Abstract
A common epitope region of enteroviruses was identified by sequence-independent single-primer amplification (SISPA), followed by immunoscreening of 11 cDNA libraries from two Korean enterovirus isolates (echoviruses 7 and 30) and a coxsackievirus B3 (ATCC-VR 30). The putative common epitope region was localized in the N terminus of VP1 when the displayed recombinant proteins from the phages were chased by the convalescent-phase sera. The genomic region encoding the common epitope region was amplified and then expressed by using the vector pGEX-5X-1. The antigenicity of the expressed recombinant protein was identified by Western blotting with guinea pig antisera for six different serotypes of enteroviruses. After successive immunization of mice with the recombinant common epitope protein, splenocytes were extracted and hybridized with P3X63-Ag8-653 cells. A total of 24 hybridomas that produced monoclonal antibodies (MAbs) against the putative common epitope of enteroviruses were selected. Four of these were immunoglobulin G1 isotypes with a kappa light chain. These MAbs recognized 15 Korean endemic serotypes and prototypes of enteroviruses in an indirect immunofluorescence assay. These results suggest that the expressed protein might be a useful antigen for producing group common antibodies and that the use of the MAbs against the putative common epitope of enteroviruses might be a valuable diagnostic tool for rapidly identifying a broad range of enteroviruses.
The genus Enterovirus, belonging to the family Picornaviridae, consists of 66 different subtypes, including polioviruses (PVs), coxsackievirus group A (CVA) and CVB, echoviruses, and the numbered enteroviruses. These viruses cause a wide variety of clinical illnesses, ranging from asymptomatic to fatal, including poliomyelitis, aseptic meningitis, myopericarditis, and respiratory, hepatic, and gastrointestinal infections (15, 23, 24). Thus, the early diagnosis of enterovirus-related infection from various clinical isolates is important for prognostic, therapeutic, and epidemiologic purposes (1, 14, 18, 26).
The primary structure and genetic organization of enteroviruses have been demonstrated recently. Conserved sequences are located in structural and nonstructural proteins of enteroviruses (12, 28, 29, 31, 35, 39). Therefore, it may be possible to use the conserved region as a common epitope for the serological diagnosis of enteroviruses. The synthetic peptides for the enterovirus group-common epitope could be used for the serological diagnosis of infections caused by a broad range of enteroviruses (3, 25). However, these approaches have not yet been proven effective for the routine laboratory diagnosis of enteroviral antigens in clinical specimens. Thus, it would be useful to develop a rapid and reliable antigen diagnostic method for enteroviral infection by using a monoclonal antibody (MAb) specific for the common epitope region.
In the present study, the genetic region of the putative common epitope of enteroviruses was determined by sequence-independent single-primer amplification (SISPA)-mediated immunoscreening analysis and expressed in Escherichia coli by a PCR-based cloning strategy. By using the expressed putative common epitope as an immunogen, highly specific MAbs against the recombinant proteins were produced and evaluated for their potential in rapid diagnostic tests that detect a broad range of enteroviruses.
MATERIALS AND METHODS
Virus and culture.
The enteroviruses used in the present study included 12 Korean isolates (E3, E4, E6, E7, E9, E25, E30, CVB1, CVB2, CVB3, CVB5, and CVB6) and three prototypes (E24 [ATCC VR-54], CVB3 [Nancy strain, ATCC VR-30], and CVB4 [ATCC VR-184]). The enteroviruses used for production of the common epitope and MAbs were E7, E30, and CVB3 (Nancy strain), and the others were used to identify an immunoreactivity of the MAbs. Viruses were inoculated onto monolayers of rhabdomyosarcoma (RD) or human larynx carcinoma (HEp2-c) cells, which were kindly provided by the World Health Organization (WHO)/Western Pacific Regional Office. Cells showing 70 to 80% of cytopathic effects were frozen-thawed three times, and cell debris was removed by centrifugation at 1,000 × g for 10 min. The supernatants were collected and used for serotyping and for viral RNA isolation. Serotyping was performed with WHO/RIVM (Rijksinstituut voor de Volksgezondheid en Milieuhygiene, Amsterdam, The Netherlands) enterovirus serum pools and/or CVB neutralization test reagents (Denka Seiken, Osaka, Japan). Procedures were performed as described in WHO guidelines (38) for enterovirus isolation.
RNA extraction and cDNA synthesis.
Viral RNAs were isolated from the supernatants of enterovirally infected HEp2-c or RD cells by the sodium dodecyl sulfate (SDS)-proteinase K method as described previously (40). Single-stranded and double-stranded cDNAs were synthesized by using random hexamers and a cDNA synthesis kit (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer's instructions.
SISPA.
The blunt-ended cDNA was ligated to AB linker and primers (30), which have EcoRI restriction sites, by using T4 DNA ligase and a >10-fold molar excess of linker and primers to cDNA. Aliquots of the AB linker-primer ligation mixtures were used directly for SISPA with a 2 μM concentration of primer A, 1.5 mM MgCl2, a 0.4 mM concentration of deoxynucleoside triphosphate, 2.5 U of Taq DNA polymerase, and 10× buffer provided by the manufacturer (Promega, Madison, Wis.). Thirty-five cycles of denaturation at 94°C for 60 s, annealing at 54°C for 90 s, and extension at 72°C for 120 s were carried out in a DNA thermal cycler 480 (Perkin-Elmer, Beaconsfield, United Kingdom). The amplified SISPA products were purified by using S-300HR MicroSpin columns (Pharmacia, Uppsala, Sweden) at 600 × g.
λgt11 library construction and immunoscreening.
The SISPA products were digested with EcoRI and cloned into a λgt11 vector (Stratagene, La Jolla, Calif.). The reaction mixture was incubated at 12.8°C for 16 h and in vitro packaged by using Gigapack III gold packaging extract (Stratagene). Approximately 2 × 105 recombinant phages were incubated with E. coli y1090r− and plated at a low density according to the manufacturer's instructions. Colonies were lifted with nitrocellulose filter papers (Protran BA 83; Schleicher & Schuell, Keene, N.H.).
An immunoscreening assay was performed with anti-E7 guinea pig antisera or antisera from patients with aseptic meningitis caused by CVB1. Anti-E7 guinea pig antisera were made by immunizing guinea pigs with purified E7 through subcutaneous injection for three times with a week interval.
PCR and direct sequence analysis of positive clones.
Positive clones selected by immunoscreening were subjected to plaque purification and directly suspended in 200 ml of sodium-magnesium buffer (pH 7.2) with 5% of chloroform. To determine the sizes of the inserts, PCR amplification was performed as follows: a 0.6-μl aliquot of positive plaque suspension was added to 29.4 μl of the PCR mixture containing a 0.4 mM concentration of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 20 pmol of primers (λgt11 forward and reverse primers), 1 U of Taq DNA polymerase (Promega), and 10× reaction buffer provided by the manufacturer. Thirty-five cycles of denaturation at 94°C for 60 s, annealing at 55°C for 60 s, and extension at 72°C for 60 s were carried out in a DNA thermal cycler 480 (Perkin-Elmer). The amplified PCR products were electrophoresed in 1% agarose gels and stained with ethidium bromide.
To characterize each positive clone, direct sequencing of the PCR products was performed. The nucleotide sequence was determined by ABI Prism dye terminator cycle sequencing ready reaction kit (Perkin-Elmer) according to the manufacturer's instructions (34).
Cloning and expression of enteroviral common epitope region.
The cDNA for cloning and expression of the enteroviral common epitope region was obtained by using reverse transcription-PCR. Oligonucleotide primers E7-271F and E7-271R, corresponding to positions 2457 to 2474 and positions 2700 to 2717 of the VP1 region of E7, were made based on the sequences of the immunopositive plaques and VP1 of enteroviruses obtained from the GenBank/EMBL database. The E7-271F primer sequence was 5′-CAAGCACTTCTGTTTCCCCGG-3′, and the E7-271R primer was sequence was 5′-ATTGTCACCATAAGCAGCCA-3′. The amplified PCR product was directly cloned into a pCR2.1 vector by using a TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.). The cloned PCR product was named pE7-271TA. After digestion with NcoI and BamHI, the E7 cDNA fragment of pE7-271TA was electrophoresed in agarose gel, purified from the gel, and treated with Klenow enzyme (Boehringer Mannheim, Hamburg, Germany) to make blunt-ended DNA. The blunt-ended E7 cDNA fragment was cloned into the SmaI site of pGEX-5X-1 vector (Pharmacia, Buckinghamshire, United Kingdom) and transformed into E. coli BL21(DE3).
To express the recombinant proteins, 3-ml aliquots of overnight culture of transformed E. coli BL21(DE3) were diluted to 1:20 in fresh Luria-Bertani medium with 50 μg of ampicillin/ml and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and then incubated for an additional 3 h at 37°C. Cells were pelleted by centrifugation at 19,000 × g for 5 min at 4°C, washed with phosphate-buffered saline (PBS; 150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4 [pH 7.3]), and disrupted by sonication. Supernatants were prepared by centrifugation at 9,800 × g for 10 min, and the expressed protein was purified by using glutathione-Sepharose 4B columns (Pharmacia). To cleave the expressed viral protein from the glutathione S-transferase (GST) carrier, the fusion protein was incubated with thrombin (Sigma, St. Louis, Mo.) at 1:500 (wt/wt) in cleavage buffer (50 mM Tris-HCl, pH 8.0; 100 mM NaCl, 2.5 mM CaCl2, 0.1% β-mercaptoethanol) for 4 h at room temperature.
SDS-PAGE and Western blotting.
Purified proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) by using the discontinuous system of Laemmli (17). Separated proteins were electrophoretically transferred to nitrocellulose membrane (0.45 μm [pore size]; Bio-Rad, Hercules, Calif.) as described by Towbin et al. (37). The membrane was then blocked with PBST (PBS containing 0.05% [vol/vol] Tween 20) containing 3% skim milk for 2 h at room temperature and incubated with anti-E7 guinea pig antisera or CVB antisera in PBST containing 3% skim milk for 2 h at room temperature. After three washes with PBST, the membrane was incubated with alkaline phosphatase-conjugated anti-guinea pig IgG or alkaline phosphatase-conjugated anti-human IgG (Sigma) for 2 h at room temperature. The conjugate was diluted 1:3,000 in 3% skim milk in PBST immediately before use. After an three additional washes with PBST, the membrane was incubated in a freshly prepared mixture of substrate solution (nitroblue tetrazolium-BCIP [5-bromo-4-chloro-3-indolylphosphate]; Promega) for 20 min at room temperature. The reaction was stopped by washing the membrane several times with distilled water.
Immunization of mice.
Aliquots (100 μl) of purified antigen in sterile physiological saline (100 μg/ml) were mixed with an equal volume of complete Freund adjuvant (Difco, Detroit, Mich.) and injected intraperitoneally into BALB/c mice. For booster immunization, 10 μg of the same antigen in Freund incomplete adjuvant (Difco) was injected into the mice on days 21 and 42. Finally, 3 days before the fusion experiment, the antigen was injected intravenously without adjuvant. Protein concentrations were measured by the method of Lowry et al. (21) with bovine serum albumin (Sigma) as the standard.
Fusion of spleen cells with myeloma cells.
Hybridization of murine spleen cells with myeloma cells (P3X63-Ag8-653, ATCC CRL 1580) was carried out as described previously (16). In brief, the immunized mice were killed, the spleen was removed aseptically, and the cells were mechanically dissociated in Dulbecco modified Eagle medium (Sigma) containing gentamicin (10 μg/ml). Spleen cells were then mixed with myeloma cells growing in the logarithmic phase at a ratio of 5:1. The cells were fused in the presence of 50% polyethylene glycol (PEG-1500; Boehringer Mannheim) while being maintained in a 37°C water bath. Afterward, polyethylene glycol was slowly diluted with cell culture medium without fetal bovine serum (Gibco-BRL, Paisley, United Kingdom). The fused cells were centrifuged at 1,500 × g for 5 min and resuspended in complete Dulbecco modified Eagle medium containing 10% fetal bovine serum. The cells were plated in 96-well tissue culture plates in 100-μl aliquots at a concentration of 2 × 105 cells/well and then incubated in a humidified CO2 incubator (5% CO2 in air) at 37°C. After 24 h of incubation, 100 μl of complete medium containing hypoxanthine, aminopterin, and thymidine (HAT medium) was added to each well. HAT medium was then added to the wells at 1:1 ratios for three consecutive days. Two more HAT medium changes were made at 3-day intervals. After this, the cells were grown in hypoxanthine and thymidine medium for the next 2 weeks, with frequent changes of the same medium. Hybridoma cells were screened for the production of antibodies by Western blot and subcloned by limiting dilution.
Production and characterization of MAbs.
For the production of MAbs, female BALB/c mice (6 weeks old) were injected intraperitoneally with 106 hybridoma cells per mouse. Ten days later, the ascitic fluid was drained by using an 18-gauge needle. The ascites were centrifuged at 1,000 × g for 10 min to remove cells, and the supernatant was rescreened for antibody production by Western blotting. MAbs were purified from collected supernatants by precipitation with 50% saturated ammonium sulfate (pH 7.0) and followed by dialysis against 0.04 M phosphate buffer (pH 6.8). The MAbs were further purified by using a protein G column (Pharmacia). The isotypes of the MAbs were determined by the mouse hybridoma isotyping kit (Boehringer Mannheim).
Immunoreactivity of the MAbs to various enteroviruses, measleviruses, and mumpsviruses.
Antigen recognition abilities of the MAbs were evaluated by indirect immunofluorescence assay (IFA). For IFA, Vero cell monolayers on microscopic slide glasses, which were infected with various enteroviruses, measleviruses, or mumpsviruses, were used as antigens. Before the complete cytopathic effect appeared, the virus-infected cells were fixed for 10 min in ice-cold acetone and stored at −20°C. The hybridomas were cultured for 3 days, and 100 μl of undiluted culture supernatant was reacted with the cells on each slide for 45 min at 37°C. The slides were washed three times with PBS. Fluorescein isothiocyanate-conjugated anti-mouse IgG (Promega) diluted in PBS at 1:20 was added, followed by incubation for 45 min at 37°C. The slides were washed three times with PBS, and the intensity of fluorescence was observed. To validate the sensitivity of MAbs, IFA was performed with four serotypes of enteroviruses: EV6, EV30, CVB3, and CVB5. The titers of reference enteroviruses were determined by the Reed-Muench endpoint procedure (20). The titrated enteroviruses as antigens were inoculated into Vero cells at 10-fold serial dilutions. The cells producing the MAbs were cultured onto 25-cm2 culture flasks. The supernatants with MAbs were then exposed to the serially diluted cells as the first antibody. Fluorescein isothiocyanate-conjugated anti-mouse IgG (Promega), which served as the second antibody, was used for the detection of virus particles inoculated into cells.
RESULTS
Determination of the common epitope region of enteroviruses.
To investigate the common epitope region of enteroviruses, cDNA libraries of Korean isolates E7 and E30 and a prototype CVB3 (Nancy strain) were constructed and immunoscreened by using anti-E7 guinea pig antisera and the antisera from patients with aseptic meningitis. Through repeated immunoscreening, 82 immunopositive cDNA clones were selected. The existence of inserts in 82 positive clones was confirmed by PCR with the λgt11 forward and reverse primers. Upon agarose gel electrophoresis of the PCR products, 61 of 82 clones produced the amplification products that were larger than 182 bp, the intact size of wild-type λgt11 phage DNA that is produced by PCR amplification with the λgt11 forward and reverse primers (data not shown). The PCR products of these 61 clones were directly sequenced, and the location of each clone on the virus genome was determined by comparing sequences of each PCR product with the sequences accessed in the GenBank, EMBL, DDBJ, and PDB databases by using a National Center for Biotechnology Information BLAST search program.
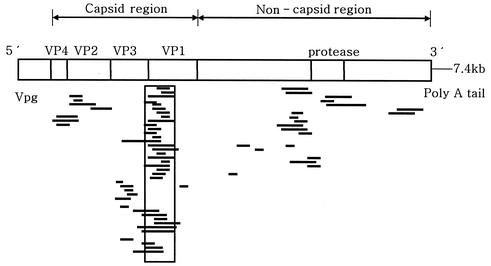
As shown in Fig. 1, 31 of 61 clones were concentrated on the region of nucleotide sequence from 2300 to 2700, which corresponds to a junction of the VP3 and VP1 regions (mainly the N terminus of VP1). Three clones were positioned in VP4, four were positioned in VP2, and eight were positioned in VP3. The remaining clones were in the regions of nonstructural proteins. These results indicate that the N terminus region of VP1 might be a common antigenic site among various serotypes of enteroviruses.
FIG. 1.
Distribution of immunoreactive λgt11 plaques on the full genome of enterovirus. A total of 31 of 61 clones (>50%) were concentrated on the upstream region of VP1 and the region of VP3-VP1 junction, as indicated by a box.
The alignment of deduced amino acid sequences of this region in E7, E11, E30, and CVB3 showed that there are several amino acid motifs that are highly conserved among different serotypes of enteroviruses (Fig. 2). In Fig. 2, the amino acid numbered 1 in the E11 sequence corresponds to the amino acid at position 474 of the enteroviral polyprotein. The immunopositive clones from four serotypes of enteroviruses, which were located at the VP3-VP1 junction, varied in their insert sizes. The clones from E11 and E30 had long inserts encoding ca. 200 amino acids, whereas those of E7 and CVB3 had short inserts encoding 129 and 58 amino acids, respectively. However, all of the clones had the conserved regions including one to two additional regions besides the regions known by others. These results demonstrate that the region of the VP3-VP1 junction would be highly antigenic and conserved.
FIG. 2.
Alignment of amino acid sequences deduced from the DNA sequence analysis of immunoreactive λgt11 clones. Amino acid sequences showing a high degree of homology are boxed. The asterisks indicate the same sequences as the ones of E11, and the dashes indicate missing sequences that were not involved within inserts of the clones.
Cloning and expression of the common epitope region.
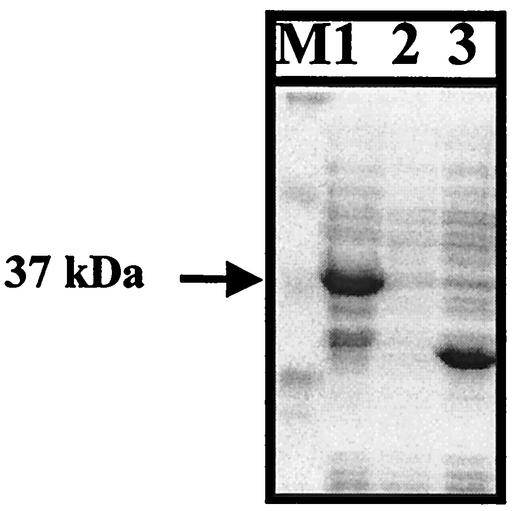
The common epitope region of enteroviruses was amplified with E7-271F and E7-271R primers. The resulting 271-bp product was cloned into a pGEX-5X-1 expression vector and transformed into E. coli BL21(DE3). The expression of cloned gene was induced by adding IPTG and then analyzed by SDS-PAGE, followed by Coomassie blue staining. The molecular mass of the expressed protein was ca. 37 kDa, which is consistent with the expected size (Fig. 3).
FIG. 3.
Recombinant proteins expressed from pE7-271. Cell lysates were separated on an SDS-PAGE gel and stained with Coomassie brilliant blue. An arrow indicates the proteins from pE7-271 induced by IPTG. Lane 1, pE7-271 induced; lane 2, pE7-271 uninduced; lane 3, pGEX-5X-1 induced; M, molecular weight markers (Novex).
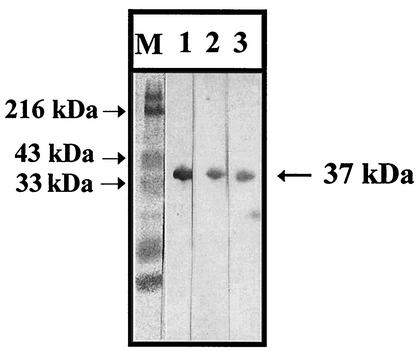
The antigenicity of the expressed protein was analyzed by Western blotting. The expressed protein reacted with guinea pig sera from three different enteroviruses, Korean isolates of E7, E30, and CVB3 (Fig. 4). Additional polyclonal antibodies against CVB2, CVB5, and a prototype CVB3 recognized the protein, whereas normal guinea pig serum or guinea pig serum acquired after RD cell infection did not. Anti-GST antibody also detected the recombinant protein of 37 kDa and the GST protein of 26 kDa. However, none of the sera from enterovirus-inoculated guinea pig reacted with the GST protein (data not shown). These results indicate that the expressed protein would be a common epitope of enteroviruses examined.
FIG. 4.
Western blot analysis of proteins expressed from pE7-271 with polyclonal antibodies against enteroviruses. The anti-enterovirus polyclonal antibodies used were antisera against E30-Korean isolate (lane 1), CVB3-Korean isolate (lane 2), and E7-Korean isolate (lane 3). M, molecular weight markers.
Production and characterization of MAbs against the common epitope.
To produce MAbs specifically reacting with the enteroviruses, mice were immunized with the purified expressed protein. After fusion and HAT selection, 109 hybridomas were derived from three fusion experiments. Of these, 29 were selected on the basis of their reactivity with the anti-E7 guinea pig antisera by Western blotting. Four of the cell lines producing these MAbs were cloned and used for further studies. The isotypes of the four MAbs (named MAb-1, MAb-2, MAb-3, and MAb-4) were IgG1, and they had a kappa light chain (data not shown).
Broad reactivity and specificity of produced MAbs.
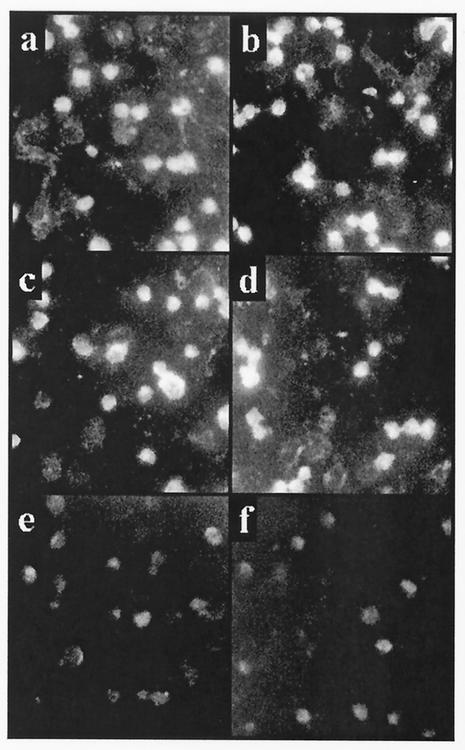
To investigate the reactivity of MAbs with 15 kinds of clinical isolates derived from patients with aseptic meningitis in Korea, IFA was performed. IFA with four MAbs was positive for reactivity with all of the 15 different serotypes of clinical isolates and prototypes of enteroviruses (Table 1). The four MAbs showed strong granular cytoplasmic fluorescence with low background fluorescence even though the intensity of fluorescence obtained with the MAbs varied according to serotypes (Fig. 5). In all cases, definite cytoplasmic staining was easily discernible in cells that were morphologically unaffected by the infection. To validate the specificity, IFA was performed with measlesviruses and mumpsviruses causing the same symptoms, at least in part, as aseptic meningitis.
TABLE 1.
Evaluation of virus recognition ability of the MAbs by IFAa
| MAb | Recognitiona of:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CVB subtype
|
Enterovirus subtype
|
Measles virus | Mumps virus | ||||||||||||||
| 1 | 2 | 3 | 4b | 5c | 5d | 6 | 3 | 4 | 6 | 7 | 9 | 24b | 25 | 30 | |||
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − |
| 3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − |
| 4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − |
Key: +, strong fluorescence; −, weak or no fluorescence.
Purchased from The American Type Culture Collection.
Isolated in 1994.
Isolated in 1997.
FIG. 5.
IFA for the recognition of enteroviral infections in Rd-cdc cells with MAbs against the putative enteroviral common epitope. Cells with light spots indicate that the MAbs could recognize the enteroviral antigens. (a) E3; (b) E9; (c) CVB1; (d) CVB5; (e) mumpsvirus; (f) negative control.
To validate the specificity of the produced MAbs, IFA was performed with measlesviruses and mumpsviruses. As shown in Table 1, the MAbs showed bright fluorescence in enterovirus-infected cells but not in mumpsvirus- and measlesvirus-infected cells. This result indicates that the activity of the MAbs would be specific to enteroviruses. In addition, the sensitivity of the MAbs was also validated through the infection of titrated enteroviruses to susceptible Vero cells. The titers of viruses were determined by the dilution factors in cells that could be recognized by the MAbs. These were determined to be 105.4, 105.8, 106.0, and 105.4 50% tissue culture infective dose(s)/100 μl for E6, E30, CVB3, and CVB5, respectively. The sensitivities of the MAbs were finally determined by using the titrated viruses. That is, the MAbs, which were involved in 100 μl of the supernatant from hybridoma cultures showing 80 to 90% confluency/slide, were reacted with Vero cells infected by the previously titrated enteroviruses, which were diluted 10-fold. We found that with E6, CVB3, and CVB5 at the dilution factor of 101 to 109, whereas the MAbs could react with E30 at 101 to 108. Taken together, these results suggest that the activity of the MAbs would be enterovirus specific and highly sensitive.
DISCUSSION
The genomic sequences conserved in enteroviruses are in the region encoding structural and nonstructural proteins, as shown by primary structure and genetic organization (2, 4, 13). Many studies using various immunological methods have been conducted to define an antigenic homology among serotypes of enteroviruses. It was found by these efforts that the genomic region responsible for the highest antigenicity (or common antigenicity) may reside in VP1, especially in the N terminus of VP1, and plays a predominant role in cross-reactivity among enteroviruses (4, 7, 9, 11, 25, 33).
Previously, computerized antigenicity prediction and selection of short linear epitopes based on the conserved genomic sequences of enteroviruses were popular for predicting the location of viral epitopes. Unfortunately, there is no evidence that either of these methods achieved a high rate of accurate prediction. Although several short linear epitopes of VP1 are apparently useful for the rise in titers of antibodies against several kinds of enteroviruses, these still result in poor reactivity. Cello et al. (3) suggested that these inconsistent results mainly originate from the conformational disadvantages of using short linear peptides.
In the present study, immunoscreening followed by construction of phage libraries with the randomly amplified SISPA products of viral genomes was used to detect a viral epitope with spatial conformation. The cDNA libraries of Korean isolates of E7 and E30 and the prototype of CVB3 were constructed and immunoscreened with anti-E7 guinea pig antisera and convalescent-phase sera from patients with aseptic meningitis to identify a common epitope region of enteroviruses. Sequence analyses of immunopositive clones showed that more than 50% of the positive clones were concentrated at the junction of the N terminus of VP1 and the C terminus of VP3. These results are in agreement with previous studies, which demonstrated that the highest titers of antibody were detected against the epitope consisting of amino acid residues 40 to 53 in the N terminus of VP1. This epitope is highly conserved among enteroviruses (4, 27, 32). Although the C terminus of VP3 also has highly conserved amino acid residues among enteroviruses and the antigenicity of this region has been proven, the common antigenicity of the region is not yet clearly defined (22, 32).
Some positive clones obtained in the present study were also found in the C terminus of VP3; however, most of these clones spanned from the C terminus of VP3 to the N terminus of VP1. Positive clones were also found in VP2, VP3, VP4, and nonstructural protein regions. These observations are in agreement with the previous study that several antigens from VP2, VP3, and VP4 regions are also capable of eliciting an IgG response, even though it has been documented very rarely (36). Thus, the common epitope of enteroviruses appears to be located in the N terminus of VP1.
The common epitope region was cloned and expressed in E. coli. The expressed protein included the amino acid sequence 598-PALTAVEGHTSQV-610 and 626-SRSESSIENFDHK-638, which has been known to be highly conserved sequences among different serotypes of enteroviruses (4, 6, 11, 33). In Fig. 2, this comparative analysis revealed the minor diversity of amino acid sequences within the boxed regions, which have been known to be conserved among enteroviruses. Variations at amino acid positions of the conserved regions, including amino acids 125 to 138 and amino acids 154 to 166, were compared in four serotypes. The amino acid substitutions in the regions, compared to the previous data published by others, occurred in two positions of 130 (E11 [V→C] and CVB3 [V→A]) and 133 (E30 [G→R]) in the sequence of amino acids 125 to 138, and in five positions of 154 (E30 [S→T]), 159 (E7 and CVB3 [S→T]), 164 (E11 and CVB3 [D→L], E30 [D→M]), 165 (E11 [H→S], E30 [D→G], CVB3 [H→C]), and 166 (E11 and CVB3 [K→R]) in the sequence of amino acids 154 to 166. Overall, these results indicate that there would be positions feasible to be substituted by other amino acids even within the conserved regions. Moreover, although the sequence analysis of the amino acids 155 to 166 of CVB3 was not carried out, it is expected that the sequence of CVB3 would be similar to those of other enteroviruses. Thus, the existing sequence databases and data in the present study support consistently that there are highly conserved sequences within the VP3-VP1 adjacent region of enteroviruses with minor modifications.
This N terminus of VP1 is homologous to the N-terminal part of VP1 of PV type 1, which primes or induces a neutralizing antibody response in experimental animals (4, 5, 7). When the PV type 1 is attached to the cell surface, the amino terminus of subsided VP1 is exposed and may be available for the host immune recognition. Similar conformational changes are observed in non-polio enteroviruses when these viruses infect humans (4, 8, 10). Furthermore, VP1 of PV is antigenic to B cells and CD4+ T cells and induces neutralizing antibodies in mice and rabbits (7, 19). Thus, this N terminus of VP1 of PV could be a candidate for a vaccine that stimulates both the cellular immune system of T cells and the humoral immune system of B cells. It is likely that the N terminus of VP1 of non-polio enteroviruses could also be a T-B epitope pair, as in PV.
The expressed recombinant protein containing the structurally stable epitope also reacted with antiviral guinea pig antisera obtained from six different enteroviruses (Korean isolates of E7, E30, CVB2, CVB3, CVB5, and an ATCC strain of CVB3). This indicates that the part of VP1 that was selected and cloned might be useful as a vaccine candidate or as a diagnostic antigen for detecting antibody against a broad range of enteroviruses.
MAbs against the putative common epitope gave strong reactions with 15 different serotypes, which have been endemic in korea during the past decade, and prototypes of enteroviruses. A major application of these MAbs in the diagnosis of enteroviruses may be a rapid identification at the genus level. Being technically very simple, relatively sensitive, and inexpensive, the use of MAbs for immunological methods, including IFA, enzyme-linked immunosorbent assay, and immunoblotting is likely to be superior to nucleic acid hybridization, which has been used conventionally for the confirmation of viruses in clinical specimens. The practical usefulness of MAbs for routine diagnostic work in virological laboratories needs to be tested further with many isolates and clinical specimens.
Acknowledgments
This work was supported by grant R01-2000-00076 from the Basic Research Program of the Korea Science and Engineering Foundation. The DNA thermal cycler used in the present study was financed by a research grant of Chung-Ang University.
REFERENCES
- 1.Abzug, M. J., M. Loeffelholz, and H. A. Rotbart. 1995. Diagnosis of neonatal enterovirus infection by polymerase chain reaction. J. Pediatr. 126:447-450. [DOI] [PubMed] [Google Scholar]
- 2.Casas, I., G. F. Palacios, G. Trallero, D. Cisterna, M. C. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138-148. [PubMed] [Google Scholar]
- 3.Cello, J., A. Samuelson, P. Stalhandske, B. Svennerholm, S. Jeansson, and M. Forsgren. 1993. Identification of group-common linear epitopes in structural and nonstructural proteins of enteroviruses by using synthetic peptides. J. Clin. Microbiol. 31:911-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow, M., R. Yabrov, J. Bittle, J. Holge, and D. Baltimore. 1985. Synthetic peptides from four separate regions of the poliovirus type 1 capsid protein VP1 induce neutralizing antibodies. Proc. Natl. Acad. Sci. USA 82:910-914; 7518-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahllund, L. J., L. Nissinen, T. Pulli, V. P. Hyttinen, Y. G. Stanway, and T. Hypia. 1995. The genome of enterovirus 11. Virus. Res. 35:215-222. [DOI] [PubMed] [Google Scholar]
- 6.Diedrich, S., G. Driesel, and E. Schreier. 1995. Sequence comparison of echovirus type 30 isolates to other enteroviruses in the 5′ noncoding region. J. Med. Virol. 46:148-152. [DOI] [PubMed] [Google Scholar]
- 7.Fiore, L., B. Ridolfi, D. Genovese, G. Buttinelli, S. Lucioli, A. Lahm, and F. M. Ruggeri. 1997. Poliovirus sabine type 1 neutralization epitopes recognized by immunoglobulin a monoclonal antibodies. J. Virol. 71:6905-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haarmann, C. M., P. L. Schwimmbeck, T. Mertens, H. P. Schultheiss, and B. E. Strauer. 1994. Identification of serotype-specific and nonserotype-specific B-cell epitopes of coxsackie B virus using synthetic peptides. Virology 200:381-389. [DOI] [PubMed] [Google Scholar]
- 9.Holge, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 299:1358-1365. [DOI] [PubMed] [Google Scholar]
- 10.Hovi, T., and M. Roivainen. 1993. Peptide antisera targeted to a conserved sequence in poliovirus capsid protein VP1 cross-react widely with members of the genus enterovirus. J. Clin. Microbiol. 31:1083-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyypia, T., T. Hovi, and G. Stanway. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins, O., J. D. Booth, P. D. Minor, and J. W. Almond. 1987. The complete nucleotide sequence of coxsackivirus B4 and its comparison to other members of the picornaviridae. J. Gen. Virol. 68:1835-1848. [DOI] [PubMed] [Google Scholar]
- 13.Jimenez-Clavero, M. A., E. Escribano-Romero, J. M. Sanchez-Vizcaino, and V. Ley. 1998. Molecular cloning, expression and immunological analysis of the capsid precursor polypeptide (P1) from swine vesicular disease virus. Virus. Res. 57:163-170. [DOI] [PubMed] [Google Scholar]
- 14.Kammerer, V., B. Kunkel, and K. Korm. 1994. Nested PCR for specific detection and rapid identification of human picornaviruses. J. Clin. Microbiol. 32:285-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschke, D. L., T. F. Jones, S. C. Buckingham, A. S. Craig, and W. Schaffner. 2002. Outbreak of aseptic meningitis associated with echovirus 13. Pediatr. Infect. Dis. 21:1034-1038. [DOI] [PubMed] [Google Scholar]
- 16.Kohler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495-497. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Leonardi, G. P., A. J. Greenberg, P. Costello, and K. Stabo. 1993. Echovirus type 30 infection associated with aseptic meningitis in Nassau County, New York, USA. Intervirology 36:53-56. [DOI] [PubMed] [Google Scholar]
- 19.Lewis, G. K., and C. Feng. 1992. Intrinsic immunogenecity of an internal VP1 T-B epitope pair of the type 1 poliovirus. Mol. Immunol. 29:1477-1485. [DOI] [PubMed] [Google Scholar]
- 20.Lipson, S. M. 1992. The neutralization test, p. 8.14.1-8.14.8. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, vol. 2. American Society for Microbiology, Washington, D.C.
- 21.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 22.Marttila, J., H. Hyoty, P. Vilja, T. Harkonen, A. Alho, M. Roivainen, T. Hyypia, and J. Ilonen. 2002. T cell epitopes in coxsackievirus B4 structural proteins concentrate in regions conserved between enteroviruses. Virology 15:217-224. [DOI] [PubMed] [Google Scholar]
- 23.Melnick, J. L. 1996. Enteroviruses: polioviruses, coxsackieviruses, ECHO viruses, and newer enteroviruses, p. 549-605. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Press Publishers, Philadelphia, Pa.
- 24.Menegus, M. A. 1985. Enteroviruses, p. 743-746. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 5th ed. American Society of Microbiology, Washington, D.C.
- 25.Mertens, T. H., U. Pika, and H. J. Eggers. 1983. Cross antigenicity among enteroviruses as revealed by immunoblot technique. Virology 129:431-442. [DOI] [PubMed] [Google Scholar]
- 26.Muir, P., F. Nicholson, M. Jhetam, S. Neogi, and J. Banatvala. 1993. Rapid diagnosis of enterovirus infection by magnetic bead extraction and polymerase chain reaction detection of enterovirus RNA in clinical specimens. J. Clin. Microbiol. 31:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palacios, G., I. Casas, A. Tenorio, and C. Freire. 2002. Molecular identification of enterovirus by analyzing a partial VP1 genomic region with different methods. J. Clin. Microbiol. 40:182-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmenberg, A. C. 1989. Sequence alignments of picornaviral capsid proteins, p. 211-241. In B. L. Semler and E. Ehrenfeld (ed.), Molecular aspects of picornavirus infection and detection. American Society for Microbiology, Washington, D.C.
- 29.Pöny, T., L. Kinnunen, T. Hyypiä, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 30.Reyes, G. R., and J. P. Kim. 1991. Sequence-independent, single-primer amplification (SISPA) of complex DNA populations. Mol. Cell. Probes 5:473-481. [DOI] [PubMed] [Google Scholar]
- 31.Rivera, V. M., J. D. Welsh, and J. V. Maizel. 1988. Comparative sequence analysis of the 5′ noncoding region of the eneroviruses and rhinoviruses. Virology 165:42-50. [DOI] [PubMed] [Google Scholar]
- 32.Roivainen, M., A. Narvanen, M. Korkolainen, M. L. Huhtala, and T. Hovi. 1991. Antigenic regions of poliovirus type 3/sabin capsid proteins recognized by human sera in the peptide scanning technique. Virology 80:99-107. [DOI] [PubMed] [Google Scholar]
- 33.Samuelson, A., B. Johansson, and M. Forsgren. 1995. Sequence analysis of echoviruses in a major antigenic region eliciting enteroviral cross-reactive antibodies. Arch. Virol. 140:2085-2091. [DOI] [PubMed] [Google Scholar]
- 34.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA Sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanway, G. 1990. Structure, function, and evolution of picornaviruses. J. Gen. Virol. 71:2483-2501. [DOI] [PubMed] [Google Scholar]
- 36.Torfason, E. G., R. Galindo, and H. L. Keyserling. 1988. Comparison of five ELISA assays for IgG antibody against coxsackievirus B1. J. Med. Virol. 25:53-60. [DOI] [PubMed] [Google Scholar]
- 37.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization. 1990. Manual for the virological investigation of poliomyelitis, p. 37-41. World Health Organization, Geneva, Switzerland.
- 39.Yosef, G. E., I. N. Brown, and J. F. Mowbray. 1987. Derivation and biochemical characterization of an enterovirus group-specific monoclonal antibody. Intervirology 28:163-170. [DOI] [PubMed] [Google Scholar]
- 40.Zoll, G. J., W. J. G. Melchers, H. Kopecka, G. Jambroes, H. J. A. van der Poel, and J. M. D. Galama. 1992. General primer-mediated polymerase chain reaction for detection of enteroviruses: application for diagnostic routine and persistent infections. J. Clin. Microbiol. 30:160-165. [DOI] [PMC free article] [PubMed] [Google Scholar]