Abstract
We conducted a meta-analysis to assess the performance of PCR for the diagnosis of smear-negative pulmonary tuberculosis (SPT) and to identify factors that account for differences in the diagnostic accuracy of different studies. Studies published before February 2002 were included if sensitivity and specificity of PCR in smear-negative respiratory or gastric-aspirate specimens could be calculated. Analysis was conducted by using summary receiver operating characteristics models. Sensitivity and specificity ranged from 9 to 100% and from 25 to 100%, respectively. Fewer than 40% of the 50 studies reported results by number of patients, reported clinical characteristics of patients, or used as a reference standard combined culture and clinical criteria. Studies that included bronchial specimens showed higher accuracy than studies that evaluated only sputum specimens or included gastric aspirates. Studies that did not report that tests were applied blindly showed higher accuracy than those reporting blind testing. Increased sensitivity due to the use of DNA purification methods was associated with decreased specificity. Studies published after 1995, using Amplicor or dUTP-UNG, were associated with an increase in specificity at the expense of lower sensitivity. We concluded that PCR is not consistently accurate enough to be routinely recommended for the diagnosis of SPT. However, PCR of bronchial specimens could be useful in highly suspicious SPT cases. Studies not reporting blind testing are likely to overestimate accuracy of PCR. Future evaluation of PCR accuracy should be conducted by patient and type of respiratory specimen, blindly, by using a reference standard that combines culture and clinical criteria and addresses the issue of how patient characteristics affect PCR accuracy.
Tuberculosis remains an important public health problem worldwide, accounting for ca. 8.0 million new cases per year (25). Smear-negative cases pose an important public health hazard and burden, accounting for as much as 17% of Mycobacterium tuberculosis transmission (11). Some patients convert to smear positivity, leading to a more severe morbidity (20). Approximately 20 to 50% of patients with pulmonary tuberculosis are smear negative, and 10% of these patients are culture negative (6, 25).
PCR reduces the time required for the identification of the Mycobacterium and may enhance the detection of smear-negative pulmonary tuberculosis (SPT) cases. However, qualitative reviews (4, 23, 27, 30, 64) and interlaboratory studies (46, 47) have pointed out the low sensitivity of PCR for the diagnosis of SPT and the significant variability in sensitivity and specificity in different studies. Proposed explanations for these findings have included differences in decontamination procedures (47), cross contamination (46), inhibition (27), sampling error (27), quality of the reference standard (27), and mixture of respiratory and other specimens (30).
Although numerous studies have contributed to our understanding of PCR performance for the diagnosis of pulmonary tuberculosis, no quantitative review has assessed its overall performance in smear-negative cases and the sources of the marked variability between studies. A meta-analytic approach is ideal for addressing these issues (31) and will contribute to the understanding of the clinical role of PCR for the diagnosis of SPT.
Therefore, the main goals of our meta-analysis were (i) to summarize diagnostic accuracy of PCR for the diagnosis of SPT in order to make recommendations for its clinical utility, (ii) to identify factors that account for differences in diagnostic accuracy of PCR, and (iii) to describe study design characteristics that should be emphasized in future studies of PCR for the diagnosis of tuberculosis (TB).
MATERIALS AND METHODS
Study selection.
To identify studies that evaluated the performance of PCR for the diagnosis of SPT, a systematic literature search, restricted to human subjects and the English or Spanish language, was performed by using MEDLINE and reference lists of all primary studies published prior to February 2002. The MEDLINE medical subject headings included “PCR” (or “polymerase chain reaction”), “Mycobacterium tuberculosis,” or “tuberculosis.” In addition, BIOSIS bibliographic database was examined for relevant abstracts published until 1 July 1999.
The publications identified from the literature search were screened by one of the investigators. Studies were accepted for further review if sensitivity and specificity of PCR for the detection of M. tuberculosis in smear-negative respiratory specimens (sputum, tracheal, or bronchial specimens) could be calculated for at least 10 patients or specimens in which M. tuberculosis was detected by the reference standard. To evaluate the reliability of the exclusion criteria, another investigator examined independently a random sample of the excluded articles.
Authors were contacted to determine whether multiple reports contained overlapping cases. In such cases, only the latest report was included. When overlap could not be determined conclusively, the study with the most inclusive information or the latest report was included.
Data extraction and study design assessment.
Two investigators abstracted data from the included studies. Disagreements were resolved by discussion with a third investigator. We abstracted 28 variables to assess study quality and determinants of PCR performance that could account for differences between studies (Table 1). We did not develop a quality score because study characteristics may affect accuracy in different directions.
TABLE 1.
Number and percentage of studies that met quality criteria for reporting and research methods to evaluate PCR as a diagnostic test
| Criterion | No. of studies (%) (n = 50)a |
|---|---|
| Type of study (prospective, retrospective) | 50 (100) |
| Study population | |
| Publication yr | 50 (100) |
| Country of study | 50 (100) |
| Age of the patients | 8 (16) |
| Clinical characteristics in the diseased groupb | 21 (42) |
| Clinical characteristics in the control groupc | 18 (36) |
| Independence of interpretation | |
| Tests reported as being applied blindly | 16 (32) |
| Unit of analysis | |
| Reported by no. of patients | 17 (34) |
| Reported by type of respiratory specimen | 24 (48) |
| Quality of reference standard | |
| Combined reference standardd | 15 (30) |
| Smear staining method | 41 (82) |
| Culture technique | 41 (82) |
| PCR procedure | |
| Sample decontamination method | 46 (92) |
| Extraction methode | 48 (96) |
| PCR sample vol | 38 (76) |
| MgCl2 concn | 20 (40) |
| No. of cycles | 48 (96) |
| Denaturation temp | 48 (96) |
| Primer annealing temp | 48 (96) |
| Primer extension temp | 48 (96) |
| Type of PCR (nested or simple) | 50 (100) |
| Target gene | 50 (100) |
| Primers | 50 (100) |
| Negative extraction controls | 11 (22) |
| Controls to monitor PCR inhibitors | 19 (38) |
| Measures to prevent ampliconsf | 35 (70) |
| Product detection method | 50 (100) |
| Analytic sensitivity | 15 (30) |
From 45 published articles (5 studies that evaluated two different target genes were analyzed independently, yielding 50 studies for the analysis).
The clinical characteristics included severity of the diseased (e.g., radiographic signs, clinical symptoms), current treatment, or response to treatment of all the patients in this group.
The clinical characteristics included history of pulmonary tuberculosis, current treatment, or definitive diagnosis of all the patients in this group.
Combined reference standard included culture and either clinical diagnosis or transbronchial biopsy or lung biopsy.
Extraction method included description of lysis and/or DNA purification methods.
Measures to prevent amplicon contamination included reporting the use of dUTP-UNG and/or description of different rooms where the PCR procedures took place.
Statistical methods.
From each study, sensitivity and specificity of PCR for the detection of M. tuberculosis were calculated on the basis of the study reference standard(s) (culture alone or a combined reference standard including culture and either clinical diagnosis or transbronchial biopsy or lung biopsy). Sensitivity and specificity reported after discrepant analysis were excluded because discrepant analysis violates the basic principle of independence for diagnostic test evaluation and, on average, overestimates the test performance (29, 40). When possible, sensitivity and specificity were computed on the basis of both the number of specimens and the number of patients. The 95% confidence intervals of sensitivity and specificity were computed by using StatXact v3.0 (StatXact-3 for Windows: Software for Exact Nonparametric Inference; Cytel Software Corp., Cambridge, Mass.).
Two methods were used to summarize and compare the diagnostic accuracy of the studies.
(i) Summary estimates of sensitivity and specificity.
We computed overall pooled estimates of the sensitivity and specificity and by strata of potential determinants of PCR performance. In addition, we computed the Cochran's Q test of homogeneity (Stata Statistical Software, release 7.0; Stata Corp., College Station, Tex.) to assess differences in sensitivity and specificity between studies.
(ii) ROC curve analysis.
Since pooled estimates of sensitivity and specificity ignore their common threshold, we conducted receiver operating characteristic (ROC) curve analyses. Initially, differences of PCR accuracy between studies were evaluated graphically by plotting sensitivity against (1 − specificity) estimates in the ROC space. Subsequently, we assessed the overall performance of PCR by using summary ROC (SROC) curves (43) (Appendix). These curves are used to summarize the results of the studies and are characterized by the maximum common value for sensitivity and specificity.
In the SROC model, the diagnostic odds ratio (DOR) is a summary measure of PCR diagnostic accuracy (31). The DOR corresponds to the odds of a positive PCR result in persons with SPT relative to the odds of a positive PCR result in patients without active TB. High values of sensitivity and specificity will lead to a high DOR. The parameter “S” represents the variation of DOR between studies due to different thresholds. A negative S value indicates that the threshold used decreases sensitivity and increases specificity, while a positive S value indicates that the threshold used increases sensitivity and decreases specificity.
To identify factors that explain differences in PCR diagnostic accuracy between studies, we added each covariate to an SROC model. The parameters of these covariates are ratios of DORs (RDORs) that quantify the relative diagnostic accuracy of each stratum. For example, when a type of specimen is evaluated, the RDOR is the ratio of the DOR of studies that included bronchial specimens to the DOR of studies that evaluated only sputum specimens. An RDOR of >1.0 indicates that studies that included bronchial specimens were more accurate than those that analyzed only sputum. Statistical analyses were conducted by using Stata version 7.0 software (Stata Statistical Software).
RESULTS
Identified studies.
Our literature search identified 1,228 reports. After our screening, 45 reports met the inclusion criteria and were selected for further review (4 abstracts and 41 published articles) (1-3, 5, 7-9, 12, 13, 15, 17-19, 21, 22, 24, 28, 32, 33, 34, 35, 36, 38, 41, 42, 44, 45, 49-52, 55, 59, 60, 61-63, 65-67; C. J. Lahti, R. J. Kilman, G. Felix, and S. M. Harvey, Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996, abstr. U-13, p. 103, 1996; B. D. Metchock, Abstr. 34th Intersci. Conf. Antimicrob. Agents Chemother., abstr. D79, 1994; N. Miller, S. Hernandez, and T. Cleary, Abstr. 95th Gen. Meet. Am. Soc. Microbiol. 1995, abstr. U-93, p. 133, 1995; M. F. Sierra, L. M. Clarke, K. G. Clarke, K. K. Young, and P. L. Hewitt, Abstr. 95th Gen. Meet. Am. Soc. Microbiol. 1995, abstr. U-85, p. 131, 1995). Most reports were excluded because PCR was not performed directly on respiratory or gastric specimens or because the type of specimens was not described (n = 544). We excluded 195 reports because of small sample size or incomplete data to compute either sensitivity or specificity. Five studies were excluded because the results were reported only after discrepant analysis (54, 56, 58; K. Kaul, S. Luke, L. Pfouts, R. Cohen, D. Schwartz, S. Muzaffar, D. Dranove, and T. C. Hu, Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996, abstr. U-17, p. 103, 1996; S. Snyder, B. Visot, C. Miller, R. Kornreich, and J. Leon, Abstr. 93rd Gen. Meet. Am. Soc. Microbiol. 1993, abstr. D-202, p. 131, 1993). Other reasons for exclusion included no smear-negative samples in the study (n = 73), review articles containing no original data or overlap of specimens between studies (n = 132), or a description of other amplification methods or PCR not done for the detection of M. tuberculosis (n = 234).
Of the 45 included reports, 5 evaluated two different target genes (12, 19, 21, 65, 67), so they were considered as 10 separate studies, providing a total of 50 studies. Exclusion of these five studies that included the same patient specimens but analyzed different target sequences did not significantly change the pooled estimates.
Assessment of quality criteria for reporting and research methods to evaluate PCR.
None of the 50 studies met all of the 28 quality criteria for reporting and research methods to evaluate PCR as a diagnostic test (Table 1). Fewer than 40% of the studies reported results by number of patients, clinical characteristics of the patients, blind testing, negative extraction controls, and analytic sensitivity and used as reference standard combined culture and clinical criteria. Only 24 (48%) of the studies reported sufficient data to compute sensitivity and specificity by the type of specimen. Four types of specimens were evaluated: sputum, bronchial specimens (bronchial washing and bronchoalveolar lavage), tracheal aspirates, and gastric aspirates.
DNA purification methods and use of dUTP-UNG for amplicon control were described or referenced in 15 and 35 studies, respectively. We assumed that if these methods were not described or referenced, DNA was not purified or dUTP-UNG for amplicon control was not used.
Forty-eight percent of the studies evaluated the automated PCR assay Amplicor (53). A total of 70% of the studies included dUTP-UNG for amplicon control, and 72% of the studies detected the amplification product by hybridization procedures.
The variables that could be evaluated as determinants of PCR accuracy were as follows: type of study design, year of publication, country where the study was conducted, tests reported to be applied blindly, type of specimen, type of reference standard, smear-staining method, culture technique, purification method, target sequence, use of dUTP-UNG, use of negative extraction controls, use of controls to monitor PCR inhibition, and PCR product detection method. Due to the small numbers within numerous categories, the decontamination procedures, lysis methods, type of primers, annealing temperature, and number of cycles could not be evaluated. Likewise, denaturation temperature, primer extension temperature, and type of PCR were insufficiently variable to examine further.
Sensitivity and specificity of PCR.
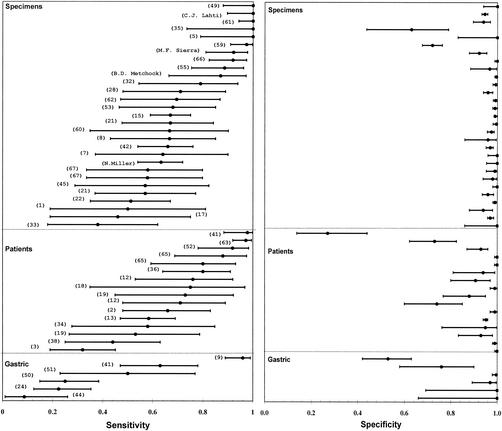
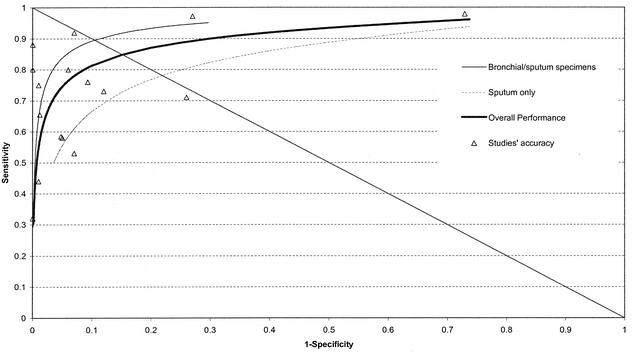
Overall, the sensitivity and specificity of PCR were highly variable and imprecise (Fig. 1). Compared to the specificity, sensitivities were more variable and lower. In 76% of the studies, sensitivities were lower than 90%, while in only 15% of the studies were the specificities lower than 90%. More than 81% of the accuracy estimates were located in the upper left quadrant of the ROC space. Only one study had a sensitivity and a specificity greater than 90% (Fig. 2).
FIG. 1.
Point estimates and 95% confidence intervals of sensitivity and specificity of PCR for detection of M. tuberculosis in smear-negative respiratory and gastric specimens. Specimen studies included studies in which respiratory specimens (sputum, tracheal aspirates, bronchial washings, and bronchoalveolar lavage) were the unit of analysis. Patient studies included studies in which the patients, whose respiratory specimens were evaluated, were the unit of analysis. Gastric studies included studies in which gastric aspirates or respiratory specimens plus gastric aspirates were evaluated. The number of studies totals 51 because 1 study reported results by both gastric aspirates and respiratory specimens. The numbers in parentheses correspond to study references.
FIG. 2.
Plot in the ROC space of accuracy estimates for PCR for the detection of M. tuberculosis in smear-negative respiratory specimens. Each of the 16 studies that were analyzed by patients is indicated by a triangle. ROC curves are shown for studies that analyzed bronchial specimens or tracheal specimens (thin line), for studies that analyzed only sputum specimens (broken line), and for all 16 studies (thick line). The intersection of the diagonal line from the upper left corner to the lower right corner of the ROC space and the SROC curve corresponds to the maximum joint sensitivity and specificity value.
Diagnostic accuracy of PCR by type of specimen.
Studies that included gastric aspirates had lower diagnostic accuracy than those that included respiratory specimens (gastric pooled sensitivity [50%] and specificity [86%]; respiratory specimen sensitivity [72%] and specificity [96%]; sputum only RDOR relative to gastric aspirates [3.7; P = 0.07] and bronchial/sputum-specimens RDOR relative to gastric aspirates [7.1; P = 0.01]). Studies that in addition to sputum included bronchial specimens showed significantly better performance than studies that included only sputum (bronchial-specimen maximum joint sensitivity and specificity value [89%] versus the sputum specimen maximum joint sensitivity and specificity value [75%]) (Fig. 2 and Table 2).
TABLE 2.
Pooled sensitivity, specificity, DOR, and adjusted RDOR values of PCR for the diagnosis of SPT by study characteristicsa
| Study characteristic | Pooledb
|
Pooled DORc
|
Adjusted RDORe
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| N | SES | SP | DOR | 95% CI | Pd | RDOR | 95% CI | P | |
| Overall | 16 | 0.72 | 0.96 | 51.11 | 27.56-94.78 | <0.01 | |||
| Sf | −30.34 | −0.56-−0.04 | 0.02 | ||||||
| Type of the study | |||||||||
| Prospective | 8 | 0.61 | 0.97 | 32.98 | 14.50-75.01 | 0.01 | 1.00 | ||
| Retrospective | 8 | 0.81 | 0.94 | 85.93 | 31.46-234.69 | <0.01 | 1.05 | 0.27-3.99 | 0.94 |
| Yr of publication | |||||||||
| 1993-1995 | 6 | 0.81 | 0.81 | 30.26 | 9.82-93.30 | <0.01 | 1.00 | ||
| 1996-2001 | 10 | 0.67 | 0.98 | 71.44 | 32.42-157.43 | <0.01 | 1.60 | 0.54-4.69 | 0.39 |
| Region of study | |||||||||
| United States | 3 | 0.66 | 0.94 | 32.07 | 7.24-142.02 | 0.06 | 1.00 | ||
| Europe, Asia, or Canada | 13 | 0.73 | 0.96 | 58.09 | 28.64-117.83 | <0.01 | 1.03 | 0.27-3.95 | 0.97 |
| Blind testing | |||||||||
| Yes | 10 | 0.62 | 0.96 | 35.07 | 18.95-64.92 | 0.01 | 1.00 | ||
| No | 6 | 0.91 | 0.93 | 118.41 | 27.87-503.11 | <0.01 | 8.81 | 2.68-28.96 | <0.01 |
| Type of specimen | |||||||||
| Sputum | 7 | 0.66 | 0.82 | 16.55 | 8.41-32.58 | 0.22 | 1.00 | ||
| Otherg | 9 | 0.76 | 0.97 | 111.32 | 48.56-255.19 | <0.01 | 2.46 | 1.02-5.92 | 0.04 |
| Reference standardh | |||||||||
| Combined | 10 | 0.73 | 0.91 | 44.17 | 20.78-93.90 | 0.01 | 1.00 | ||
| Culture only | 6 | 0.70 | 0.98 | 79.30 | 22.37-281.10 | <0.01 | 1.00 | 0.33-3.02 | 0.99 |
| Smear staining methodi | |||||||||
| ZN or Kinyoun | 4 | 0.77 | 0.88 | 25.11 | 6.58-95.72 | 0.01 | 1.00 | ||
| Other | 8 | 0.59 | 0.97 | 45.52 | 23.88-86.78 | 0.05 | 1.20 | 0.27-5.56 | 0.82 |
| Type of culturej | |||||||||
| Solid and liquid media | 8 | 0.61 | 0.95 | 31.97 | 15.19-67.29 | 0.01 | 1.00 | ||
| Solid medium only | 3 | 0.80 | 0.92 | 60.04 | 22.39-161.05 | 0.29 | 3.42 | 0.91-12.8 | 0.07 |
| Purification methodk | |||||||||
| No | 10 | 0.67 | 0.97 | 40.68 | 17.53-94.41 | <0.01 | 1.00 | ||
| Yes | 4 | 0.92 | 0.75 | 70.36 | 27.87-177.66 | 0.15 | 2.96 | 0.74-11.88 | 0.13 |
| Target sequencel | |||||||||
| Other | 5 | 0.88 | 0.72 | 24.59 | 9.08-66.57 | 0.03 | 2.00 | 0.41-9.83 | 0.39 |
| IS6110 | 5 | 0.77 | 0.96 | 87.03 | 21.18-357.66 | 0.01 | 1.00 | ||
| Amplicor, Cobas Amplicor | 6 | 0.56 | 0.98 | 75.49 | 25.30-225.22 | <0.01 | 0.85 | 0.2-3.37 | 0.81 |
| No | 3 | 0.58 | 0.96 | 34.30 | 11.26-104.53 | 0.07 | 1.00 | ||
| Yes | 3 | 0.52 | 1.00 | 288.04 | 28.39-2,922.5 | 0.04 | 0.78 | 0.51-88.22 | 0.91 |
| Negative extraction controls | |||||||||
| Yes | 5 | 0.68 | 0.90 | 21.14 | 8.16-54.83 | 0.03 | 1.00 | ||
| No | 11 | 0.73 | 0.97 | 83.84 | 38.22-183.88 | <0.01 | 1.34 | 0.39-4.63 | 0.64 |
| Controls to monitor inhibitionm | |||||||||
| Yes | 8 | 0.65 | 0.99 | 52.93 | 19.24-145.63 | <0.01 | 1.43 | 0.46-4.46 | 0.53 |
| No | 8 | 0.80 | 0.91 | 53.25 | 22.21-127.64 | <0.01 | 1.00 | ||
| dUTP-UNG | |||||||||
| Yes | 10 | 0.64 | 0.98 | 80.63 | 34.38-189.10 | <0.01 | 2.28 | 0.54-9.67 | 0.26 |
| No | 6 | 0.88 | 0.76 | 27.99 | 10.25-76.44 | 0.01 | 1.00 | ||
| Detection method | |||||||||
| Hybridization | 10 | 0.65 | 0.96 | 41.29 | 20.09-84.87 | <0.01 | 1.00 | ||
| Gel electrophoresis | 6 | 0.83 | 0.90 | 75.26 | 22.95-246.83 | 0.01 | 1.56 | 0.34-3.66 | 0.86 |
Included 16 studies analyzed by number of patients that analyzed only respiratory specimens. Studies that included gastric aspirates were excluded.
Pooled estimates of sensitivity (SES) and specificity (SP) obtained by using random-effects models.
Pooled estimates of DOR and 95% confidence intervals (95% CI) by using random-effects models.
P value of the test of homogeneity of the studies' DORs.
RDOR and 95% confidence intervals (95% CI) obtained by using random-effects models. Models are adjusted by threshold (S) and report of blind testing.
Threshhold (S).
“Other” included studies that reported results without discriminating by type of respiratory specimen (sputum, tracheal aspirates, bronchial washings, and bronchoalveolar lavage).
Combined reference standards included culture and either clinical diagnosis or transbronchial biopsy or lung biopsy.
In four studies the smear staining method was not reported. This model is not adjusted for blind testing due to the small sample size. ZN, Ziehl-Neelsen; “other” includes fluorescence microscopy.
In five studies the type of culture was not reported; this model is not adjusted for blind testing due to the small sample size.
In two studies purification was done on selected specimens that contained inhibitors and were excluded.
“Other” target sequences included 38Kda, PPH 7301 clone, MPB 70, MPB 64, IS986, and IS6110/PAB.
Inhibitors could be assessed by using a positive inhibitor controls that were internal, concurrent, or in parallel.
Effects of study design characteristics and reference standard.
Diagnostic accuracy of PCR was higher among studies that did not report blind testing compared to studies that reported blind testing (Table 2). Blind testing was a strong confounder; therefore, all of the models were adjusted for it.
We hypothesized that diagnostic accuracy of PCR would appear greater if PCR was compared to a less sensitive reference standard. In fact, in studies that used a less sensitive culture method, only the use of solid media was more sensitive than studies that used both solid and liquid media. No differences in diagnostic accuracy were found when combinations of reference standard were compared to culture alone.
Effects of PCR characteristics.
Studies that purified DNA were more sensitive than those that did not (Table 2). However, this increase was associated with a significant and positive shift in threshold (S) (P = 0.04), indicating that the improved sensitivity was at the expense of reduced specificity.
Studies that were published after 1995 that used Amplicor, and noncommercial PCRs that used dUTP-UNG were associated with a negative shift in threshold, suggesting that specificity increased at the expense of a reduction in sensitivity (P values for year of publication, target sequence, and use of dUTP-UNG were 0.09, 0.01, and 0.002, respectively).
SROC models among studies analyzed by specimens.
In the 29 studies with results only by specimens and that included only respiratory specimens, we assessed the effects of all of the variables presented in Table 2, except for the type of study and “gold standard,” because few studies were conducted prospectively or used a combined reference standard. Most of these factors followed similar patterns, as in studies analyzed by the number of patients (data not shown). Blind testing and type of specimen were associated with an increase in diagnostic accuracy. Furthermore, studies that used chemiluminescence or radioactive methods were more accurate than those using colorimetric methods or gel electrophoresis.
Effect of unit of analysis.
To examine the effect of unit of analysis, we compared the six studies that reported their results by both number of patients and number of specimens (3, 18, 38, 44, 51, 52). Sensitivity tended to be higher when the results were analyzed by patient (pooled sensitivity, 72%; specificity, 88%; DOR, 26.1) compared to analysis by specimen (pooled sensitivity, 59%; specificity, 88%; DOR, 18.0) but was not statistically significant (P = 0.27)
Comparison between culture and PCR.
The sensitivity and specificity of PCR and culture could be validly compared only in the four studies that used a combined reference standard and reported their results by number of patients (3, 18, 38, 52). For PCR, sensitivity ranged from 32 to 92% and specificity ranged from 93 to 100%. The sensitivity of culture ranged from 43 to 72%, and the specificity was uniformly 100%. The pooled estimates of sensitivity for culture (51.4%) and PCR (59.8%) did not differ significantly.
DISCUSSION
This meta-analysis summarized the accuracy of PCR for the diagnosis of SPT from 50 studies and described the effects of study design and PCR characteristics on diagnostic accuracy for the first time. Sensitivity estimates were more variable and frequently lower than the specificity. Differences in diagnostic accuracy between studies were best explained by the type of specimen and report of blind testing. The greatest diagnostic accuracy was found among studies that included bronchial specimens, whereas the lowest accuracy was found when specimens that included gastric aspirates were evaluated. Reporting blind testing was associated with less accuracy. DNA purification was associated with greater sensitivity at the expense of lower specificity. Studies conducted after 1995 that used Amplicor or that used dUTP-UNG in noncommercial kits had greater specificity but less sensitivity.
Differences in diagnostic accuracy were explained in large part by the type of specimen. This finding supports previous recommendations to use bronchial specimens in suspected cases of SPT (36). Because many of the studies that included bronchial specimens also evaluated sputum specimens, the difference between these studies and those that assessed only sputum specimens was probably underestimated.
Purification of DNA prior to amplification has the potential of eliminating inhibitors and concentrating scarce primer targets in the sample and therefore has been utilized to increase PCR sensitivity (16). Our findings supported this potential but also demonstrated a concerning trade-off of lower specificity. It is likely that the increased sample manipulation that occurred during DNA extraction resulted in amplicon contamination or cross-contamination between specimens with M. tuberculosis. Such potential for contamination should be monitored with negative extraction controls (whose sputa are known not to contain M. tuberculosis), and laboratories that purify sample DNA should take extra precautions to prevent amplicon contamination.
Studies that failed to report blind testing were likely to overestimate accuracy, particularly sensitivity, thus underscoring the need to improve quality of design and reporting. This interpretation is in accord with those of other meta-analyses (26, 37) and blinded multicenter studies. However, blind testing could simply have been a marker of study design quality. In fact, in our analysis, report of blind testing was strongly correlated with the study being conducted prospectively (r = 0.77).
The reference standard to evaluate PCR performance for pulmonary TB is imperfect. Although culture is known to have low sensitivity in SPT cases (48), it was the only reference standard in 35 of the 50 studies. Because we found that the culture media affected the estimates of PCR accuracy, these estimates should be generalized only to settings when similar media are used. Furthermore, the large variation in reference standard criteria could explain why no differences in accuracy were found when combined reference standards were compared to culture alone. Therefore, we recommend explicitly describing the reference standard.
To best predict the clinical utility of PCR, results should be reported by patients providing detailed information about current treatment, clinical and radiological evidence of TB, and prevalence of TB in their settings. Our finding of a trend of higher diagnostic accuracy when PCR was reported by the number of patients compared to by the number of specimens agrees with the findings of previous studies (19, 50) and indicates that, for SPT, sensitivity will increase if multiple specimens per patient are evaluated because not all of the samples contained a detectable amount of DNA.
As in any meta-analysis, our analysis could be influenced by publication bias (10). The test for publication bias suggested a bias toward publishing studies with high specificity estimates (studies including bronchial specimens [P value for sensitivity of 0.25, P value for specificity of 0.10, and P value for DOR of 0.01] versus the respective P values for sputum [i.e., 0.35, 0.04, and 0.55]).
On the basis of our findings and clinical considerations, we do not recommend that PCR replace culture nor that it be used routinely for the diagnosis of SPT. However, PCR of bronchial specimens could be useful in cases in which there is a high suspicion of TB on the basis of clinical and radiological findings. For example, if the pretest probability of TB was 50%, based on average pooled estimates for studies including bronchial specimens (sensitivity, 76%; specificity, 97%) (Table 2), the average positive predictive value of PCR would be 96%. Hence, a positive PCR would provide enough certainty to initiate treatment and reduce the need for further diagnostic evaluations. However, a negative PCR would provide less clinical certainty because the negative predictive value would be only 80%. Due to the high variability in diagnostic performance of PCR, clinicians who chose to use PCR for the diagnosis of SPT should be aware of the PCR technique and its sensitivity and specificity in their clinical laboratories.
Although PCR is a promising diagnostic test, more rigorous studies are required to better define its role in the diagnosis of SPT clinical practice. Future studies of PCR performance should report the number of patients and type of respiratory specimen, tests should be applied blindly, and should use as a reference standard a combination of culture and clinical diagnosis. The question of which SPT cases are best diagnosed with PCR should be addressed further by reporting PCR accuracy by clinical characteristics.
Acknowledgments
This work was supported by the National Institute for Allergy and Infectious Diseases (1-R01-AI41917). W.C.M. received support from the Clinical Associate Physician Program of the General Clinical Research Center (RR00046), Division of Research Resources, National Institutes of Health.
We thank Roy L. Hopfer, who provided valuable comments on the manuscript.
APPENDIX
Summary receiver operating characteristic models. The method used to construct the SROC curve (39, 43) estimates the parameters of the curve by using an weighted linear regression model of the form: D = A + βS + E, where D (dependent variable) represents the logarithm of the DOR computed for each study (37). The “A” (intercept) represents the common DOR of the corresponding test (the higher the intercept, the closer the curve will be to the upper left hand corner) and “S” [independent variable: logit(TPR) + logit(FPR)] represents the variation of DOR between studies due to different thresholds. The SROC model takes into consideration possible heterogeneity of study results attributed to different thresholds. To assess the effect of study and PCR characteristics on the threshold, we compared the means of S for categories of each characteristic.
To explore sources of heterogeneity, other than the threshold, between studies, we included covariates in the SROC model. The fit of the model was evaluated by computing the residual sum of square (57). If the model was insufficient to account for the observed heterogeneity, we used the random-effects model (14). Due to a poor fit, the models presented in these analyses are random-effects models.
REFERENCES
- 1.Abe, C. H. 1993. Detection of Mycobacterium tuberculosis in clinical specimens by polymerase chain reaction and Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test. J. Clin. Microbiol. 31:3270-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeda, J., A. Garcia, J. Gonzalez, L. Quinto, P. J. Ventura, R. Vidal, G. Rufi, J. A. Martinez, M. T. Jimenez de Anta, A. Trilla, and P. L. Alonso. 2000. Clinical evaluation of an in-house IS6110 polymerase chain reaction for diagnosis of tuberculosis. Eur. J. Clin. Microbiol. Infect. Dis. 19:859-867. [DOI] [PubMed] [Google Scholar]
- 3.Al Zahrani, K., H. Al Jahdali, L. Poirier, P. Rene, and D. Menzies. 2001. Yield of smear, culture and amplification tests from repeated sputum induction for the diagnosis of pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 5:855-860. [PubMed] [Google Scholar]
- 4.American Thoracic Society Workshop. 1997. Rapid diagnostic tests for tuberculosis: what is the appropriate use? Am. J. Respir. Crit. Care Med. 155:1804-1814. [DOI] [PubMed] [Google Scholar]
- 5.Amicosante, M., L. Richeldi, G. Trenti, G. Paone, M. Campa, A. Bisetti, and C. Saltini. 1995. Inactivation of polymerase inhibitors for Mycobacterium tuberculosis DNA amplification in sputum by using capture resin. J. Clin. Microbiol. 33:629-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anonymous. 1993. Centers for Disease Control and Prevention. 1991 Tuberculosis statistics in the United States. U.S. Department of Health and Human Services, Atlanta, Ga.
- 7.Araj, G. F., R. S. Talhouk, L. Y. Itani, W. Jaber, and G. W. Jamaleddine. 2000. Comparative performance of PCR-based assay versus microscopy and culture for the direct detection of Mycobacterium tuberculosis in clinical respiratory specimens in Lebanon. Int. J. Tuberc. Lung Dis. 4:877-881. [PubMed] [Google Scholar]
- 8.Arimura, M., T. Ohuchi, Y. Suzuki, A. Hishinuma, S. Oikawa, J. Sato, and T. Ieiri. 1996. Clinical significance of direct detection of Mycobacterium tuberculosis in respiratory specimens by polymerase chain reaction. Dokkyo J. Med. Sci. 23:143-148. [Google Scholar]
- 9.Bahrmand, A. R., V. V. Bakayev, and M. H. Babaei. 1996. Use of polymerase chain reaction for primary diagnosis of pulmonary tuberculosis in the clinical laboratory. Scand. J. Infect. Dis. 28:469-472. [DOI] [PubMed] [Google Scholar]
- 10.Begg, C., and M. Mazumdar. 1994. Operating characteristics of a rank correlation test for publication bias. Biometrics 50:1088-1101. [PubMed] [Google Scholar]
- 11.Behr, M. A., S. A. Warren, H. Salamon, P. C. Hopewell, A. P. de Leon, C. L. Daley, and P. M. Small. 1999. Transmission of Mycobacterium tuberculosis from patients smear-negative for acid-fast bacilli. Lancet 353:444-449. [DOI] [PubMed] [Google Scholar]
- 12.Beige, J., J. Lokies, T. Schaberg, U. Finckh, M. Fischer, H. Mauch, H. Lode, B. Kohler, and A. Rolfs. 1995. Clinical evaluation of a Mycobacterium tuberculosis PCR assay. J. Clin. Microbiol. 33:90-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennedsen, J., V. O. Thomsen, G. E. Pfyffer, G. Funke, K. Feldmann, A. Beneke, P. A. Jenkins, M. Hegginbothom, A. Fahr, M. Hengstler, G. Cleator, P. Klapper, and E. G. Wilkins. 1996. Utility of PCR in diagnosing pulmonary tuberculosis. J. Clin. Microbiol. 34:1407-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berkey, C. S., D. C. Hoaglin, F. Mosteller, and G. A. Colditz. 1995. A random effects regression model for meta-analysis. Stat. Med. 14:395-411. [DOI] [PubMed] [Google Scholar]
- 15.Bogard, M., J. Vincelette, R. Antinozzi, R. Alonso, T. Fenner, J. Schirm, D. Aubert, C. Gaudreau, E. Sala, M. J. Ruiz-Serrano, H. Petersen, L. A. B. Oostendrop, and H. J. Burkardt. 2001. Multicenter study of a commercial, automated polymerase chain reaction system for the rapid detection of Mycobacterium tuberculosis in respiratory specimens in routine clinical practice. Eur. J. Clin. Microbiol. Infect. Dis. 20:724-731. [DOI] [PubMed] [Google Scholar]
- 16.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cartuyvels, R., C. De Ridder, S. Jonckheere, L. Verbist, and J. Van Eldere. 1996. Prospective clinical evaluation of Amplicor Mycobacterium tuberculosis PCR test as a screening method in a low-prevalence population. J. Clin. Microbiol. 34:2001-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin, D. P., D. M. Yajko, W. K. Hadley, C. A. Sanders, P. S. Nassos, J. J. Madej, and P. C. Hopewell. 1995. Clinical utility of a commercial test based on the polymerase chain reaction for detecting Mycobacterium tuberculosis in respiratory specimens. Am. J. Respir. Crit. Care Med. 151:1872-1877. [DOI] [PubMed] [Google Scholar]
- 19.Cohen, R. A., S. Muzaffar, D. Schwartz, S. Bashir, S. Luke, L. P. McGartland, and K. Kaul. 1998. Diagnosis of pulmonary tuberculosis using PCR assays on sputum collected within 24 hours of hospital admission. Am. J. Respir. Crit. Care Med. 157:156-161. [DOI] [PubMed] [Google Scholar]
- 20.Cowie, R. L., M. E. Langton, and B. C. Escreet. 1985. Diagnosis of sputum smear- and sputum culture-negative pulmonary tuberculosis. S. Afr. Med. J. 68:878. [PubMed] [Google Scholar]
- 21.Dalovisio, J. R., J. S. Montenegro, S. A. Kemmerly, C. F. Genre, R. Chambers, D. Greer, G. A. Pankey, D. M. Failla, K. G. Haydel, L. Hutchinson, M. F. Lindley, B. M. Nunez, A. Praba, K. D. Eisenach, and E. S. Cooper. 1996. Comparison of the amplified Mycobacterium tuberculosis (MTB) direct test, Amplicor MTB PCR, and IS6110-PCR for detection of MTB in respiratory specimens. Clin. Infect. Dis. 23:1099-1106. [DOI] [PubMed] [Google Scholar]
- 22.D'Amato, R. F., A. A. Wallman, L. H. Hochstein, P. M. Colaninno, M. Scardamaglia, E. Ardila, M. Ghouri, K. Kim, R. C. Patel, and A. Miller. 1995. Rapid diagnosis of pulmonary tuberculosis by using Roche Amplicor Mycobacterium tuberculosis PCR test. J. Clin. Microbiol. 33:1832-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Della-Latta, P. 1999. The mycobacteriology milestones: journeying to the new millennium. Lab. Med. 30:408-417. [Google Scholar]
- 24.Devallois, A., E. Legrand, and N. Rastogi. 1996. Evaluation of Amplicor MTB test as adjunct to smears and culture for direct detection of Mycobacterium tuberculosis in the French Caribbean. J. Clin. Microbiol. 34:1065-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. Raviglione. 1999. Global burden of tuberculosis: estimated incidence, prevalence and mortality by country. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 26.Fahey, M., L. Irwig, and P. Macaskill. 1995. Meta-analysis of pap test accuracy. Am. J. Epidemiol. 141:680-689. [DOI] [PubMed] [Google Scholar]
- 27.Forbes, B. A. 1997. Critical assessment of gene amplification approaches on the diagnosis of tuberculosis. Immunol. Investig. 26:105-116. [DOI] [PubMed] [Google Scholar]
- 28.Forbes, B. A., and K. E. Hicks. 1993. Direct detection of Mycobacterium tuberculosis in respiratory specimens in a clinical laboratory by polymerase chain reaction. J. Clin. Microbiol. 31:1688-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hadgu, A. 1997. Bias in the evaluation of DNA amplification tests for detecting Chlamydia trachomatis. Stat. Med. 16:1391-1399. [DOI] [PubMed] [Google Scholar]
- 30.Ieven, M., and H. Goossens. 1997. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin. Microbiol. Rev. 10:242-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irwig, L., A. N. Tosteson, C. Gatsonis, J. Lau, G. Colditz, T. C. Chalmers, and F. Mosteller. 1994. Guidelines for meta-analyses evaluating diagnostic tests. Ann. Intern. Med. 120:667-676. [DOI] [PubMed] [Google Scholar]
- 32.Jan, I. S., P. R. Hsueh, L. J. Teng, L. N. Lee, P. C. Yang, and K. T. Luh. 1998. Evaluation of an automatic polymerase chain reaction assay for identification of Mycobacterium tuberculosis in respiratory specimens. J. Formos. Med. Assoc. 97:204-209. [PubMed] [Google Scholar]
- 33.Kearns, A. M., R. Freeman, M. Steward, and J. G. Magee. 1998. A rapid polymerase chain reaction technique for detecting M. tuberculosis in a variety of clinical specimens. J. Clin. Pathol. 51:922-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kocagoz, T., E. Yilmaz, S. Ozkara, S. Kocagoz, M. Hayran, M. Sachedeva, and H. F. Chambers. 1993. Detection of Mycobacterium tuberculosis in sputum samples by polymerase chain reaction using a simplified procedure. J. Clin. Microbiol. 31:1435-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kox, L. F. F., D. Rhienthong, A. M. Miranda, N. Udomsantisuk, K. Ellis, J. van Leeuwen, S. van Heusden, S. Kuijper, and A. H. J. Kolk. 1994. A more reliable PCR for detection of Mycobacterium tuberculosis in clinical samples. J. Clin. Microbiol. 32:672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liam, C. K., Y. C. Chen, S. F. Yap, P. Srinivas, and P. J. Poi. 1998. Detection of Mycobacterium tuberculosis in bronchoalveolar lavage from patients with sputum smear-negative pulmonary tuberculosis using a polymerase chain reaction assay. Respirology 3:125-129. [DOI] [PubMed] [Google Scholar]
- 37.Lijmer, J., B. Mol, S. Heisterkamp, G. Bonse, M. Prins, J. van der Meulen, and P. Bossuyt. 1999. Empirical evidence of design related bias in studies of diagnostic tests. JAMA 282:1061-1066. [DOI] [PubMed] [Google Scholar]
- 38.Lim, T. K., A. Gough, N. K. Chin, and G. Kumarasinghe. 2000. Relationship between estimated pretest probability and accuracy of automated Mycobacterium tuberculosis assay in smear-negative pulmonary tuberculosis. Chest 118:641-647. [DOI] [PubMed] [Google Scholar]
- 39.Littenberg, B., and E. Moses. 1993. Estimating diagnostic accuracy from multiple conflicting reports: a new meta-analytic method. Med. Decision Making 13:313-321. [DOI] [PubMed] [Google Scholar]
- 40.Miller, W. C. 1998. Can we do better than discrepant analysis for new diagnostic test evaluation? Clin. Infect. Dis. 27:1186-1193. [DOI] [PubMed] [Google Scholar]
- 41.Mitarai, S., K. Oishi, M. Fukasawa, H. Yamashita, T. Nagatake, and K. Matsumoto. 1995. Clinical evaluation of polymerase chain reaction DNA amplification method for the diagnosis of pulmonary tuberculosis in patients with negative acid-fast bacilli smear. Tohoku J. Exp. Med. 177:13-23. [DOI] [PubMed] [Google Scholar]
- 42.Moore, D. F., and J. I. Curry. 1995. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by Amplicor PCR. J. Clin. Microbiol. 33:2686-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moses, L., and D. Shapiro. 1993. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Stat. Med. 12:1293-1316. [DOI] [PubMed] [Google Scholar]
- 44.Munoz, C., A. Gene, I. Perez, A. Mira, J. Roca, and C. Latorre. 1997. Diagnosis of tuberculosis in children: evaluation of the technique of polymerase reaction. An. Espan. Pediatr. 47:353-356. [PubMed] [Google Scholar]
- 45.Nolte, F. S., B. Metchock, J. E. McGowan, Jr., A. Edwards, O. Okwumabua, C. Thurmond, P. S. Mitchell, B. Plikaytis, and T. Shinnick. 1993. Direct detection of Mycobacterium tuberculosis in sputum by polymerase chain reaction and DNA hybridization. J. Clin. Microbiol. 31:1777-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noordhoek, G. T., A. H. Kolk, G. Bjune, D. Catty, J. W. Dale, P. E. Fine, P. Godfrey-Faussett, S. N. Cho, T. Shinnick, S. B. Svenson, S. Wilson, andJ. D. A. Van Embden. 1994. Sensitivity and specificity of PCR for detection of Mycobacterium tuberculosis: a blind comparison study among seven laboratories. J. Clin. Microbiol. 32:277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noordhoek, G. T., J. D. A. Van Embden, and A. H. J. Kolk. 1996. Reliability of nucleic acid amplification for detection of Mycobacterium tuberculosis: an international collaborative quality control study among 30 laboratories. J. Clin. Microbiol. 34:2522-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palacios, J. J., J. Ferro, M. Telenti, S. G. Roces, P. Prendes, J. M. Garcia, A. I. Nicolas, J. Rodrigues, H. Villar, and J. F. de Quiros. 1996. Comparison of solid and liquid culture media with polymerase chain reaction for detection of Mycobacterium tuberculosis in clinical samples. Eur. J. Clin. Microbiol. Infect. Dis. 15:478-483. [DOI] [PubMed] [Google Scholar]
- 49.Patnaik, M., K. Liegmann, and J. B. Peter. 2001. Rapid detection of smear-negative Mycobacterium tuberculosis by PCR and sequencing for rifampin resistance with DNA extracted directly from slides. J. Clin. Microbiol. 39:51-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierre, C., C. Olivier, D. Lecossier, Y. Boussougant, P. Yeni, and A. J. Hance. 1993. Diagnosis of primary tuberculosis in children by amplification and detection of mycobacterial DNA. Am. Rev. Respir. Dis. 147:420-424. [DOI] [PubMed] [Google Scholar]
- 51.Piersimoni, C., A. Callegaro, D. Nista, S. Bornigia, F. De Conti, G. Santini, and G. De Sio. 1997. Comparative evaluation of two commercial amplification assays for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J. Clin. Microbiol. 35:193-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Querol, J. M., M. A. Farga, D. Granda, C. Gimeno, and J. Garcia de Lomas. 1995. The utility of polymerase chain reaction (PCR) in the diagnosis of pulmonary tuberculosis. Chest 107:1631-1635. [DOI] [PubMed] [Google Scholar]
- 53.Rajalahti, I., P. Vuorinen, M. M. Nieminen, and A. Miettinen. 1998. Detection of Mycobacterium tuberculosis complex in sputum specimens by the automated Roche Cobas Amplicor Mycobacterium Tuberculosis Test. J. Clin. Microbiol. 36:975-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reischl, U., N. Lehn, H. Wolf, and L. Naumann. 1998. Clinical evaluation of the automated COBAS AMPLICOR MTB assay for testing respiratory and nonrespiratory specimens. J. Clin. Microbiol. 36:2853-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez, J. C., E. Fuentes, and G. Royo. 1997. Comparison of two different PCR detection methods: application to the diagnosis of pulmonary tuberculosis. APMIS 105:612-616. [DOI] [PubMed] [Google Scholar]
- 56.Rosenstraus, M., Z. Wang, S. Y. Chang, D. Debonville, and J. P. Spadoro. 1998. An internal control for routine diagnostic PCR: design, properties, and effect of clinical performance. J. Clin. Microbiol. 36:191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rothman, K., and S. Greenland. 1998. Modern epidemiology. J. B. Lippincott Publishers, Philadelphia, Pa.
- 58.Shibuya, Y., T. Shiozaki, M. Hayashi, and Y. Sugiyama. 2000. Efficacy of Amplicor PCR for the diagnosis of tuberculosis in respiratory specimens other than sputum. Tuberc. Lung Dis. 80:209-215. [DOI] [PubMed] [Google Scholar]
- 59.Shim, M. S., S. Y. Lee, S. H. Cho, Y. K. Park, G. H. Bai, and S. J. Kim. 1993. Efficiency of different primers in polymerase chain reaction to detect Mycobacterium tuberculosis in clinical specimens. J. Kor. Soc. Microbiol. 28:391-395. [Google Scholar]
- 60.Smith, M. B., J. S. Bergmann, S. L. Harris, and G. L. Woods. 1997. Evaluation of the Roche AMPLICOR MTB assay for the detection of Mycobacterium tuberculosis in sputum specimens from prison inmates. Diagn. Microbiol. Infect. Dis. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 61.Tan, M. F., W. C. Ng, S. H. Chan, and W. C. Tan. 1997. Comparative usefulness of PCR in the detection of Mycobacterium tuberculosis in different clinical specimens. J. Med. Microbiol. 46:164-169. [DOI] [PubMed] [Google Scholar]
- 62.Tevere, V. J., P. L. Hewitt, A. Dare, P. Hocknell, A. Keen, J. P. Spadoro, and K. K. Y. Young. 1996. Detection of Mycobacterium tuberculosis by PCR amplification with pan-Mycobacterium primers and hybridization to an M. tuberculosis-specific probe. J. Clin. Microbiol. 34:918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong, C. F., W. W. Yew, C. Y. Chan, L. Y. Au, S. W. Cheung, and A. F. Cheng. 1998. Rapid diagnosis of smear-negative pulmonary tuberculosis via fibreoptic bronchoscopy: utility of polymerase chain reaction in bronchial aspirates as an adjunct to transbronchial biopsies. Respir. Med. 92:815-819. [DOI] [PubMed] [Google Scholar]
- 64.Woods, G. L. 2001. Molecular techniques in mycobacterial detection. Arch. Pathol. Lab. Med. 125:122-126. [DOI] [PubMed] [Google Scholar]
- 65.Yam, W. C., K. Y. Yuen, and W. H. Seto. 1998. Direct detection of Mycobacterium tuberculosis in respiratory specimens using an automated DNA amplification assay and a single tube nested polymerase chain reaction (PCR). Clin. Chem. Lab. Med. 36:597-599. [DOI] [PubMed] [Google Scholar]
- 66.Yuen, K. Y., K. S. Chan, C. M. Chan, B. S. W. Ho, L. K. Dai, P. Y. Chau, and M. H. Ng. 1993. Use of PCR in routine diagnosis of treated and untreated pulmonary tuberculosis. J. Clin. Pathol. 46:318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuen, K. Y., W. C. Yam, L. P. Wong, and W. H. Seto. 1997. Comparison of two automated DNA amplification systems with a manual one-tube nested PCR assay for diagnosis of pulmonary tuberculosis. J. Clin. Microbiol. 35:1385-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]