Abstract
To determine whether confirmatory tests for extended-spectrum β-lactamase (ESBL) production in Escherichia coli are necessary, we selected 131 E. coli isolates that met the National Committee for Clinical Laboratory Standards (NCCLS) screening criteria for potential ESBL production from the Project ICARE (Intensive Care Antimicrobial Resistance Epidemiology) strain collection. For all 131 isolates, the broth microdilution (BMD) MIC of at least one extended-spectrum cephalosporin was ≥2 μg/ml. For 21 of 131 (16%) isolates, the ESBL confirmatory test was positive; i.e., the BMD MICs of ceftazidime or cefotaxime decreased by ≥3 doubling dilutions in the presence of clavulanic acid (CA) or the disk diffusion zone diameters increased by ≥5 mm around ceftazidime or cefotaxime disks in the presence of CA. All 21 isolates were shown by PCR to contain at least one of the genes blaTEM, blaSHV, and blaOXA, and in isoelectric focusing (IEF) tests, all isolates demonstrated at least one β-lactamase band consistent with a TEM, SHV, or OXA enzyme. Of the 21 isolates, 3 showed a CA effect for cefotaxime by BMD but not by disk diffusion testing. A total of 59 (45%) of the 131 isolates demonstrated decreased susceptibility to cefpodoxime alone (MIC = 2 to 4 μg/ml), and none had a positive ESBL confirmatory test result. These were classified as false positives according to ESBL screen test results. For the remaining 51 (39%) isolates, the cefpodoxime MICs ranged from 16 to >128 μg/ml and the MICs for the other extended-spectrum cephalosporins were highly variable. All 51 isolates gave negative ESBL confirmatory test results. Most showed IEF profiles consistent with production of both a TEM and an AmpC β-lactamase, and representative isolates of several phenotypic groups showed changes in porin profiles; these 51 isolates were considered true negatives. In all, only 16% of 131 E. coli isolates identified as potential ESBL producers by the current NCCLS screening criteria were confirmed as ESBL producers. Thus, changing the interpretation of extended-spectrum cephalosporins and aztreonam results from the susceptible to the resistant category without confirming the presence of an ESBL phenotype would lead to a large percentage of false resistance results and is not recommended. However, by increasing the cefpodoxime MIC screening breakpoint to ≥8 μg/ml, 45% of the false-positive results could be eliminated. NCCLS has incorporated this change in the cefpodoxime screening breakpoint in its recent documents.
Extended-spectrum β-lactamases (ESBLs) are enzymes produced by some gram-negative bacilli that mediate resistance to extended-spectrum cephalosporins and aztreonam (4, 6, 14). ESBLs are most commonly recognized in Klebsiella spp. and Escherichia coli but have also been detected in a variety of Enterobacteriaceae and Pseudomonas aeruginosa isolates (6; G. A. Jacoby and K. Bush website [http://www.lahey.org/studies/webt.htm]). Resistance mediated by ESBLs can be difficult to detect, depending on the antimicrobial agents tested (4, 29, 31). The National Committee for Clinical Laboratory Standards (NCCLS) recommends using one or more of the antimicrobial agents aztreonam, cefpodoxime, ceftazidime, cefotaxime, and ceftriaxone (20-22) to screen for ESBL activity. The presence of ESBL activity in an isolate that is positive by the screening test should be confirmed by testing the isolates with cefotaxime and ceftazidime in the presence and absence of clavulanic acid (CA) to determine whether enzymatic hydrolysis of the cephalosporin tested is inhibited in the presence of CA (20, 21). While the term ESBL traditionally referred just to TEM and SHV β-lactamases, there is a growing number of β-lactamases, including OXA-type and CTX-M-type enzymes, that can also hydrolyze extended-spectrum cephalosporins and are inhibited by CA (4, 14, 19, 32).
The family of AmpC β-lactamases includes the chromosomal enzymes of E. coli, Enterobacter spp., Citrobacter spp., Morganella spp., Proteus spp., P. aeruginosa, and other species (5, 10, 12, 13, 24) in addition to a growing number of plasmid-mediated β-lactamases related to the above enzymes (3, 6, 14, 27). These β-lactamases also mediate resistance to extended-spectrum cephalosporins and aztreonam in addition to cephamycins such as cefoxitin and cefotetan (6; Jacoby and Bush [http://www.lahey.org/studies/webt.htm]). Unlike ESBLs, however, AmpC β-lactamases are not inhibited by CA (6, 14). Thus, testing isolates in the presence of cefotaxime and ceftazidime with and without CA can usually differentiate ESBL-producing strains from AmpC-producing strains, provided that ESBL-containing strains don't harbor additional β-lactamases or porin changes that mask the CA effect. Survey data from laboratories participating in CDC's Foodnet program, however, suggest that many laboratories do not perform the ESBL confirmation tests (9). When the specificity of the NCCLS ESBL screening breakpoints (i.e., ≥2 μg/ml for cefpodoxime, ceftazidime, ceftriaxone, cefotaxime, or aztreonam) is high, as has recently been shown for Klebsiella pneumoniae (29), omitting confirmatory testing may be acceptable. However, to our knowledge, such specificity data have not been reported for E. coli. Thus, we selected 131 E. coli isolates from the Project ICARE (Intensive Care Antimicrobial Resistance Epidemiology) (1, 11) strain collection that met the NCCLS screening criteria for potential ESBL production and characterized their β-lactamases by isoelectric focusing (IEF) tests and their β-lactamase gene profiles by PCR assays to determine the specificity of the NCCLS ESBL screening breakpoints to ascertain whether the CA tests were, indeed, necessary.
MATERIALS AND METHODS
Bacterial strains.
E. coli isolates for which the MIC of at least one extended-spectrum cephalosporin or aztreonam was ≥2 μg/ml were collected from 26 Project ICARE laboratories during a 4-year period. Organisms were identified using standard biochemical reactions (2) or Vitek GNI or GNI+ cards (bioMérieux, Durham, N.C.).
Antimicrobial susceptibility testing.
All isolates were stored at −70°C until used. Each isolate was grown in subcultures on Trypticase soy agar containing 5% sheep blood (BD BioSciences, Sparks, Md.) two times prior to testing. Using MIC trays made in house according to the NCCLS procedure (20), organisms were tested by the NCCLS broth microdilution (BMD) reference method described in document M7-A5 (20). MIC trays contained ceftazidime, cefotaxime, ceftriaxone, cefpodoxime, and aztreonam, both alone and with 4 μg of CA/ml. Organisms were also tested using the NCCLS disk diffusion reference method M2-A7 (21), and the results were interpreted using the M100-S10 standard (22). Confirmation of ESBL activity was undertaken by testing cefotaxime and ceftazidime in the presence and absence of CA as described previously by the NCCLS (20, 21). Isolates for which MICs of cefotaxime or ceftazidime decreased by ≥3 twofold dilutions when tested in the presence of CA or zone diameters increased by ≥5 mm in the presence of CA were considered positive for ESBLs. Quality-control organisms included E. coli ATCC 25922, K. pneumoniae ATCC 700603, Staphylococcus aureus ATCC 29213, and S. aureus ATCC 25923.
IEF.
IEF tests were performed on cell extracts as previously described (7, 17). β-Lactamase bands within the following isoelectric point (pI) ranges were categorized as consistent with the following enzymes (4, 12): pI 5.2 to 5.7, TEM; pI 7.4 to 8.2, SHV; pI 7.0 to 7.2, OXA; and pI 8.3 to 8.6, AmpC. As previously noted (29), these ranges are not exclusive of other β-lactamases but served as a screening method for detection of ESBLs in conjunction with PCR results.
PCR assays.
PCR assays to determine the presence of the blaTEM and blaSHV β-lactamase genes were performed as described previously (15, 28). The PCR assays used for blaOXA and blaCTX-M β-lactamase genes were those described by Steward et al. (29).
PCR amplification of ampC promoter and attenuator regions.
Primers AB1 (5′-GATCGTTCTGCCGCTGTG-3′) and ampC2 (5′-GGGCAGCAAATGTGGAGCAA-3′) were used to amplify the E. coli ampC promoter and attenuator regions as previously described (8). The forward and reverse sequences of the amplification products were determined using products from two independent PCRs as templates.
OMP profile determination.
Outer-membrane proteins (OMPs) were isolated from selected strains as previously described (28). Briefly, cells were grown to mid-log phase in nutrient broth and harvested by centrifugation. Cell pellets were washed and resuspended in sodium phosphate buffer (pH 7.0), and the cell walls were disrupted by treatment with lysozyme and sonication. Total membrane proteins were separated from cell debris by differential centrifugation. Sarkosyl-insoluble OMPs were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis through 10% polyacrylamide-8 M urea gels. E. coli strains C600, MH513 (OmpF−), and MH225 (OmpC−) (which were the generous gift of Thomas Silhavy) were used as controls.
PFGE.
Using the restriction enzyme BlnI, pulsed-field gel electrophoresis (PFGE) was performed on subsets of isolates from the same institution as previously described (29). The gels were run on a CHEF DR-III apparatus (Bio-Rad Laboratories, Hercules, Calif.) under the following conditions: pulse time, 2.5 to 60 s; run time, 21.5 h; temperature, 14°C; voltage, 6 V/cm. Banding pattern interpretation was based on published criteria (30).
RESULTS
Screening for ESBLs.
One hundred thirty-one isolates of E. coli were tested by using the NCCLS ESBL confirmation procedures (20, 21). For each of the 131 isolates, the MICs of at least one extended-spectrum cephalosporin or aztreonam were ≥2 μg/ml (i.e., the isolates were ESBL screening test positive by NCCLS criteria). A total of 21 (16%) isolates (with 18 different PFGE profiles) from 12 different hospitals showed a more than threefold doubling dilution decrease in the BMD MIC of either cefotaxime or ceftazidime or both when tested in the presence of CA (i.e., a CA effect). Of the 21 isolates, 18 also showed a ≥5-mm increase in the diameter of the zone of inhibition around the cefotaxime or ceftazidime disk. All 21 isolates were positive for blaTEM, blaSHV, or blaOXA or a combination of those genes by PCR, and according to the results of IEF testing, all demonstrated at least one β-lactamase band consistent with the presence of a TEM, SHV, or OXA enzyme. The patterns of the PCR and IEF results for these isolates are shown in Table 1. According to IEF test results, a total of 11 isolates contained multiple β-lactamases.
TABLE 1.
PCR and IEF profiles of isolates that demonstrate a CA effect (ESBL producers)
| No. of isolates (n = 21) | PCR result | IEF profile(s) (no. of isolates) |
|---|---|---|
| 8 | blaTEM + blaSHV positive | 5.4, 5.6, 6.6, 8.1 (4) |
| 5.4, 6.6, 8.1 (1) | ||
| 5.3, 8.1 (2) | ||
| 7.7 (1) | ||
| 1 | blaTEM + blaSHV + blaOXA positive | 5.3, 6.5, 8.0 (1) |
| 8 | blaTEM positive only | 5.6 (3) |
| 5.3-5.4 (5) | ||
| 1 | blaTEM + blaOXA positive | 5.2, 5.5, 8.4 (1) |
| 1 | blaSHV + blaOXA positive | 7.1, 8.1 (1) |
| 2 | blaOXA positive only | 7.2, 7.6 (1) |
| 7.1 (1) |
Of 21 ESBL-producing isolates, 3 (of which demonstrated a different PFGE profile) showed a CA effect for cefotaxime by BMD but not by disk diffusion testing. For isolate EC1013, the cefpodoxime, cefotaxime, ceftazidime, and ceftriaxone MICs were 4, 2, 0.25, and 0.5 μg/ml, respectively (data not shown). Although the ceftazidime MIC decreased by only a single dilution when tested with CA, the cefotaxime MIC decreased from 2 to 0.25 μg/ml with CA, a threefold doubling dilution change. However, there was no increase in the zone sizes around ceftazidime and cefotaxime disks when tested in the presence of CA. This isolate was positive by PCR for blaTEM and showed a single β-lactamase band of pI 5.4 by IEF testing. The other two isolates contained β-lactamases with pIs of 7.2 and 7.6 (EC3521) and 7.1 (EC3871), were positive only with blaOXA primers, and demonstrated ceftazidime MICs of 1.0 and 0.5 μg/ml, ceftriaxone MICs of 0.5 and 1.0 μg/ml, and cefotaxime MICs of 2 and 8 μg/ml, respectively. A CA effect was noted only for cefotaxime for both strains; in each case, the cefotaxime MICs decreased by 3 dilutions in the presence of CA. This demonstrates that CA confirmation test results can show inconsistencies among both the drugs and methods used. However, regardless of the method, NCCLS requires only one of the ESBL tests to be positive for an organism to be designated an ESBL producer. Since 18 of 21 (86%) ESBL-positive isolates also demonstrated nonsusceptible results for cefoxitin (i.e., MICs ≥ 16 μg/ml), cefoxitin MIC results could not be used to differentiate between those isolates harboring ESBLs and those harboring AmpC β-lactamases or other mechanisms of cephalosporin resistance.
Isolates for which cefpodoxime alone showed an increased MIC that do not show a CA effect.
For fifty-nine (45%) isolates showing a wide variety of PFGE profiles, the cefpodoxime MICs were elevated just enough to make them ESBL screen test positive (at 2 to 4 μg/ml), while the ceftazidime and cefotaxime MICs remained low or unchanged (≤1 μg/ml) from levels typically observed for susceptible E. coli strains (25). No CA effect was observed with cefotaxime or ceftazidime for any of these isolates. A total of 33 isolates were positive by PCR for blaTEM only; one of those isolates showed three β-lactamase bands by IEF testing. Six isolates were positive by PCR for blaOXA, but none of the 59 isolates was positive for blaSHV. The PCR and IEF patterns of the isolates are shown in Table 2. One of the 20 isolates that were negative for blaTEM, blaOXA, and blaSHV by PCR also failed to demonstrate the presence of a β-lactamase by IEF testing and appeared to lack changes in its OMPs (26). Thus, the mechanism of decreased susceptibility to cephalosporins remains unclear.
TABLE 2.
PCR and IEF profiles of isolates that do not demonstrate a CA effect but show decreased susceptibility to cefpodoxime only
| No. of isolates (n = 59) | PCR result | IEF profile(s) (no. of isolates) |
|---|---|---|
| 33 | blaTEM positive only | 5.4, 5.7, 6.8 (1) |
| 5.3-5.5 (32) | ||
| 20 | Negative for blaTEM, blaSHV, and blaOXA | 8.4-8.6 (18) |
| 5.6 (1) | ||
| No β-lactamase detected (1) | ||
| 6 | blaOXA positive only | 7.1-7.2 (6) |
Isolates for which cefotaxime and ceftazidime showed increased MICs that do not show a CA effect.
The remaining 51 (39%) isolates, which also failed to show a CA effect by BMD testing with cefotaxime and ceftazidime, constituted a very heterogeneous group of isolates with widely divergent PFGE profiles. The cefpodoxime MICs ranged from 16 to >128 μg/ml, and ceftazidime MICs ranged from 4 to 128 μg/ml. The β-lactamase profiles of these isolates were similar to those of isolates showing decreased susceptibility to cefpodoxime alone (Table 3). A total of 31 of the isolates produced both a TEM β-lactamase (pI 5.3 to 5.5) and the E. coli AmpC chromosomal β-lactamase (pI ≥ 8.3); all 31 were positive by PCR for blaTEM. Of those isolates, 10 were negative by PCR for blaTEM, blaSHV, and blaOXA, although one showed an IEF profile consistent with the presence of enzymes other than an AmpC β-lactamase. Five isolates had IEF and PCR results consistent with the presence of OXA-type β-lactamases.
TABLE 3.
PCR and IEF profiles of isolates that do not demonstrate a CA effect but show resistance to multiple extended-spectrum cephalosporins
| No. of isolates (n = 51) | PCR result | IEF profile(s); p1 result(s) (no. of isolates) |
|---|---|---|
| 36 | blaTEM positive only | 5.3-5.5; 8.3-8.6 (31) |
| 5.4, 5.7, 7.0 (1) | ||
| 5.5, 5.7, 7.1 (1) | ||
| 5.3-5.4 (3) | ||
| 10 | Negative for blaTEM, blaSHV, and blaOXA | 8.3, 8.5 (1) |
| 8.5 (4) | ||
| 8.3 (3) | ||
| 5.4, 8.4 (1) | ||
| 5.6, 6.5, 6.9 (1) | ||
| 5 | blaOXA positive only | 7.1-7.2; 8.4 (5) |
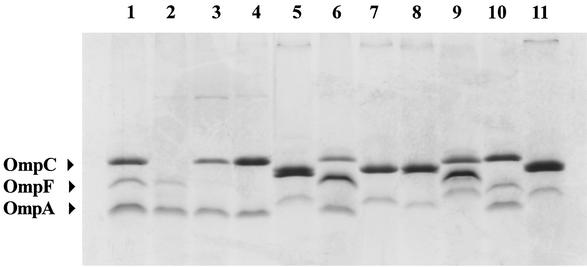
The ampC promoter and attenuator regions of 35 isolates that had an antibiogram result consistent with enhanced AmpC expression (i.e., cefpodoxime MICs ≥ 64 μg/ml, ceftazidime MICs ≥ 8 μg/ml, cefotaxime MICs ≥ 8 μg/ml, and cefoxitin MICs ≥ 32 μg/ml) and produced a β-lactamase band with a pI of ≥8.3 in IEF testing were sequenced to determine whether mutations in these regions were responsible for enhanced resistance to cephalosporins (8). The DNA sequence results fell into five groups: (i) 7 isolates showed a previously observed 2-bp insertion between the −35 and −10 regions and five base changes in the attenuator loop, including several that have been associated with enhanced expression of ampC; (ii) 10 isolates displayed novel as well as previously reported modifications in both the promoter and attenuator loops; (iii) 3 isolates showed no modifications in the attenuator loop but displayed either five or six base changes in the promoter which are thought to increase transcription of ampC; (iv) 6 isolates displayed several novel modifications in the promoter region but none in the attenuator loop; and (v) 9 isolates showed no changes in the promoter or attenuator loop when compared to those harboring the wild-type E. coli sequence. The MIC profiles of the extended-spectrum cephalosporins were not consistent within the groups showing the same mutational profiles. This suggested that changes in OMPs may have contributed to or been responsible directly for the elevated extended-spectrum cephalosporin MICs in these isolates. Thus, eight isolates representing the various promoter and attenuator sequence groups were examined for changes in porin profiles. All eight isolates showed alterations in the porin profiles compared to E. coli C600 (Fig. 1), suggesting that alteration in OmpF and OmpC contributed to the enhanced resistance of these isolates to extended-spectrum cephalosporins.
FIG. 1.
Polyacrylamide gel showing OMP preparations of three E. coli control strains (lanes 1 to 3) and eight E. coli clinical isolates (lanes 4 to 11), the latter of which show decreased susceptibility to cephalosporins that is not reversed by CA. The ampC promoter and attenuator sequence group (see text) for each isolate is indicated. Lane 1, E. coli C600 (wild type); lane 2, E. coli MH225 (OmpC−); lane 3, E. coli MH513 (OmpF−); lane 4, isolate 578 (group 5); lane 5, 792 (group 1); lane 6, 1340 (group 3); lane 7, 3100 (group 1); lane 8, 3283 (group 4); lane 9, 3331 (group 3); lane 10, 3516-2 (group 5); lane 11, 3977 (group 2).
DISCUSSION
In this study of 131 E. coli that gave positive ESBL screen test results according to NCCLS criteria, only 21 (16%) of the isolates actually contained an ESBL, as shown by performing the NCCLS confirmation tests by BMD and disk diffusion with and without CA. Of these 21 isolates, 5 produced an OXA β-lactamase, either alone or in addition to a TEM or SHV enzyme, which did not mask the CA effect (6, 14). Three of the isolates produced conflicting ESBL confirmation test results by BMD and disk diffusion testing. For these three, the BMD tests showed a CA effect with cefotaxime only; neither of the disk tests with cefotaxime or ceftazidime was positive. For all three isolates, the cefotaxime MICs were ≤8 μg/ml and the ceftazidime and ceftriaxone MICs were ≤1 μg/ml. Based on NCCLS guidelines, these isolates were considered to be ESBL producers, since at least one ESBL confirmatory test result with CA was positive (20).
The poor specificity of the ESBL screening test for E. coli is in contrast to the previous findings of Steward et al. with K. pneumoniae, which showed that approximately 84% of isolates that gave positive ESBL screen test results showed a CA effect by either BMD or disk diffusion testing and contained an identifiable ESBL (29). In that study, the 16% of isolates that were not confirmed to be ESBL producers appeared to contain a variety of novel β-lactamases.
In the present study, 45% of the E. coli isolates showed decreased susceptibility to cefpodoxime only and none gave a positive ESBL confirmatory test result. The low MICs of the other extended-spectrum cephalosporins (≤1 μg/ml) suggested that a 2- μg/ml breakpoint for cefpodoxime is too nonspecific to be used as an indicator for potential ESBL activity in E. coli. Recent results demonstrated that several mechanisms of resistance contribute to this phenotype, including changes in OMP profiles (such as loss of OmpF and OmpC), the presence of β-lactamases not inhibited by CA (such as OXA-type enzymes), and low-level expression of the E. coli AmpC chromosomal β-lactamase (26). Given the low levels of cephalosporin resistance seen in these isolates and the lack of clinical data suggesting that patients fail therapy when their infections are treated with these drugs, NCCLS recently raised the cefpodoxime ESBL screening breakpoint from 2 to 8 μg/ml to increase the specificity of this test and reduce the number of false-positive results (23). This also reaffirms that the use of ESBL confirmation tests for E. coli is critically important to prevent reporting false resistance results.
A total of 31 of the isolates that were positive by ESBL screen testing but did not show a CA effect harbored both TEM and AmpC β-lactamases and demonstrated a resistance profile consistent with enhanced production of the E. coli AmpC β-lactamase (6, 14, 24; Jacoby and Bush [http://www.lahey.org/studies/webt.htm]). While this resistance phenotype has often been attributed to mutations in the ampC promoter and attenuator regions that enhance ampC expression (8, 14, 18, 24), our sequence analysis of these regions from 35 isolates suggests that the presence of such mutations alone does not explain the enhanced resistance profiles of many of the E. coli isolates examined. Rather, it is probable that changes in porin profiles (such as loss of OmpF) in conjunction with the promoter and attenuator mutations and expression of blaTEM together result in cephalosporin MICs that can be either in the moderate range (8 to 32 μg/ml) or high (≥64 μg/ml). While efflux pumps such as AcrAB may play a role in cephalosporin resistance (16, 18), we did not examine this possibility in our study.
Of the E. coli isolates in this study, 16 contained blaOXA genes. Although 14 isolates had IEF profiles that included bands of pI 7.0 to 7.2, the other isolates demonstrated bands with pIs of 8.0 to 8.4. This reaffirms that IEF testing serves only as an indicator of potential β-lactamase activity and is not a definitive test for specific β-lactamase production (29). Other isolates in the study that were negative with our three blaOXA primer sets demonstrated IEF bands of pI 7.0 to 7.2. While these may yet turn out to be OXA-type β-lactamases, it serves to remind us that there are a variety of β-lactamases and β-lactamase genes yet to be discovered.
In summary, confirmation of ESBL production in E. coli isolates that give positive results with the NCCLS ESBL screening tests is critical if the laboratory is to avoid inappropriately changing the classification of results from penicillin and cephalosporin testing of isolates we believe to be susceptible from the susceptible to the resistant category. Whether to change the interpretations for isolates showing other mechanisms of resistance remains an open question. In this study, the 55 isolates with high cephalosporin MICs that did not show a CA effect were considered as true negatives, i.e., they did not contain ESBLs. This study also raises the issue of whether NCCLS should establish further criteria, such as having at least one indicator extended-spectrum cephalosporin in the nonsusceptible range, before interpretations are changed to the resistant category. This may prevent laboratories from erroneously changing penicillin and cephalosporin interpretations from the susceptible to the resistant category for isolates that (due simply to TEM-1 production and minor changes in outer-membrane proteins) appear to be ESBL producers but remain clinically susceptible to β-lactam compounds. No doubt the reporting of ESBLs, AmpCs, and other β-lactamases should continue to challenge microbiologists for years to come.
Acknowledgments
We thank Gregory Anderson for assistance with the DNA sequencing studies and Christine Steward and Kelley Brittain for helpful discussions and critical review of the manuscript. We also thank the microbiology personnel at Project ICARE hospitals for sending isolates of E. coli.
Phase IV of Project ICARE is supported in part by grants to the Rollins School of Public Health of Emory University by the following companies: Astra-Zeneca Pharmaceuticals, Wilmington, Del.; Bayer Corporation, Pharmaceuticals Division, West Haven, Conn.; Cubist Pharmaceuticals, Lexington, Mass.; Elan Pharmaceuticals, San Diego, Calif.; Pharmacia Corporation, Bedminster, N.J.; and Roche Laboratories, Nutley, N.J.
Use of trade names is for identification purposes only and does not constitute endorsement by the Public Health Service or the U.S. Department of Health and Human Services.
REFERENCES
- 1.Archibald, L., L. Phillips, D. Monnet, J. E. McGowan, Jr., F. Tenover, and R. Gaynes. 1997. Antimicrobial resistance in isolates from inpatients and outpatients in the United States: the increasing importance of the intensive care unit. Clin. Infect. Dis. 24:211-215. [DOI] [PubMed] [Google Scholar]
- 2.Bopp, C. A., F. W. Brenner, J. G. Wells, and N. A. Strockbine. 1999. Escherichia, Shigella, and Salmonella, p. 459-474. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 3.Bou, G., A. Oliver, M. Ojeda, C. Monzon, and J. Martinez-Beltran. 2000. Molecular characterization of FOX-4, a new AmpC-type plasmid-mediated β-lactamase from an Escherichia coli strain isolated in Spain. Antimicrob. Agents Chemother. 44:2549-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A. 2001. Extended-spectrum β-lactamases. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bret, L., C. Chanal-Claris, D. Sirot, E. B. Chaibi, R. Labia, and J. Sirot. 1998. Chromosomally encoded AmpC-type β-lactamase in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 42:1110-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush, K., and S. B. Singer. 1989. Effective cooling allows sonication to be used for liberation of β-lactamases from Gram-negative bacteria. J. Antimicrob. Chemother. 24:82-84. [DOI] [PubMed] [Google Scholar]
- 8.Caroff, N., E. Espaze, I. Bérard, H. Richet, and A. Reynaud. 1999. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum β-lactamase production. FEMS Microbiol. Lett. 173:459-465. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2000. Laboratory capacity to detect antimicrobial resistance, 1998. Morb. Mortal. Wkly. Rep. 48:1167-1171. [PubMed] [Google Scholar]
- 10.Coudron, P. E., E. S. Moland, and K. S. Thomson. 2000. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a Veterans Medical Center. J. Clin. Microbiol. 38:1791-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fridkin, S. K., C. D. Steward, J. R. Edwards, E. R. Pryor, J. E. McGowan, Jr., L. K. Archibald, R. P. Gaynes, F. C. Tenover, and Project Intensive Care Antimicrobial Resistance Epidemiology Hospitals. 1999. Surveillance of antimicrobial use and antimicrobial resistance in U.S. hospitals: Project ICARE Phase 2. Clin. Infect. Dis. 29:245-252. [DOI] [PubMed] [Google Scholar]
- 12.Galleni, M., F. Lindberg, S. Normark, S. Cole, S. Honore, B. Joris, and J. Frère. 1988. Sequence and comparative analysis of three Enterobacter cloacae ampC β-lactamase genes and their product. Biochem. J. 250:753-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindberg, F., and S. Normark. 1986. Sequence of the Citrobacter freundii OS60 chromosomal AmpC β-lactamase gene. Eur. J. Biochem. 156:441-445. [DOI] [PubMed] [Google Scholar]
- 14.Livermore, D. M. 1995. β-Lactamases in the clinical laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mabilat, C., and S. Goussard. 1993. PCR detection and identification of genes for extended-spectrum β-lactamases, p. 553-559. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 16.Mallea, M., J. Chevalier, C. Bornet, A. Eyraud, A. Davin-Regli, C. Bollet, and J. M. Pages. 1998. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology 144:3003-3009. [DOI] [PubMed] [Google Scholar]
- 17.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical iso-electric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 18.Mazzariol, A., G. Cornaglia, and H. Nikaido. 2000. Contributions of the AmpC β-lactamase and the AcrAB multidrug efflux system in intrinsic resistance of Escherichia coli K-12 to β-lactams. Antimicrob. Agents Chemother. 44:1387-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naas, T., and P. Nordmann. 1999. OXA-type β-lactamases. Curr. Pharm. Des. 5:865-879. [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests. Approved standard—7th ed.; M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial susceptibility testing. NCCLS approved standard M100-S10. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing. NCCLS approved standard M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Nelson, E. C., and B. G. Elisha. 1999. Molecular basis of AmpC hyperproduction in clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 43:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neu, H. C. 1982. The new beta-lactamase-stable cephalosporins. Ann. Intern. Med. 97:408-419. [DOI] [PubMed] [Google Scholar]
- 26.Oliver, A., L. M. Weigel, J. K. Rasheed, J. E. McGowan, Jr., and F. C. Tenover. 2002. Mechanisms of decreased susceptibility to cefpodoxime in Escherichia coli. Antimicrob. Agents Chemother. 46:3829-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasheed, J. K., C. Jay, B. Metchock, F. Berkowitz, L. Weigel, J. Crellin, C. Steward, B. Hill, A. A. Medeiros, and F. C. Tenover. 1997. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steward, C. D., J. K. Rasheed, S. K. Hubert, J. W. Biddle, P. M. Raney, G. J. Anderson, P. P. Williams, K. L. Brittain, A. Oliver, J. E. McGowan, Jr., and F. C. Tenover. 2001. Characterization of clinical isolates of Klebsiella pneumoniae from 19 laboratories using the National Committee for Clinical Laboratory Standards extended-spectrum β-lactamase detection methods. J. Clin. Microbiol. 39:2864-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenover, F. C., J. M. Mohammed, T. S. Gorton, and Z. F. Dembeck. 1999. Detection and reporting of organisms producing extended-spectrum β-lactamases: a survey of laboratories in Connecticut. J. Clin. Microbiol. 37:4065-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]