Abstract
We describe a rapid and reproducible PCR assay for quantitation of the Candida albicans ribosomal DNA (rDNA) in clinical blood samples based on the TaqMan principle (Applied Biosystems), in which a signal is generated by cleavage of a template-specific probe during amplification. We used two fluorogenic probes based on universal, fungus-specific primers, one for the detection of C. albicans species DNA and one for the detection of all Candida genus DNA. C. albicans blastoconidia mixed with whole blood in a titration experiment yielded a linear PCR signal over a range of 3 orders of magnitude. The TaqMan-based PCR assay for C. albicans exhibited a low limit of detection (5 CFU/ml of blood) and an excellent reproducibility (96 to 99%). While the C. albicans species-specific probe had 100% specificity for C. albicans, all Candida genus-specific probes cross-reacted with other organisms likely to coinfect patients with C. albicans infections. On the basis of these data, we determined the C. albicans loads with a species-specific probe from 122 blood samples from 61 hematology or oncology patients with clinically proven or suspected systemic Candida infections. Eleven positive samples exhibited a wide range of C. albicans loads, extending from 5 to 100,475 CFU/ml of blood. The sensitivity and specificity of the present assay were 100 and 97%, respectively, compared with the results of blood culture. These data indicate that the TaqMan-based PCR assay for quantitation of C. albicans with a species-specific probe provides an attractive alternative for the identification and quantitation of C. albicans rDNA in pure cultures and blood samples.
Invasive candidiasis, a common cause of nosocomial infection, is a leading cause of infections among patients receiving bone marrow transplantations or those with leukemia or other cancers and is also associated with a high rate of morbidity and mortality (10, 18, 22). Given the rapidly fatal course of candidiasis, there is a need for improved methods for early diagnosis and subsequent initiation of antifungal therapy in order to have a significant impact on the death rate (1, 12). One such improvement is the application of rapid and sensitive methods based on PCR that enable the amplification and quantitation of a broad range of fungal pathogens directly from specimens and pure cultures.
Two quantitative real-time PCR systems have been described for Candida detection. An assay based on the PCR LightCycler system (Roche Molecular Diagnostics Systems, Indianapolis, Ind.) was applied by Löeffler et al. (15) for quantification of Candida albicans DNA in blood to which known numbers of blastoconidia had been added. Those investigators also used the assay to determine the fungal burdens in a small number of blood samples taken from patients with hematological malignancies. More recently, Guiver et al. (7) and Bowman et al. (3) described the automated detection of fungal DNA by using the TaqMan assay (Applied Biosystems, Foster City, Calif.) with clinical isolates and murine tissues, respectively. This assay, which takes advantage of the 5′-3′ nucleolytic activity of Taq polymerase (9), has been evaluated to quantitatively estimate the C. albicans species burdens in various mouse tissues, to monitor disease progression, and to measure antifungal drug efficacy (3). However, to our knowledge this assay has not yet been adapted to determination of the fungal load in clinical blood samples.
Here we describe a rapid and reproducible method for determination of the C. albicans ribosomal DNA (rDNA) loads in blood samples by using the TaqMan technology combined with the GeneAmp 5700 real-time sequence detection system (Applied Biosystems) (6, 8). The PCR assay based on the TaqMan principle generates a signal by cleaving a template-specific hybridization probe during amplification (9, 13). We first evaluated the sensitivity of this assay by adding C. albicans blastoconidia to blood, extracting total DNA, and demonstrating a linear response over a range of 3 orders of magnitude with a low limit of detection. We also evaluated the specificities of two fluorogenic probes (one C. albicans species-specific probe and one all Candida genus-specific probe) and the reproducibility and linear range of the TaqMan-based PCR assay. Finally, this assay was tested with template DNA prepared directly from 122 blood samples from hematology or oncology patients with either disseminated candidiasis or suspected disseminated candidiasis, and the results were compared with those of standard blood culture and identification methods.
MATERIALS AND METHODS
Fungal culture.
C. albicans strain ATCC 24433 was cultured on Sabouraud-glucose-agar for 72 h at 30°C. Serial dilutions of fungal cells were prepared with sterile saline suspensions that were adjusted to a 0.5 McFarland standard (which is approximately 106 CFU/ml).
Clinical samples.
During the study period, a total of 122 clinical blood specimens from 61 patients were analyzed. Samples were obtained from patients hospitalized in our institution with clinically proven or suspected systemic Candida infection. The patients' symptoms and characteristics included persistent fever, unresponsiveness to broad-spectrum antibiotic therapy, specimen positivity by histology, the isolation of Candida from blood or other sterile sites, colonization of multiple sites with Candida, and repeated computed tomography and ultrasound scans suggesting a mycotic lesion (e.g., in the liver, spleen, or lung). All blood cultures were evaluated in the BACTEC 9600 blood culture system (Becton Dickinson Diagnostic Instrument Systems, Sparks, Md.), which detected microbial growth by continuous monitoring. Blood (8 to 10 ml) was inoculated into two culture vial types, BACTEC Plus aerobic/F (enriched soybean-casein digest broth) and BACTEC Plus anaerobic/F (prereduced enriched soybean-casein digest broth), for aerobic and anaerobic cultures, respectively. EDTA-anticoagulated whole-blood specimens (5 ml) for PCR assay were taken in parallel from each patient. Blood cultures were considered negative if they remained negative after 14 days of incubation in the automated BACTEC system. Thirty-six control blood samples from patients without clinical evidence of an invasive fungal infection were also tested.
Sensitivity of TaqMan assay.
For the testing of sensitivity, whole blood from healthy volunteers was artificially spiked in a titration experiment with C. albicans blastoconidia to final concentrations of 103, 102, 101, and 5 cells per ml (in serial dilutions). DNA was extracted from the samples and analyzed.
DNA extraction.
The DNA extraction procedure used was a modification of a method described previously (5); resuspension and incubation of the fungal pellet in 50 mM NaOH and neutralization with 1 M Tris-HCl (pH 7) were not necessary, and the rates of centrifugation for the leukocyte lysis and spheroplast genesis steps were reduced to 1,500 and 2,000 × g, respectively. Briefly, after the hypotonic lysis of erythrocytes in 5 ml of EDTA-anticoagulated blood samples with erythrocyte lysis buffer (10 mM Tris [pH 7.6], 5 mM MgCl2, 10 mM NaCl) and the enzymatic lysis of leukocytes with leukocyte lysis buffer (10 mM Tris [pH 7.6], 10 mM EDTA, 50 mM NaCl, 0.2% sodium dodecyl sulfate, 200 μg of proteinase K [Roche Diagnostics, Mannheim, Germany] per ml), the spheroplasts were directly generated by incubation of the pellets with 500 μl of lyticase buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 0.2% β-mercaptoethanol, 1 U of recombinant lyticase [ICN Biomedicals, Aurora, Ohio] per 100 μl) for 45 min at 37°C. Finally, spheroplasts lysis and DNA extraction were accomplished with the QIAmp Tissue kit (Qiagen, Hilden, Germany), according to the instructions of the manufacturer. The DNA recovered in 100 μl of elution buffer was immediately analyzed or stored at −20°C until testing.
Amplification and quantitation of rDNA by the TaqMan assay.
The universal fungal amplification primers ITS86 and ITS4 (21) are complementary to conserved sequences in 5.8S rDNA and 28S rDNA, respectively. Two fluorogenic probes designed by Shin et al. (20) were used in this study: CA-FAM (where FAM is 6-carboxyfluorescein) for the detection of C. albicans species DNA and All-CAN-TET (where TET is tetrachloro-6-craboxyfluorescein), an all Candida genus-specific probe for the detection of the DNA of the five major Candida species. All of these oligonucleotides were synthesized by Applied Biosystems.
DNA samples were analyzed by using the GeneAmp 5700 sequence detection system (Applied Biosystems). Each 25 μl of the TaqMan PCR mixture consisted of a 5-μl aliquot of sample DNA, 1× PCR buffer, 3.5 mM MgCl2, 0.2 μM primer ITS86, 0.2 μM primer ITS4, each deoxyribonucleoside triphosphate at a concentration of 0.2 mM, 0.2 μM CA-FAM or All-CAN-TET probe, 2 U of Taq DNA polymerase (Roche Diagnostics), and 0.5 μl of 6-carboxy-“x”-rhodamine (ROX) passive reference dye (Invitrogen, Groningen, The Netherlands). Thermal cycling conditions consisted of heating at 94°C for 10 min, which preceded a two-stage temperature profile of 30 s at 95°C and 1 min and 30 s at 60°C for 40 cycles. Reactions are characterized by the time during cycling when a threshold of baseline fluorescence (CT) is exceeded; CT predicts the quantity of input target (8).
In each reaction run, C. albicans species genomic DNA was serially diluted over a range of 4 logs (1,000 to 0.05 pg DNA, corresponding to 105 to 5 CFU) to enable the generation of a standard curve. The latter is generated by plotting the CT values versus log10(N), where N is the concentration of the standard. The C. albicans DNA level in each blood sample is determined by locating its CT on the standard curve. Negative controls were tested by using the same PCR mixture under the amplification conditions described above but without template DNA.
Genomic DNA.
Genomic DNA for use in the TaqMan PCR specificity studies was extracted from C. albicans, Candida glabrata, Candida guilliermondii, Candida krusei, Candida norvegensis, Candida parapsilosis, Candida tropicalis, Cryptococcus neoformans, Saccharomyces cerevisiae, Aspergillus flavus, Aspergillus fumigatus, Aspergillus nidulans, Aspergillus niger, Penicillium spp., Fusarium spp., Scedosporium apiospermum, Alternaria spp., Curvularia spp., Hortaea werneckii, Auxarthron spp., Chaetomium spp., Phialophora verrucosa, Epiconum purpurescens, Haemophilus influenzae, Klebsiella spp., Proteus mirabilis, Morganella morganii, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia. The blood lysis step described above was omitted from the DNA extraction steps for yeasts and molds. Isolation of genomic DNA from bacterial cultures was accomplished with the QIAmp Tissue kit according to the instructions of the manufacturer. The specificities of the C. albicans species-specific and the Candida genus-specific probes were also tested with purified DNA from a human lymphocyte cell line (RAJI) (11) and DNA extracted with the QIAmp DNA Blood Mini kit (Qiagen) from blood samples from cytomegalovirus-positive patients.
RESULTS
Sensitivity of detection of known numbers of C. albicans blastoconidia.
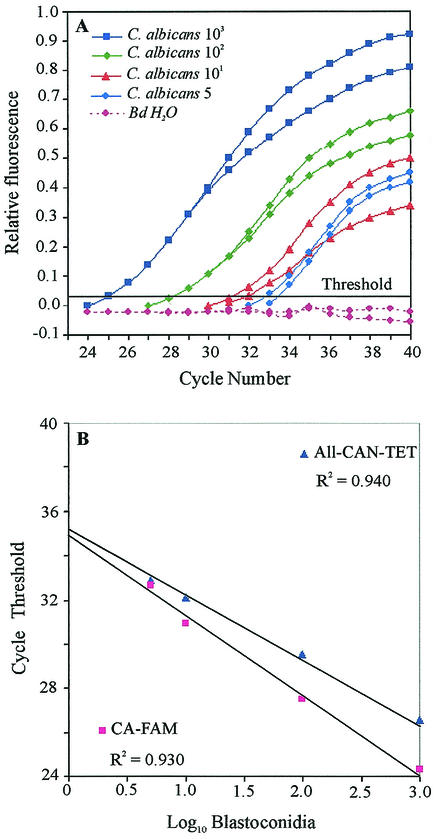
To determine the sensitivity limit of the TaqMan PCR assay with a pair of universal fungal primers in conjunction with either a C. albicans species-specific probe (CA-FAM) or an all Candida genus-specific probe (All-CAN-TET), whole blood from healthy volunteers was artificially mixed with C. albicans blastoconidia (103 to 5 cells per ml). The genomic DNA extracted from these samples was then used as a template, as described in Materials and Methods. Figure 1A shows the levels at which the CT values were determined by using the CA-FAM probe.
FIG. 1.
Quantitation of C. albicans blastoconidia in seeded human blood specimens and comparative sensitivities of DNA detection by using the CA-FAM and All-CAN-TET probes. Serially diluted C. albicans blastoconidia were mixed with blood, and the extracted DNA was used in duplicate as the template in the TaqMan assay. (A) The relative fluorescence refers to the increase in the fluorescence emission of the reporter dye relative to that of the passive reference dye. The threshold fluorescence was determined by using the CA-FAM probe. No signal was obtained when double-distilled water was used as a negative control. (B) Log10 concentration of blastoconidia plotted against the mean CT values generated from the amplification plots in panel A (CA-FAM probe) and those generated with the All-CAN-TET probe.
In this titration series, the signal generated from both probes during these PCRs was linear over a range of 3 orders of magnitude (Fig. 1B). At Candida concentrations above 5 CFU/ml of blood, CT values for the CA-FAM probe were lower than those for the All-CAN-TET probe. Moreover, when the latter probe was used, almost all genomic DNA extracts prepared from unspiked samples used as negative controls displayed a signal in the TaqMan assay (calculated mean ± standard deviation CT value for whole blood from 15 healthy volunteers, 36.60 ± 1.50). In contrast, no signal was detected when the CA-FAM probe was used in these reactions with the negative controls (CT values, 40). The higher baseline value calculated for the All-CAN-TET probe resulted in an identical limit of detection for each fluorescent probes, which was 5 CFU/ml of blood. This corresponded to the sensitivity of amplification of fungal DNA with the LightCycler system (15).
Specificities of the C. albicans species-specific and Candida genus-specific probes.
The specificities of the two DNA fluorescent probes were tested in a TaqMan assay with a range of Candida strains and other fungal, bacterial, and viral pathogens; a human lymphocyte cell line was also analyzed. The CA-FAM probe correctly identified 100% of the corresponding strains without cross-reaction with DNA purified from heterologous species (Table 1). In contrast, the All-CAN-TET probe identified all Candida species analyzed, S. cerevisiae, A. nidulans, Penicillium spp., A. flavus, A. fumigatus, Curvularia spp., Fusarium spp., Alternaria spp., H. werneckii, P. verrucosa, E. purpurescens, A. niger, Auxarthron spp., C. neoformans, S. apiospermum, and Chaetomium spp. No signal was obtained in the TaqMan PCR when bacterial, viral, or human DNA was used as the template.
TABLE 1.
Comparative specificity of DNA detection by using the C. albicans species-specific and all-Candida genus-specific fluorescent probes
| Genomic DNAa |
CTb
|
|
|---|---|---|
| CA-FAM | All-CAN-TET | |
| S. cerevisiae | − | 16.91 ± 0.14 |
| C. parapsilosis | − | 20.22 ± 0.20 |
| C. albicans | 17.54 ± 0.11 | 20.23 ± 0.27 |
| A. nidulans | − | 20.31 ± 0.26 |
| Penicillium spp. | − | 20.92 ± 0.07 |
| C. krusei | − | 21.28 ± 0.10 |
| C. guilliermondii | − | 21.29 ± 0.18 |
| C. glabrata | − | 21.57 ± 0.03 |
| C. tropicalis | − | 21.64 ± 0.29 |
| C. norvegensis | − | 21.67 ± 0.21 |
| A. flavus | − | 21.84 ± 0.13 |
| A. fumigatus | − | 22.73 ± 0.24 |
| Curvularia spp. | − | 22.74 ± 0.01 |
| Fusarium spp. | − | 23.20 ± 0.19 |
| Alternaria spp. | − | 24.05 ± 0.07 |
| H. werneckii | − | 24.45 ± 0.16 |
| P. verrucosa | − | 24.54 ± 0.21 |
| E. purpurescens | − | 24.77 ± 0.16 |
| A. niger | − | 25.30 ± 0.14 |
| Auxarthron spp. | − | 26.20 ± 0.11 |
| C. neoformans | − | 26.36 ± 0.18 |
| S. apiospermum | − | 26.39 ± 0.22 |
| Chaetomium spp. | − | 32.19 ± 0.01 |
| Klebsiella spp. | − | − |
| H. influenzae | − | − |
| M. morganii | − | − |
| S. maltophilia | − | − |
| P. mirabilis | − | − |
| P. aeruginosa | − | − |
| Human lymphocyte line | − | − |
| Cytomegalovirus | − | − |
One nanogram of purified DNA was analyzed.
Values are expressed as means ± standard deviations. Data are from triplicate samples from two to three experiments. A negative value was assigned for all CT values equal to 40.
Reproducibility.
Amplification of DNA from C. albicans was performed with 1,000, 5, 0.50, 0.25, and 0.05 pg of target DNA per reaction (corresponding to 105, 500, 50, 25, and 5 CFU, respectively) in six parallel experiments. The mean ± standard deviation CT values for these experiments with DNA from identical extractions were as follows: for 1,000 pg, 20.27 ± 0.41; for 5 pg, 28.18 ± 0.44; for 0.50 pg, 32.17 ± 0.49; for 0.25 pg, 34.04 ± 0.51; and for 0.05 pg, 35.62 ± 0.54. These are equivalent to standard errors of 0.17, 0.18, 0.20, 0.21, and 0.22, respectively. The same concentrations of C. albicans genomic DNA used as external standards (1,000 to 0.05 pg) were used for the quantitation of DNA in clinical samples.
Linear range.
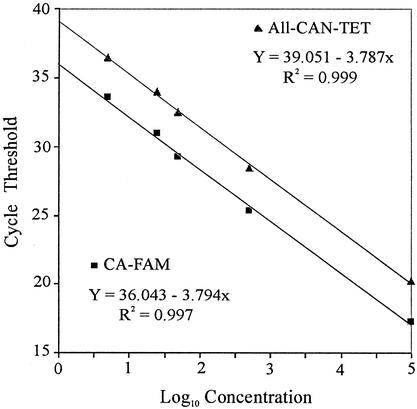
The linear range of the TaqMan system for the quantitation of C. albicans was determined by analysis of two identical dilution series of C. albicans genomic DNA that had been amplified by using either the CA-FAM or the All-CAN-TET probe. Both probes yielded different regression lines: the slopes were 3.794 and 3.787, respectively; the intercepts were 36.043 and 39.051, respectively; and the correlation coefficients (R2 values) of the curves were 0.997 and 0.999, respectively (Fig. 2). Because the slopes were, on average, equivalent and the difference between the two intercepts was approximately 3, detection of the DNA standard with the All-CAN-TET probe within the linear range was overestimated by a factor of approximately 7 (0.84 log10). This result indicates that species-specific probes provide the basis for the very accurate quantitation of C. albicans DNA by the TaqMan assay.
FIG. 2.
Linear range for the TaqMan assay for C. albicans by using the CA-FAM and All-CAN-TET probes. Two identical dilution series of C. albicans DNA (1,000 to 0.05 pg DNA, corresponding to 105 to 5 CFU) were used as external standards. Each dilution was tested in triplicate. Linear regression analysis was performed on the mean CT values for each dilution series plotted against the log10 concentration of standard DNA.
Quantitation of DNA in clinical samples.
The ability of the TaqMan assay to measure the C. albicans rDNA loads in 122 blood samples from 61 patients was assessed. Eleven samples tested displayed a signal by TaqMan analysis (Table 2). The C. albicans rDNA load ranged between 5 and 100,475 CFU/ml of blood when the results were compared to those for a DNA standard by using the CA-FAM probe. Six positive samples had weak to moderate C. albicans loads (<30 CFU/ml of blood), three had elevated C. albicans loads (>100 CFU/ml of blood), and two had highly elevated C. albicans loads (>1,000 CFU/ml of blood). Finally, 36 control blood samples from patients without clinical evidence of an invasive fungal infection tested negative for C. albicans by the TaqMan assay with the CA-FAM probe. Thirteen culture bottles with blood from these patients showed that the patients had bacteremia (caused by Lactobacillus spp. [n = 1], Enterococcus spp. [n = 2], Fusobacterium spp. [n = 3], and Staphylococcus epidermidis [n = 7]).
TABLE 2.
Efficiency of TaqMan-based detection with CA-FAM probe of C. albicans DNA from patient blood samples
| Diagnosisa | TaqMan result (CFU/ml) | Blood culture result | Blood originb |
|---|---|---|---|
| AML | 5 | Negativec | PAC |
| AML | 5 | Positive | PER |
| SCLC | 6 | Positive | PER |
| Gastric tumor | 7 | Positive | PER |
| AML and breast tumor | 17 | Negative | PER |
| AML | 27 | Negativec | PER |
| AML | 120 | Negativec | PAC |
| Colon tumor | 339 | Positive | Unknown |
| Colon tumor | 525 | Positive | PAC |
| Pancreas tumor | 3,760 | Positive | CVC |
| Metastatic breast tumor | 100,475 | Positive | PAC |
AML, acute myeloid leukemia; SCLC, small-cell lung cancer.
PAC, Port-a-cath; PER, peripheral blood; CVC, central venous catheter.
The three blood culture samples were from the same patient.
Three blood specimens, which were culture negative, obtained from a single patient were identified as containing C. albicans by the TaqMan assay (5, 27, and 120 CFU/ml of blood; Table 2). This patient had received an allogeneic bone marrow transplant, had been severely neutropenic for several weeks, and had received prophylactic fluconazole, which may suppress Candida growth but not completely eradicate the organism. Of the patients who exhibited very high C. albicans loads (3,760 and 100,475 CFU/ml of blood) and in whom candidemia was documented by culture, one had a documented catheter infection with a large number of positive blood cultures and a larger number of colonies from catheter samples compared to the colony counts from peripheral blood (Isolator quantitative culture), and the second had extremely high C. albicans loads and a documented catheter infection, but only one of four blood cultures was positive.
Fifteen (12%) of the 122 blood cultures were positive for Candida spp.: 6 for C. albicans, 3 for C. glabrata, 2 for C. tropicalis, 1 for C. guilliermondii, 1 for Candida lipolytica, 1 for C. lusitaniae, and 1 for a mixture of C. albicans and C. glabrata. Eleven (9%) EDTA-anticoagulated blood specimens were positive by the TaqMan assay with the CA-FAM probe; six of these were culture positive for C. albicans, one was culture positive for C. albicans and C. glabrata, and four remained negative by blood culture. Among the 44 (36%) blood specimens that were TaqMan positive with the All-CAN-TET probe, 29 were culture negative for Candida spp. and 15 were culture positive for Candida spp. Compared with the results of blood culture, the sensitivity and specificity of the TaqMan PCR assay with the CA-FAM probe observed with blood samples were 100 and 97%, respectively; those for the TaqMan PCR assay with the All-CAN-TET probe were 100 and 72%, respectively. Finally, for both probes, the TaqMan assay had a very high negative predictive value of 100%.
DISCUSSION
Most commercial or in-house fungal DNA quantitative methods are based on DNA amplification with conventional thermocyclers, followed by hybridization with biotin- or digoxigenin-labeled probes. These methods use end-point measurement to quantify the amount of amplified product. At any given cycle within the exponential phase of PCR, but not during the plateau, the amount of product is proportional to the initial amount of template. In contrast, the real-time TaqMan PCR assay for the quantitation of C. albicans rDNA enables accurate calculation of the amount of PCR product at a point in the early exponential phase of the reaction (CT) and eliminates the concern that a reaction plateau will be reached at different cycles. This allows determination of the initial fungal rDNA load from the amount of PCR product by reference to an external standard.
By using the TaqMan assay, the smallest amount of C. albicans DNA tested and reliably quantified is 5 CFU/ml of blood with either the species-specific or the genus-specific probe. In practice, however, a relatively high fluorogenic signal was displayed by unspiked whole blood from healthy volunteers. This drawback was associated solely with the all Candida genus-specific probe and can result in false-positive reactions. To circumvent this, the All-CAN-TET probe was assigned a higher baseline value, which, following PCR, enables differentiation of PCR-specific and nonspecific products. This result was in accordance with those reported by Shin et al. (20), who applied a 5′ exonuclease assay to quantify medically significant species of Candida from blood culture bottles using fluorescent DNA probes and luminescence spectrometric detection. The investigators used a positive cutoff value for the All-CAN-TET probe approximately five times higher than the cutoff value for the CA-FAM probe.
The TaqMan assay for C. albicans quantitation exhibited a good reproducibility of 96% when the CT values for dilution series of six different extracts of purified C. albicans DNA in the range of 1,000 to 0.05 pg were analyzed. Moreover, triplicate amplifications with each dilution performed in each assay allow us to calculate a high reproducibility of 99%. A similar reproducibility was exhibited by another recently described quantitative test for Candida with the LightCycler instrument (15).
The specificities of the probe sets used in real-time PCR assays for C. albicans detection are often established by using closely related species and DNA of human origin (7, 15). In our study, the CA-FAM probe had 100% specificity for C. albicans. In contrast, the All-CAN-TET probe cross-hybridized with DNA purified from other species such as S. cerevisiae, A. fumigatus, A. flavus, and Fusarium spp. We recently reported similar cross-reactivities between this digoxigenin-labeled all Candida genus-specific probe (by PCR-enzyme-linked immunosorbent assay) and DNA from Fusarium spp. using colorimetric detection of amplified products from clinical blood samples (19). In addition, in a 5′ exonuclease assay for Candida, the All-CAN-TET probe was shown to hydridize with products amplified from S. cerevisiae, A. fumigatus, and A. flavus (20). The investigators recommended further analysis with S. cerevisiae- and Aspergillus-specific probes to differentiate between these species and Candida species. Hence, a positive PCR result with the all Candida genus-specific probe does not exclude the presence of species other than the common Candida species, and the concomitant use of species-specific probes is necessary.
Among the most important factors that limit the direct application of the all Candida genus-specific probe to blood samples is its design from the highly conserved 5.8S region of rDNA (16, 20). Recently, human DNA-derived sequences were detected in blood specimens from healthy individuals by using real-time PCR with a probe targeting conserved regions of bacterial 16S rDNA (17). In our study with blood samples from individuals without clinical signs or symptoms suggesting infection or other disease, the threshold was generally exceeded during later cycles of amplification with the all Candida genus-specific probe. A preliminary search of GenBank showed a region of similarity between the sequence of this probe and that of 5.8S human rDNA (data not shown). We are in the process of creating clone libraries from PCR products amplified from blood. When the sequences of these clones match human DNA-derived sequences, redesign of a new all Candida-specific probe permitting sole detection of the major clinically important species of Candida can be achieved by use of the sequence from the less conserved internal transcribed spacer 2 region, between the 5.8S rRNA gene and the 28S rRNA gene. This region is distinctive and has great potential for use as a taxonomic tool (4, 21). The redesign of all Candida genus-specific probes can also be performed by use of the recently commercialized TaqMan Minor Groove Binder (Applied Biosystems), which enhances the melting temperature of the oligonucleotide, resulting in shorter probe sequences.
The greatest impact of the real-time PCR assay on routine diagnostic assays may be the saving of time from the time of sample collection to the time of a final diagnosis. As the fungal DNA extraction step is still a limiting factor, requiring more than half of a working day, this PCR assay reduces the time to achieve a final result by several hours, allowing a report to be made on the same day that the test is performed. The detection of fungemia by conventional culture methods can be difficult. It is generally attributed to very small fungal loads in clinical samples (2, 14). Recently, in a preliminary study that used the LightCycler PCR assay, quantitation of fungal DNA showed that two of nine PCR-positive patients with hematological malignancies had fungal loads of more than 100 CFU/ml of blood (15). In our study, the species-specific probe correctly determined C. albicans loads of more than 100 CFU/ml of blood in 5 of 11 PCR-positive hematology or oncology patients. It is interesting that the two samples exhibiting the highest C. albicans loads (>1,000 CFU/ml of blood) were both from patients with documented catheter infections, one of the most common conditions associated with fungemia. Moreover, the positive “gold standard” for C. albicans detection is difficult to define, because most disseminated infections are not detected until death and cultures might be falsely negative, for instance, because of the use of antifungal agents. In the present study, all four samples with positive PCR results and negative culture results were obtained from patients while they were receiving antifungal treatment. Given its high sensitivity, a prospective clinical study of the TaqMan-based PCR assay for the identification of patients at risk for invasive fungal infections is under way.
REFERENCES
- 1.Beck-Sagué, C., and W. R. Jarvis, et al. 1993. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980-1990. J. Infect. Dis. 167:1247-1251. [DOI] [PubMed] [Google Scholar]
- 2.Borst, A., M. A. Leverstein-Van Hall, J. Verhoef, and A. C. Fluit. 2001. Detection of Candida spp. in blood cultures using nucleic acid sequence-based amplification (NASBA). Diagn. Microbiol. Infect. Dis. 39:155-160. [DOI] [PubMed] [Google Scholar]
- 3.Bowman, J. C., G. K. Abruzzo, J. W. Anderson, A. M. Flattery, C. J. Gill, V. B. Pikounis, D. M. Schmatz, P. A. Liberator, and C. M., Douglas. 2001. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob. Agents Chemother. 45:3474-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y. C., J. D. Eisner, M. M. Katar, S. L. Rassoulian-Barrett, K. LaFe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeast using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Einsele, H., H. Hebart, G. Roller, J. Löeffler, I. Rothenhöfer, C. A. Muller, A. Bowden, J. A. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelfand, D. H., P. M. Holland, R. K. Saiki, and R. M. Watson. May 1993. U.S. patent 5,210,015.
- 7.Guiver, M., K. Levi, and B. A. Oppenheim. 2001. Rapid identification of Candida species by TaqMan PCR. J. Clin. Pathol. 54:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heid, C., J. Stevens, K. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 9.Holland, P. M., R. D. Abramson, R. M. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn, R., B. Wong, T. Kiehn, and D. Armstrong. 1985. Fungemia in a cancer hospital: changing frequency, early onset and results of therapy. Rev. Infect. Dis. 7:646-655. [DOI] [PubMed] [Google Scholar]
- 11.Hovanes, K., T. W. Li, and M. L. Waterman. 2000. The human LEF-1 gene contains a promoter preferentially active in lymphocytes and encodes multiple isoforms derived from alternative splicing. Nucleic Acids Res. 28:1994-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis, W. R. 1995. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin. Infect. Dis. 20:1526-1530. [DOI] [PubMed] [Google Scholar]
- 13.Livak, K. J., S. J. A. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 14.Löeffler, J., H. Hebart, S. Magga, D. Schmidt, L. Klingspor, J. Tollemar, U. Schumacher, and H. Einsele. 2000. Identification of rare Candida species and other yeasts by polymerase chain reaction and slot blot hybridization. Diagn. Microbiol. Infect. Dis. 38:207-212. [DOI] [PubMed] [Google Scholar]
- 15.Löeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfert and the Light Cycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lott, T. J., R. J. Kuykendall, and E. Reiss. 1993. Nucleotide sequence analysis of the 5.8S rDNA and adjacent ITS2 region of Candida albicans and related species. Yeast 2:1199-1206. [DOI] [PubMed] [Google Scholar]
- 17.Nikkari, S., I. J. McLaughlin, W. Bi, D. E. Dodge, and D. A. Relman. 2001. Does blood of healthy subjects contain bacterial ribosomal DNA? J. Clin. Microbiol. 39:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittet, D., and R. P. Wenzel. 1995. Nosocomial bloodstream infections. Arch. Intern. Med. 155:1177-1184. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Villalobos, H., M. Aoun, C. Heymans, J. M. De Bruyne, V. Duchateau, J. M. Verdebout, and F. Crokaert. 2002. Cross reaction between a pan-Candida genus probe and Fusarium spp. in a fatal case of Fusarium oxysporum pneumonia. Eur. J. Clin. Microbiol. Infect. Dis. 21:149-152. [DOI] [PubMed] [Google Scholar]
- 20.Shin, J. H., F. S. Nolte, B. P. Holloway, and C. J. Morrison. 1999. Rapid identification of up to three Candida species in a single reaction tube by a 5′ exonuclease assay using fluorescent DNA probes. J. Clin. Microbiol. 37:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turenne, C. Y., S. E. Sanche, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whimbey, E., T. Kiehn, P. Brannon, A. Blevins, and D. Armstrong. 1987. Bacteremia and fungemia in a patient with neoplastic diseases. Am. J. Med. 82:723-730. [DOI] [PubMed] [Google Scholar]