Myerson, and T. Booley. 1997. Association of specific cytomegalovirus genotypes with death from myelosuppression after marrow transplantation. Blood 90:2097-2102. [PubMed] [Google Scholar]
- 28.Vogelberg, C., U. Meyer-König, F. T. Hufert, G. Kirste, and D. von Laer. 1996. Human cytomegalovirus glycoprotein B genotypes in renal transplant recipients. J. Med. Virol. 50:31-34. [DOI] [PubMed] [Google Scholar]
- 29.Wada, K., S. Mizuno, K. Kato, T. Kamiya, and K. Ozawa. 1997. Cytomegalovirus glycoprotein B sequence variation among Japanese bone marrow transplant recipients. Intervirology 40:215-219. [DOI] [PubMed] [Google Scholar]
- 30.Zipeto, D., C. Hong, G. Gerna, M. Zavattoni, D. Katzenstein, T. C. Merigan, and L. Rasmussen. 1998. Geographic and demographic differences in the frequency of human cytomegalovirus gB genotypes 1-4 in immunocompromised patients. AIDS Res. Hum. Retrovir. 14:533-536. [DOI] [PubMed] [Google Scholar]