Abstract
We report a strategy for encapsulating and condensing DNA. When T5 phage binds to its membrane protein receptor, FhuA, its double stranded DNA (120,000 bp) is progressively released base pair after base pair in the surrounding medium. Using cryoelectron microscopy, we have visualized the structures formed after T5 phage DNA is released into neutral unilamellar proteoliposomes reconstituted with the receptor FhuA. In the presence of spermine, toroidal condensates of circumferentially wrapped DNA were formed. Most significantly, the sizes of these toroids were shown to vary, from 90 to 200 nm in their outer diameters, depending on the number of DNA stands transferred. We have also analyzed T5 DNA release in bulk solution containing the detergent-solubilized FhuA receptor. After DNA release in a spermine containing solution, huge DNA condensates with a diameter of about 300 nm were formed containing the DNAs from as many as 10–20 capsids. At alkaline pH, the condensates appeared as large hollow cylinders with a diameter of 200 nm and a height of 100–200 nm. Overall, the striking feature of our experiments is that, because of the progressive release of DNA from the phage capsid, the mechanism of toroid formation is fundamentally different from that in the classical studies in which highly dilute, “naked” DNA is condensed by direct addition of polyvalent cations; as a consequence, our method leads to toroids of arbitrary size.
Keywords: polyamines, condensation, FhuA, toroids
The condensation of DNA, i.e., the collapse of an extended worm-like random coil DNA molecule into a compact ordered state, has received considerable attention in diverse areas of science (1–3). In biology, it represents a process by which genetic information is packaged and protected. In medicine, it provides a promising means for gene therapy applications through new forms of condensed genes to be delivered to target cells. In polymer and condensed matter physics, DNA condensation is an example of a coil-globule transition or of a self-assembly process in which aggregation of two or more DNA molecules are involved.
In vitro condensation of DNA by different chemical agents has provided useful insights into the physical factors governing folding and packaging of DNA. Among these condensing agents, much interest has focused on polycations that modify electrostatic interactions between DNA segments through neutralization of their charges (4) and/or mediation of attractive forces between them (5). These agents, which induce collapse, aggregation, or complexation by interacting directly with DNA molecules, include small ions (polyamines, cobalt hexamine), cationic polypeptides (polylysine, polyhistidine), or proteins (histones) (6–11).
When condensation is induced by addition of polyamines (spermine, spermidine) to very dilute aqueous solution of DNA, striking toroidal structures are the most common morphologies observed by electron microscopy (12–14). Here DNA is circumferentially wrapped as a toroid with a well defined hole in the middle and an outer diameter of about 90 ± 10 nm. One of the most surprising findings to emerge from these condensation studies is that the toroids fall into a remarkably narrow range of sizes independent of the length of DNA being condensed (from 400 to 50,000 bp), of its genetic information (e.g., for DNA from virus, bacterial plasmid or calf thymus), and of whether it is circular or linear (1).
However, in all of the above studies, to avoid extensive intermolecular DNA aggregation, the measurements must be conducted at extremely low DNA concentrations (≈1 μg DNA/ml), much lower than the prevailing DNA concentrations in biological cells. In addition, this requirement, together with the need to mix DNA and polyamines quickly without shearing of DNA, has caused technical problems that limit attempts to work with longer DNA or to follow the very rapid kinetics of toroid formation. Thus, despite extensive studies on DNA condensation, using a variety of techniques, understanding of the mechanisms involved in DNA toroid formation is still a matter of debate.
In the present paper, we demonstrate the potential of a new strategy for DNA delivery and for investigating the spermine-induced condensation of several long double stranded DNAs. Our strategy relies on the observation that the binding of bacteriophage T5 to its bacterial membrane receptor FhuA triggers the release of its genome, a double strand of 120,000 bp, linearly into the surrounding medium (15). Moreover, when FhuA is reconstituted in the membrane of a liposome, T5 phage bound to FhuA transfers its DNA directly into the liposome (16–17).
Using cryoelectron microscopy, we have analyzed the structures formed after phage DNA ejection either in small liposomes or in bulk solution containing different amounts of spermine. A striking feature of this study is that phage DNA transferred into liposomes is condensed into toroidal structures whose sizes increase significantly with the number of DNA strands injected into the proteoliposome. This demonstrates that toroids of arbitrary and controllable size can be formed and is discussed in comparison with previous literature on spermine-induced DNA condensation.
Materials and Methods
Materials.
Egg phosphatidylcholine of the highest purity was purchased from Avanti Polar lipids, N,N-dimethyldodecylamine N-oxide from Fluka, spermine-4HCl from Sigma, and DNase I Bovine Pancreas (DNase) from Pharmacia. All other reagents were of analytical grade.
Preparation of Proteoliposomes.
Hexahistidine-tagged FhuA protein was purified from an Escherichia coli overproduced strain as described (18). Reconstitution of FhuA into phosphatidylcholine liposomes was performed essentially as described (16–17) using polystyrene beads for detergent removal (19). In all experiments, the buffer composition was 150 mM NaCl, 20 mM Hepes⋅Tris (pH 7), and spermine-4HCl at the desired concentration.
DNA Transfer into Proteoliposomes.
T5 phages (1011 particles) were added to the proteoliposomes such that the ratio of the phage particles to FhuA molecule was approximately 1:100. In all experiments, DNase (1 μg) and MgSO4 (5 mM) were added to digest DNA occasionally released in the external medium. The mixtures were incubated at 37°C for 1 h before observation by electron microscopy.
DNA Release in the External Medium.
T5 phages were added to a solution of FhuA solubilized in 0.1% N,N-dimethyldodecylamine N-oxide such that the ratio of the phage particles to FhuA molecule was 1:100. Aliquots were taken at different time intervals and immediately were frozen in liquid ethane. DNA release was studied at 37°C in a buffer containing 20 mM Hepes⋅Tris (pH 7), or Tris⋅HCl (pH 8), 150 mM NaCl, and spermine-4HCl at the desired concentration.
Cryoelectron Microscopy.
Samples were deposited on a holey carbon grid and were rapidly frozen in liquid ethane as described by Dubochet et al. (20). Micrographs were recorded on a Philips CM120 electron microscope operating at 120 kV with a magnification of 45,000× and a nominal defocus of 1 μm.
Results
Transfer of T5 Genome into Proteoliposomes.
To avoid any interaction between tetravalent spermine and phospholipids, the membrane protein FhuA was reconstituted into proteoliposomes made of neutral phosphatidylcholine. Fig. 1 shows representative cryoelectron micrographs of proteoliposomes resulting from such reconstitutions. The liposomes appeared unilamellar with diameters ranging from 70 to 300 nm. When T5 phages were incubated with the FhuA-containing proteoliposomes, they bound to the vesicle surfaces by the distal end of their tails, indicating a good reconstitution of the receptor into the membrane.
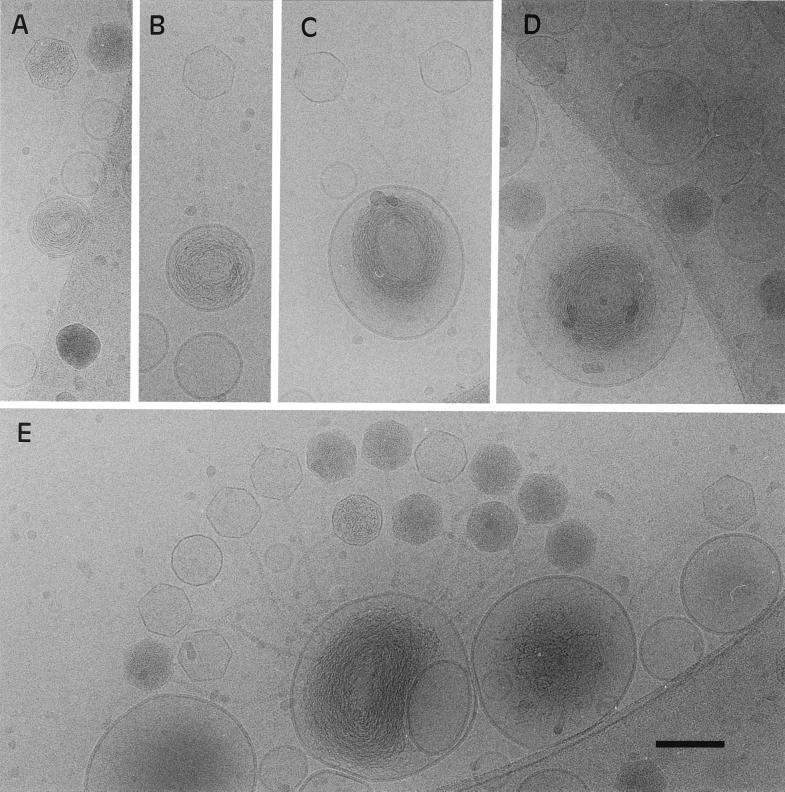
Figure 1.
Transfer of T5 genome into proteoliposomes containing different amounts of spermine. (A) Without spermine, the viral genome is only partially transferred. (B) A single DNA strand is fully entrapped in a vesicle containing 20 mM spermine. (C) Two DNA strands transferred into a vesicle containing 30 mM spermine. The circular arrangement of DNA suggests that DNA has been organized in a toroid during the early stage of DNA transfer. (D) At 50 mM spermine, the fully transferred DNA strand is condensed into a toroid occupying a volume smaller than the liposomal volume. At lower spermine concentration, although the amount of entrapped DNA increases with the spermine concentration, the strand is not condensed into a toroid but rather occupies all of the internal space of the liposome. (Bar = 100 nm.)
In the absence of encapsulated spermine, many of the capsids of phages attached to the proteoliposomes appeared partially filled with DNA, as shown by their lower electron densities. The proteoliposomes associated with these partially empty capsids contained DNA strands that have been transferred from the capsids into the liposomes (Fig. 1A). It has to be stressed that, in the absence of spermine, although the entrapped DNA appeared densely packed, it filled all of the intraliposomal volume without adopting an organized structure.
When proteoliposomes were reconstituted in the presence of spermine, all of the T5 genome could be now transferred into the liposome. For spermine concentrations of 20 and 30 mM (Fig. 1 B and C, respectively), entrapped DNA strands still filled the whole volume of the vesicles. In the presence of 50 mM spermine-containing liposomes, the DNA strands transferred from the phage capsids were condensed into compact structures occupying a volume smaller than the internal volume of the liposome (Fig. 1D).
At the highest (50 mM) spermine concentration, different structures were observed, depending on the number of phages that had transferred their genome and on the size of the liposome. Fig. 2A shows a liposome 90 nm in diameter in which the almost complete transfer of one genome has led to a pseudotoroidal structure. For larger liposomes, 130–200 nm in diameter, the transfer of one phage DNA led to the formation of characteristic toroidal structures with DNA strands clearly wrapped circumferentially around a central hole and with an outer diameter of 120–130 nm (Fig. 2B; see also Fig. 1D). Increasing the number of DNA strands transferred into the proteoliposomes did not change the number of toroids in the liposomes but increased significantly the size of the toroid formed. For example, a toroid composed of two DNA strands ejected from two different phages had an outer diameter of 150–160 nm (Fig. 2 C and D) whereas a toroid composed of six DNA strands had a diameter of about 210 nm (Fig. 2E). The size increased up to 230 nm when about 10 phages have transferred their DNA into one liposome (data not shown).
Figure 2.
Toroids of arbitrary size induced by DNA transfer into 50 mM-spermine-containing liposomes. (A) Toroid made of part of 120,000-bp T5 genome. (B) Toroid made of a single DNA strand. (C) Toroid made of two DNA strands. (D) Toroid of two to three DNA strands. (E) Toroid made of more than six DNA strands. (Bar = 100 nm.)
DNA Condensation After T5 DNA Release in Bulk Solution.
We have also analyzed the process of DNA condensation after DNA release from T5 phages bound to isolated FhuA molecules in a detergent-containing solution. In the presence of 1 mM to 4 mM spermine, DNA strands released from the phage capsid appeared as long disentangled filaments that were progressively condensed into very long bundles up to 100 μm in length (data not shown), longer than the fully extended 120,000 bp of T5 phage DNA that span about 40 μm. In the presence of 5 mM spermine, DNA strands ejected from the capsids appeared organized into very large toroidal structures. The electron micrograph shown in Fig. 3C indicated that the toroids formed were rather homogenous in size, with an outer diameter of 300 nm, and that a comparatively constant number of phages (10–20) are involved in each structure.
Figure 3.
Release of T5 genome in 5 mM spermine bulk solution. (A and B) Typical electron micrographs of the annular structures (70–100 nm in diameter) formed 1–2 min after adding T5 phages to FhuA. (C) Large toroids are formed after 15 min of incubation at pH 7. (D) DNA released in solution at pH 8. DNA condensates appear as hollow cylinders exhibiting highly ordered DNA strands. On circular view, DNA is disposed in concentric arcs with a 26-Å spacing (black arrow). On tilted view (marked by an asterisk), a regular disposition of DNA strands is visible (arrowhead). (Bar = 100 nm.)
The kinetics of the spermine-induced DNA condensation process, difficult to analyze inside proteoliposomes, was followed by analyzing samples at different incubation times after addition of T5 phages to solubilized FhuA. It is known that, once the phage has been added to a FhuA-containing solution, DNA is released within a few tens of seconds (15). After 1–2 min of incubation, the shortest time that can be actually analyzed in our experimental conditions, small annular structures (70–100 nm in outer diameter and 30–40 nm in inner diameter) could be visualized (Fig. 3 A and B). These structures were generated from several DNA strands partially released from all of the phages around one toroid, indicating that their formation occurs very rapidly and does not require full DNA release from the phages. After 5 min of incubation, all phages appeared empty, and very large structures could be observed, similar to those depicted in Fig. 3C after 15 min of incubation.
Toroid formation in the presence of 5 mM spermine was analyzed as a function of the pH of the solution. Tore formation could only be detected between pH 7 and 9. However, at pH 8 and 9, the DNA condensates appeared significantly more tightly condensed and organized in hollow cylinders as opposed to the toroidal structures observed at pH 7. Indeed, although the most common view of these condensates was a circular one (probably attributable to the most frequent orientation within the ice layer), intermediate views clearly corresponded to tilted cylinders. From the two tilted structures seen in Fig. 3D, the straight height of a cylindrical condensate was estimated to be about 100–200 nm. In addition, the DNA strands in these structures appeared highly ordered in small areas. In circular views, DNA was clearly disposed in concentric arcs with a spacing corresponding to a distance of 26 Å, whereas on tilted views these distances varied from 27 to 35 Å. Elsewhere, the DNA arrangement looked more fuzzy.
Discussion
In the present manuscript, we report on the use of a strategy for forming spermine-induced DNA condensates. DNA release in bulk solution or into small unilamellar liposomes was controlled by the binding of T5 phage to its membrane receptor FhuA. Unlike previous experiments developed for DNA condensation studies, our system provides the unique advantage that long and intact DNA strands, 120,000 base pairs long, can be delivered progressively, base pair after base pair, into a medium containing the desired spermine concentration. Using this strategy, we have been able to form and visualize by cryoelectron microscopy spermine-induced toroidal structures inside neutral unilamellar liposomes. In this way, we made toroids of arbitrary size, with considerably larger dimensions than those previously reported, shedding new light on the understanding of DNA organization within toroidal condensates and the kinetics of their formation.
Reconstitution of FhuA into the membrane of a liposome allows T5 phages to bind to their membrane receptors and to transfer their genome into the internal volume of the proteoliposomes. In the absence of encapsulated spermine, phage DNA transfer was never complete, and the partially transferred DNA strands appeared densely packed but filled all of the internal volume of the liposome. In the presence of 50 mM encapsulated spermine, the amount of transferred DNA increased considerably, as seen by completely empty capsids, indicating that an entire genome could be transferred into the proteoliposomes. The presence of internal spermine also drastically modified the DNA packaging within the liposome, leading to characteristic toroidal structures. When a single DNA has been transferred into a liposome, it appears wrapped as a single toroid of outer diameter 120–140 nm, slightly larger than the dimensions reported in the literature (1, 21). Increasing the number of phages bound to their receptors resulted in larger, multimolecular toroids whose size increased with the number of DNA strands transferred. Thus, toroidal structures with outer diameters as large than 200 nm could be visualized in the small internal volume of the liposome.
This large range of DNA packaging represents an original feature of our system, which has to be related to the fact that, in our experimental strategy, the DNA is progressively released from the capsid, inducing DNA condensates of one or several DNA strands. When the linear DNA strand enters the liposome, it wraps into a toroid that then grows in size. Indeed, Fig. 2A clearly shows that DNA wrapping starts before the complete transfer of DNA is achieved within the vesicle. In addition, our results demonstrate that a single toroid could be formed within each liposome, even when several (6–10) phages have transferred their DNA. This indicates that the single toroid has collected many DNA strands that have been delivered either independently or in a synchronized manner.
Another particularity of our study on spermine-induced DNA condensation into liposomes is that, as opposed to experiments in bulk solution, the liposomes are closed systems and spermine is encapsulated with a fixed concentration. In the presence of 50 mM spermine, a liposome with 140 nm in diameter has encapsulated about 42,500 spermine molecules, which represent 170,000 positive charges. This amount of spermine is sufficient to observe a toroid of one DNA strand containing 240,000 phosphate charges, in a good agreement with the calculation of Wilson and Bloomfield (22) for DNA collapse using Manning's counterion condensation theory (4). When these neutralization conditions for DNA collapse are not fulfilled, DNA strands occupied the maximal internal volume of the liposomes. Thus, at low spermine concentration, we propose that DNA is condensed at the beginning of DNA transfer (8); but then further DNA transfer leads to a progressive depletion of spermine and the condition for DNA collapse is no longer satisfied, provoking DNA decondensation. Such a DNA decondensation process has already been described in solution as a fast process induced either by increasing the amount of DNA (23) or by diluting the condensing agent (7).
As a step toward a better understanding of the possibility of forming toroids of arbitrary size, we have also analyzed the structures formed on phage DNA release in bulk solution. In solution containing 5 mM spermine, the DNA released from the phage capsids was condensed into very large individual toroids. The size of these large condensates was relatively constant with a mean outer diameter of 300 nm. Again, the progressive base pair after base pair release of phage DNA allowed the packaging of a large amount of DNA that corresponds to a final DNA bulk concentration of about 0.4 mg/ml. Furthermore, our results indicate that the process of toroid formation is rapid because the large toroidal structures are completely formed within 5 min of starting the reaction. After 1–2 min, however, electron micrographs show DNA condensed into much smaller toroids. These small structures, produced in the early stage of DNA release, likely constitute a nucleus for further toroid condensation on continued DNA release.
Our data are important in the framework of theories developed recently for understanding the apparently constant size of condensed DNA toroids. As mentioned in the introduction, an abundant literature suggests that the size of polyamine-induced toroids is largely independent of the DNA length, type, and genetic information; the fundamental packing unit for DNA is a toroidal structure, 90 nm in outside diameter with a 30-nm-diameter hole. This constant size of condensed DNA particles is characteristic of the classical studies of toroid formation induced by adding polyvalent cations to highly dilute solutions of DNA. Basically, two theoretical approaches have been pursued to address this issue. In the first, the finite size of a toroid is argued to be the thermodynamically stable state of circumferentially wound DNA, reflecting the competition between the bending energy of the DNA chains and their mutual attraction in the presence of topologically unavoidable defects (24). Here it is predicted that the free energy density of a toroid begins to increase at a certain volume threshold that happens to correspond to a few tens of thousands of base pairs. This volume is where the topological defects have accumulated to the point at which their contribution to the free energy density begins to overwhelm that from cohesive interactions. [The predictions for longer (more than 50,000 bp) molecules are hard to test in the usual highly dilute solutions because of DNA degradation by shear forces arising in their handling.] In the second theoretical approach, the thermodynamically stable state of condensed DNA is argued to be an infinite (macroscopic) crystal rather than a finite-sized bundle of hexagonally packed, parallel molecules (25). These authors consider that the size of the toroids is limited by a kinetic barrier that cannot be surmounted by thermal fluctuations. This barrier arises from the fact that the interaction between strands is repulsive at large separation distances and minimized by mutually perpendicular orientation whereas the polyvalent cation-induced attraction is operative only at short distances and for parallel configurations. Accordingly, each added molecule must have enough energy to rotate and become aligned with those strands already condensed in order for the attractive forces to be felt. In our experiments, the viral ejection of DNA allows it to add to the condensate in a progressive and concerted manner, with its orientation automatically aligned with those in the growing toroid, thereby avoiding the kinetic barrier. Accordingly, it is possible to form toroids of essentially unlimited size containing an arbitrary number of base pairs. This mechanism of condensation into toroids is clearly fundamentally different from that in the classical studies and it is this difference that allows for new control of toroid formation.
In addition to these important features, a final observation concerns the striking effect of pH on the morphology of the condensates. Although at pH 7 DNA condensates resemble the classical toroidal structure, they evolve at pH values of 8 and 9 toward cylindrical structures, 200 nm in diameter and 200 nm in height. Furthermore, a regular packaging of DNA as revealed by a repeat spacing in the hollow cylinders is only visible at these alkaline pH values. In these regular arrays, the spacing varies from 26 to 35 Å, which could reflect different orientations of a hexagonal interhelical packing (26–27). Because the regular disposition is only visible in part of the structure, it is difficult to infer that this organization exists throughout the whole cylinder. These structures could be easily distorted because of a high flexibility or because of the presence of topological defects. It is also possible that different crystalline phases could be induced by a local variation of spermine concentration within the DNA condensates (28).
Besides the consequences of our study in the analysis of spermine-induced DNA condensation, we believe that our approach may have important applications in biological contexts. DNA condensation could be studied by using other types of polyamines or globular, cationic proteins like histones, either encapsulated in liposomes or in solution. Because phage (T5 here) represents a stable supply of DNA that can be opened with a specific key (FhuA), this strategy seems well suited for studying other types of DNA-protein interactions as well. From a practical point of view, the results can be important for gene therapy applications because one can make DNA condensates of any desired size either, in bulk solution or encapsulated into neutral liposomes. The DNA-containing proteoliposomes produced in this way could serve as alternative vehicles for the transfer of foreign genomes into eukaryotic cells because: (i) as opposed to cationic liposomes, disruptive electrostatic interactions between lipids and DNA, and between liposomes and cell membranes, are unlikely in our system; (ii) the entrapped DNA is of far larger size than the plasmids commonly used; (iii) DNA can be strongly condensed, as demonstrated in this study, and can thereby be better protected from degradation; and (iv) the lipid bilayer comprising the “membrane” of the encapsulated DNA can be easily functionalized to optimize targeting, without in any way affecting the toroid formation process.
Acknowledgments
We thank G. Moeck and L. Plançon for providing FhuA and G. Pehau-Arnaudet for his technical assistance. This work was supported by a grant of the Centre National de la Recherche Scientifique, program “Physique et Chimie du Vivant.” W.M.G. thanks the Institut Curie for its hospitality during the Spring of 1999, when he visited there as Rothschild Professor.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130187297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130187297
References
- 1.Bloomfield V A. Biopolymers. 1991;31:1471–1481. doi: 10.1002/bip.360311305. [DOI] [PubMed] [Google Scholar]
- 2.Bloomfield V A. Curr Opin Struct Biol. 1996;6:334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 3.Bloomfield V A. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 4.Manning G S. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 5.Gronbech-Jensen N, Mashl R J, Bruisma R F, Gelbart W M. Phys Rev Lett. 1997;78:2477–2480. [Google Scholar]
- 6.Gosule L C, Schellman J A. J Mol Biol. 1978;121:311–326. doi: 10.1016/0022-2836(78)90366-2. [DOI] [PubMed] [Google Scholar]
- 7.Widom J, Baldwin R L. J Mol Biol. 1980;144:431–453. doi: 10.1016/0022-2836(80)90330-7. [DOI] [PubMed] [Google Scholar]
- 8.Pelta J, Livolant F, Sikorav J L. J Biol Chem. 1996;271:5656–5662. doi: 10.1074/jbc.271.10.5656. [DOI] [PubMed] [Google Scholar]
- 9.Laemmli U K. Proc Natl Acad Sci USA. 1975;72:4288–4292. doi: 10.1073/pnas.72.11.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Ramirez M, Subirana J. Biopolymers. 1994;34:285–292. [Google Scholar]
- 11.Leforestier A, Fudaley S, Livolant F. J Mol Biol. 1999;290:481–494. doi: 10.1006/jmbi.1999.2895. [DOI] [PubMed] [Google Scholar]
- 12.Chattoraj D K, Gosule L C, Schellman J A. J Mol Biol. 1978;121:327–337. doi: 10.1016/0022-2836(78)90367-4. [DOI] [PubMed] [Google Scholar]
- 13.Arscott P G, Li A Z, Bloomfield V A. Biopolymers. 1990;30:619–630. doi: 10.1002/bip.360300514. [DOI] [PubMed] [Google Scholar]
- 14.Plum G E, Arscott P G, Bloomfield V A. Biopolymers. 1990;30:631–643. doi: 10.1002/bip.360300515. [DOI] [PubMed] [Google Scholar]
- 15.Boulanger P, le Maire M, Bonhivers M, Dubois S, Desmadril M, Letellier L. Biochemistry. 1996;35:14216–14224. doi: 10.1021/bi9608673. [DOI] [PubMed] [Google Scholar]
- 16.Plançon L, Chami M, Letellier L. J Biol Chem. 1997;272:16868–16872. doi: 10.1074/jbc.272.27.16868. [DOI] [PubMed] [Google Scholar]
- 17.Lambert O, Plançon L, Rigaud J L, Letellier L. Mol Microbiol. 1998;30:761–765. doi: 10.1046/j.1365-2958.1998.01107.x. [DOI] [PubMed] [Google Scholar]
- 18.Moeck G S, Tawa P, Xiang H, Ismail A A, Turnbull J L, Coulton J W. Mol Microbiol. 1996;22:459–471. doi: 10.1046/j.1365-2958.1996.00112.x. [DOI] [PubMed] [Google Scholar]
- 19.Rigaud J L, Levy D, Mosser G, Lambert O. Eur Biophys J. 1998;27:305–319. [Google Scholar]
- 20.Dubochet J, Adrian M, Chang J-J, Homo J-C, Lepault J, McDowall A W. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 21.Hud N V, Allen M J, Downing K H, Lee J, Balhorn R. Biochem Biophys Res Commun. 1993;193:1347–1354. doi: 10.1006/bbrc.1993.1773. [DOI] [PubMed] [Google Scholar]
- 22.Wilson R W, Bloomfield V A. Biochemistry. 1979;18:2192–2196. doi: 10.1021/bi00578a009. [DOI] [PubMed] [Google Scholar]
- 23.Porschke D. Biochemistry. 1984;23:4821–4828. doi: 10.1021/bi00316a002. [DOI] [PubMed] [Google Scholar]
- 24.Park S Y, Harries D, Gelbart W M. Biophys J. 1998;75:714–720. doi: 10.1016/S0006-3495(98)77561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ha B-Y, Liu A J. Europhys Lett. 1999;46:624–630. [Google Scholar]
- 26.Schellman J A, Parthasarathy N. J Mol Biol. 1984;175:313–329. doi: 10.1016/0022-2836(84)90351-6. [DOI] [PubMed] [Google Scholar]
- 27.Ubbink J, Odijk T. Biophys J. 1995;68:54–61. doi: 10.1016/S0006-3495(95)80158-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelta J, Durand D, Doucet J, Livolant F. Biophys J. 1996;71:48–63. doi: 10.1016/S0006-3495(96)79232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]