Abstract
Randomly amplified polymorphic DNA (RAPD) has been successfully used to detect genetic variations among isolates of Paracoccidioides brasiliensis. However, the usefulness of this technique for assessing important parasitic properties is still unconfirmed. In the present work we further investigated the applicability of RAPD in revealing important intrinsic and extrinsic features of this fungus associated with geographical origin, time of isolation, source of clinical specimen, clinical forms of human disease and also in vitro and in vivo susceptibility to antimicrobial and antifungal drugs. The RAPD patterns allowed us to distinguish all of the analyzed strains, which included 26 clinical isolates, 2 animal isolates, and 1 environmental isolate of P. brasiliensis obtained from different geographic regions, confirming the strong discriminating power of this technique. A phenetic tree, build from the RAPD data, showed that although the two nonclinical Brazilian strains were set together the majority of the clinical Brazilian strains were randomly distributed through different sub-branches of a major cluster without any correlation to any of the parameters analyzed. A second major cluster, however, has grouped isolates from Mato Grosso and Roraima (Brazil) that not only were susceptible in vitro to trimethoprim-sulfamethoxazole but also produced a good in vivo response. These results open new vistas for epidemiological and clinical studies of P. brasiliensis.
Paracoccidioides brasiliensis is a dimorphic fungus alternating between the mycelial form observed in room temperatures and the yeast form found in infected tissues or in culture at 35 to 37°C. The human paracoccidioidomycosis caused by this fungus is characterized by a range of clinical manifestations from benign or asymptomatic forms to severe and disseminated disease that is often fatal. The exact causes of the clinical pleomorphism are not understood, but it is reasonable to presume that factors associated with both the fungus and the patient are involved. In relation to the fungus, although wide biological and biochemical diversity has been observed, no clear correlation with pathogenesis has been demonstrated (3, 9, 12, 13, 17, 20, 23). Until now very little has been known about the genome sequences of P. brasiliensis; hence, the use of molecular approaches for isolate typing has been difficult.
The development of the randomly amplified polymorphic DNA (RAPD) technique represented a landmark in the molecular characterization of diverse organisms, especially for microbial strain identifications when very few genomic sequences are available. In relation to other molecular techniques, RAPD analysis offers the advantage of requiring small amounts of DNA, simpler procedures, and the use of arbitrary primers (26). Due to these characteristics, RAPD has been widely used for human diseases to detect genomic variations among isolates of fungi such as Histoplasma capsulatum (27), Cryptococcus neoformans (19), Aspergillus fumigatus (1), and P. brasiliensis (3, 9, 12, 13, 23). Several authors have used the RAPD technique to investigate possible associations between P. brasiliensis fungus banding patterns and geographical distribution (3), clinical or animal origin (21), virulence and pathology (12, 13), and atypical isolates not presenting with in vitro dimorphism (9) but, until now, little if any correlation was observed. Moreover, in the few cases in which some association could be found, the data in the literature are controversial. For instance, when characterizing by RAPD analysis seven isolates of P. brasiliensis (five from Brazil and two from Ecuador), Soares et al. (23) were not able to detect any correlation with geographical origins. Calcagno et al. (3) later used this technique to cluster 33 P. brasiliensis strains into five major groups closely related to geographical origin (Venezuela, Brazil, Peru, Colombia, and Argentina) but not to pathological features of the disease. Molinari-Madlum et al. (12) have shown that RAPD patterns of P. brasiliensis isolates from Brazil and Ecuador were reasonably well correlated with the degree of virulence in mice. However, Motta et al. (13) could not find any association between RAPD profiles of 35 strains of P. brasiliensis (also with the majority isolated from Brazil) either with virulence for experimentally infected mice or with clinical forms of human disease.
In the present work we review the usefulness of the RAPD technique for discriminating P. brasiliensis isolates in relation to important intrinsic and extrinsic features such as geographical origin, time of isolation, source of clinical specimen, clinical forms of human disease, and also in vitro and in vivo susceptibility to antimicrobial and antifungal drugs.
MATERIALS AND METHODS
Fungal strains.
We analyzed 29 isolates from different geographical areas that are listed in Table 1.
TABLE 1.
P. brasiliensis isolates analyzed in this study
| Strain | Yr of isolation | Region, country of isolation | Sex/age (yr)a | Occupation | Localization or type of lesions in patient/origin | Clinical form | Trimethoprim-sulfamethoxazole MIC (μg/ml) | Amphotericin B MIC (μg/ml) |
|---|---|---|---|---|---|---|---|---|
| Pb-73 (ATCC 32071) | 1970 | Antioquia, Colombia | U/U | Unknown | Unknown | Unknown | 40.0 | 0.125 |
| Pb-9 (ATCC 36324) | 1960 | Caracas, Venezuela | U/U | Unknown | Lymph nodes and mucocutaneous lesions | Unknown | 5.00 | 0.500 |
| Pb-306 | 1991 | Trujillo, Venezuela | U/U | Unknown | Mouth, lung | Unknown | 0.62 | 1.000 |
| Pb-333 | 1996 | Lima, Peru | U/U | Unknown | Subcutaneous abscess | Unknown | 40.0 | 0.062 |
| Pb-Tatu | Pará, Brazil | U/U | Armadillo | 0.62 | 0.500 | |||
| Pb-Pinguim | Antarctica, Uruguay | U/U | Penguin feces | 2.50 | 0.500 | |||
| Pb-262 | Minas Gerais, Brazil | U/U | Dog food | 0.62 | 0.500 | |||
| Pb-JT-1 (ATCC 90659) | 1990 | Minas Gerais, Brazil | U/U | Unknown | Unknown | Chronic | 1.25 | 0.500 |
| Pb-JT-2 | 1992 | Minas Gerais, Brazil | F/18 | Unknown | Biopsy | Chronic | NTb | NT |
| Pb-JT-4 | 1993 | Minas Gerais, Brazil | U/U | Chronic | 20.0 | 0.125 | ||
| Pb-JT-5 | 1993 | Minas Gerais, Brazil | F/56 | House maid | Generalized lymphadenopathy | Chronic | 2.50 | 0.125 |
| Pb-339 | São Paulo, Brazil | U/U | Unknown | Chronic | 5.00 | 0.250 | ||
| Pb-18 | São Paulo, Brazil | U/U | Unknown | Unknown | Chronic | 40.0 | 0.250 | |
| Pb-265 | São Paulo, Brazil | U/U | Unknown | Unknown | Unknown | 40.0 | 0.250 | |
| Pb-SN | São Paulo, Brazil | U/U | Unknown | Unknown | Chronic | 40.0 | 0.250 | |
| Pb-1087 | 2000 | Rio G. do Sul, Brazilc | M/60 | Retired | Sputum | Chronic | 10.0 | 0.125 |
| Pb-686 | 2000 | Rio G. do Sul, Brazil | M/55 | Chicken farmer | Sputum | Chronic | 1.25 | 0.125 |
| Pb-1430 | 1998 | Rio G. do Sul, Brazil | M/U | Bronchial washing | Chronic | NT | NT | |
| Pb-218 | 2000 | Mato Grosso, Brazil | M/40 | Farm laborer | Ganglion aspirate | Chronic | 10.0 | 0.250 |
| Pb-YRJ | 1999 | Mato Grosso, Brazil | M/22 | Carpenter | Lymph node biopsy | Subacute | 320 | 1.000 |
| Pb-263 | 2001 | Mato Grosso, Brazil | M/49 | Farm laborer | Sputum | Chronic | 20.0 | 0.250 |
| Pb-283 | 1999 | Mato Grosso, Brazil | F/23 | Housewife | Ganglion aspirate | Acute | 0.62 | 0.250 |
| Pb-286 | 1999 | Mato Grosso, Brazil | M/60 | Gardener | Sputum | Chronic | 40.0 | 0.031 |
| Pb-717 | 2000 | Mato Grosso, Brazil | M/58 | Farm laborer | Ganglion aspirate | Chronic | 20.0 | 0.031 |
| Pb-769 | 2000 | Roraima, Brazil | M/48 | Farm laborer | Oral mucus membrane | Chronic | 0.62 | 0.062 |
| Pb-369 | 2000 | Mato Grosso, Brazil | F/25 | Housewife | Ganglion aspirate | Acute | 2.50 | 0.250 |
| Pb-84 | 2001 | Mato Grosso, Brazil | M/59 | Map maker | Inferior periodontal lesion | Chronic | 10.0 | 1.000 |
| Pb-133 | 2001 | Mato Grosso, Brazil | M/40 | Farm laborer | Ear lobe | Chronic | 40.0 | 0.031 |
| Pb-351 | 1999 | Mato Grosso, Brazil | M/47 | Farm laborer | Ganglion aspirate | Chronic | 10.0 | 0.500 |
U, unknown; M, male; F, female.
NT, not tested.
Rio G. do Sul, Rio Grande do Sul.
DNA preparation.
For DNA preparation P. brasiliensis strains were maintained on solid Fava-Neto medium at 35°C (5) with two to three subcultivations every 7 days to obtain the yeast-like form of the microorganism in the exponential growth phase (11). A loop full of cells from each strain was then collected and suspended in 500 μl of sorbitol citrate buffer (1.1 M sorbitol, 0.1 M sodium citrate; pH 5.6). To the cell suspension, we added 1 ml of extracting buffer (Tris-spermidine; 40 mM Tris-HCl [pH 8.0], 4 mM spermidine, 10 mM EDTA, 0.1 M NaCl, 10 mM β-mercaptoethanol, 0.1% sodium dodecyl sulfate) supplemented with 50 mg of glucanase (Glucanex-Novo Nordisk). Afterward, the mixture was incubated at 37°C under agitation in a thermoshaker operated at 35 turns min−1 for 3 h. DNA was obtained after two phenol-chloroform extractions and ethanol precipitation. Finally, DNA quantification was performed by comparison with DNA standards electrophoresed in agarose gels (2).
RAPD analysis.
For the RAPD analysis, we tested 10 different primers among the stocks available in our laboratory. The primers OPA-1 (CAGGCCCTTC), OPA-2 (TGCCGAGCTG), OPA-3 (AGTCAGCCAG), OPA-4 (AATCGGCGTG), and OPG-14 (GGATGAGACC), obtained from Operon Biotechnology, yielded the best results. These oligonucleotides were selected based on high-intensity bands, hypervariability, and good definition of amplified DNA fragments and were used for molecular characterization of different isolates of P. brasiliensis. RAPD analysis was carried out basically as described by Williams et al. (26) with minor modifications. The RAPD reaction mixture contained 1 ng of genomic DNA, 1 pmol of primer; 500 μM concentrations of each deoxynucleoside triphosphate, and 0.3 U of Taq DNA polymerase (Gibco-BRL) in a final volume of 10 μl of the PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 3.5 mM MgCl2). The PCRs were conducted in a Perkin-Elmer GeneAmp PCR system 2400 according to the following parameters: denaturation at 95°C for 30 s, followed by 40 cycles at 94°C for 1 min, annealing at 40°C for 2 min, and extension at 72°C for 2 min. In the last cycle the extension step time was increased to 7 min. Reproducibility was checked by PCR, repeating the analysis at least three times. RAPD products were analyzed by electrophoresis on a 8% polyacrylamide gel in Tris-borate-EDTA (pH 8.0 buffer) and visualized by silver staining as described by Santos et al. (22).
For RAPD data analysis, the relative mobility position of all bands present in each analyzed P. brasiliensis strain was calculated and transformed in a data matrix in which the character “1” means the presence of a specific band and “0” represents its absence. We used the Nei and Li algorithm (15) contained in the TREECON computer package program (25) to calculate the genetic distances between the strains. Unrooted phenograms were constructed by UPGMA (unweighted pair group with arithmetic mean) method, and the robustness of the tree topology was assessed by 1,000 bootstrap resampling (4, 6, 24-26).
Drug susceptibility tests.
For in vitro drug susceptibility tests, the yeast cells were obtained in the exponential phase of growth in McVeigh and Morton (MVM; pH 7.0) (18) medium after at least three subcultures. Susceptibility was evaluated to both antimicrobial (trimethoprim-sulfamethoxazole; FIOCRUZ-RJ [Brazil]) and antifungal (amphotericin B; Sigma Chemical Co., St. Louis, Mo.) drugs. The MIC for each strain was determined by using a broth macrodilution procedure according to NCCLS criteria (standard M38-P), with minor modifications (8, 10, 14). Stock solutions of amphotericin B (50 mg ml−1) and trimethoprim-sulfamethoxazole (250 mg ml−1) were freshly prepared in dimethyl sulfoxide. Serial twofold solutions were made with MVM liquid medium as the diluent to yield final drug concentrations ranging from 4.0 to 0.007 μg ml−1 for amphotericin B and 640 to 0.31 μg ml−1 for trimethoprim-sulfamethoxazole. Drug-free and titrated solubilizing vehicle (dimethyl sulfoxide) controls were included. Inocula were determined spectrophotometrically by using a yeast suspension in sterile 0.85% saline that gave 70% transmittance at 520 nm. The yeast cells were collected from the solid medium (MVM); diluted (1:10) in a counting solution containing 0.9% NaCl, 4% formaldehyde, and 4% Tween 20; vortexed to disperse the aggregated cells; and counted in a Neubauer chamber. An initial inoculum density of 105 cells ml−1 was obtained (16). A 0.1-ml aliquot of this suspension was then added to 0.9 ml of MVM broth, giving the desired dilutions of the drug and a final concentration of 104 cell ml−1 (8, 14). Strains were grown at 35°C under agitation in a thermoshaker. The MIC of amphotericin B was read after 5 to 7 days and was defined as the lowest drug concentration exhibiting no visible growth compared to a drug-free control tube. For the trimethoprim-sulfamethoxazole, MIC was defined as the lowest concentration that resulted in a visual turbidity of 80% inhibition or less compared to that produced by the growth control (8, 14). All assays were performed in triplicate.
For the Mato Grosso (Brazil) patients group, we could also evaluate the therapeutic response to the trimethoprim-sulfamethoxazole. In these cases the patients were treated for 60 days with 160 mg plus 800 mg twice daily (attack phase). The cure criteria used were the absence of clinical symptomatology, as well as negative micological and radiological tests.
RESULTS
The P. brasiliensis strains and the parameters considered in the present study are listed in Table 1. These parameters included the isolation period of the strains; the geographic origin; patient occupation, sex, and age; the clinical specimen; the clinical form of the disease; and the MICs of amphotericin B and trimethoprim-sulfamethoxazole.
Drug susceptibility data.
We investigated the in vitro susceptibility of 27 of 29 isolates in relation to trimethoprim-sulfamethoxazole and amphotericin B. The MICs varied from 0.62 to 320 μg ml−1 for trimethoprim-sulfamethoxazole and from 0.031 to 1.0 μg ml−1 for amphotericin B. For amphotericin B, 0.031 and 0.125 μg ml−1 were the drug concentrations able to inhibit, respectively, 50 and 90% of the analyzed isolates. Higher concentrations, i.e., 2.5 and 40 μg of trimethoprim-sulfamethoxazole ml−1, were necessary to inhibit 50 and 90% of the same strains (Table 1).
Phylogenetic data.
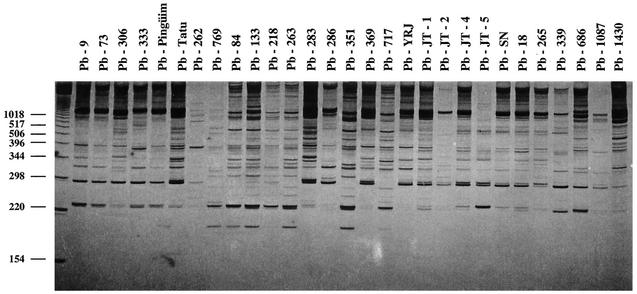
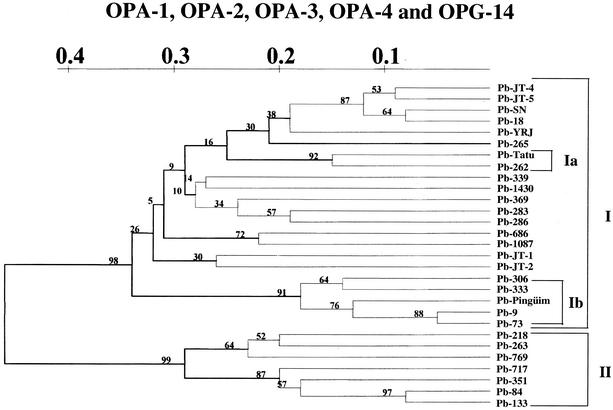
A total of 29 P. brasiliensis strains, including 26 clinical isolates, 1 animal isolate, and 2 environmental isolates (primer OPA-4), are shown in Fig. 1. Similar results were obtained for all of the other primers used (data not shown). We detected 11.57 ± 4.144 (average ± the standard deviation) bands for each strain varying from 154 to 1018, depending on the primer and the strain analyzed, and no two strains exhibited identical RAPD patterns when each primer was considered separately or in combination. RAPD data were used for phylogenetic analysis of the Paracoccidioides group. Figure 2 shows the UPGMA tree obtained from the data matrix constructed with the five combined RAPD primers. Despite the great diversity observed for the group, all analyzed P. brasiliensis strains were clustered in two major branches, strongly supported by 98% of the bootstrapped trees.
FIG. 1.
RAPD profiles showing polymorphism among the 29 analyzed P. brasiliensis isolates. The primer used was OPA-4.
FIG. 2.
Phenogram of P. brasiliensis based on the UPGMA method derived from RAPD assays generated by using combined primers.
The major group I (the upper branch in Fig. 2) encompassed the majority of the clinical isolates from the northern to the southern parts of South America, together with an animal strain (Pb-Tatu) and two environmental strains (Pb-262 from dog food and Pb-Pinguim from penguin feces). Inside this major branch, a clear separation of the Brazilian and non-Brazilian strains could be seen. The strains from Venezuela, Colombia, Peru, and Uruguayan Antarctica were clustered together in a sub-branch denoted Ib in Fig. 2. Within this subgroup the two analyzed Venezuelan strains were set apart from each other. Among the Brazilian strains, the nonclinical isolates (Pb-Tatu and Pb-262) were subclustered in a separated sub-branch (denoted Ia in the Fig. 2) supported by a bootstrap value of 92%. Fifteen clinical strains isolated from different states of Brazil were randomly distributed through all other sub-branches of the major group I. No association between the relative positions of them and any one of the investigated parameters was found.
A completely different profile was observed with the strains belonging to the major group II (lower major branch in the Fig. 2). Six of them were recently isolated in the state of Mato Grosso (central-western region of Brazil) and one from a patient from Roraima, a state in northern part of Brazil. In all cases the patients with presented the chronic form of untreated paracoccidioidomycosis and showed clinical and also in vitro susceptibility to both trimethoprim-sulfamethoxazole and amphotericin B drugs.
DISCUSSION
We evaluated here the usefulness of RAPD analysis as a tool for determining different intrinsic and extrinsic parameters of the P. brasiliensis strains. However, as already found in previous studies, we did not observe any association between the RAPD profiles of the isolates and most of the investigated parameters such as the isolation period of the strains; patient occupation, sex, or age; and the clinical form (acute or chronic) of the disease (3, 9, 12, 13, 23).
Similar results were also observed in relation to the geographic origin of the analyzed strains. Although we detected the clustering of the isolates from Peru, Colombia, and Venezuela, all of them localized in the northern part of South America; the strains isolated from Brazil showed greater genetic diversity and could not be grouped together. This was even more notable when we compared the strains isolated from just one state of Brazil, in this case Mato Grosso. Part of the strains (4 of 10) were set in major group I and part (6 of 10) in major group II, with fewer than 65.09% of shared bands between them. These results contrast with those of Calcagno et al. (3), who could separate 33 strains from Argentina, Brazil, Colombia, Peru, and Venezuela into five groups arranged according to geographical zones when the OPG-14 arbitrary primer was used. Even when we used this primer we could not detect the clusterization in either the 15 Brazilian strains or the 2 analyzed isolates from Venezuela.
With regard to the source of the isolates, we observed the grouping of two of the three nonclinical strains: a strain isolated from dog food (Pb-262 [Minas Gerais, southern Brazil]) and a strain isolated from an armadillo (Pb-Tatu [Pará, northern Brazil]). The strain Pb-Pinguim, isolated from penguin feces in Antarctic Uruguay, was set apart, together with all of the other non-Brazilian strains. However, this is a singular strain, representing the unique specimen of P. brasiliensis already isolated from penguins (7). Although we have studied very few environmental and/or animal isolates, our findings are very interesting and stimulate us to further investigate the possibility of using RAPD to distinguish clinical from nonclinical P. brasiliensis strains.
Puzzling results were observed when we investigated possible correlations of the RAPD profiles and the therapeutic success or in vitro susceptibility to the currently used antimicrobial drugs: amphotericin B and trimethoprim-sulfamethoxazole. We could not find any association between the RAPD profiles and the MICs for both drugs. In our in vitro assay all analyzed strains were susceptible to both compounds except for strain Pb-YRJ, which was resistant to trimethoprim-sulfamethoxazole (10). However, we detected a interesting association between the strains isolated in Mato Grosso (Brazil) and those set in major group II in our tree, which were all isolated from patients that presented good therapeutic responses to trimethoprim-sulfamethoxazole with the usual recommended doses (960 mg twice daily). These findings contrast with other strains isolated from the same state, at the same time, and set into major group I, which were obtained from patients who presented relapses or failed therapeutics with trimethoprim-sulfamethoxazole (although three of them were susceptible to these drugs in vitro). Although they are very preliminary, these results indicate that in vivo susceptibility or resistance to drugs may be more associated with parasite genetics than originally suspected, thus opening new perspectives for epidemiological and clinical studies of P. brasiliensis. Further studies are necessary to validate these findings that acquire greater relevance when associated with the difficulties encountered in treating paracoccidiodomycosis, whether related to the choice of drug for each form and phase of the disease or the socioeconomic conditions and the competence of the immune system of a particular patient.
Acknowledgments
We thank Gioconda San-Blás (IVIC, Caracas, Venezuela) for providing strains Pb-306 and Pb-333 (ATCC 32071 and 36324), Patrícia Cisalpino (UFMG [Brazil]) for providing strains Pb-Tatu and Pb-262, and Sydney Hartz Alves (UFSM [Brazil]) for strains Pb-686 and Pb-1087.
This research was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, the Conselho Nacional de Desenvolvimento Científico e Tecnológico, and the Fundação de Amparo à Pesquisa do Estado de Minas Gerais.
REFERENCES
- 1.Aufauvre-Brown, A., J. Cohen, and D. W. Holden. 1992. Use of randomly amplified polymorphic DNA markers to distinguish isolates of Aspergillus fumigatus. J. Clin. Microbiol. 30:2991-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borges, M. J., M. O. Azevedo, R. Bonatelli, M. S. S. Felipe, and S. Astolfi-Filho. 1990. A practical method for the preparation of DNA from filamentous fungi. Fungal Genet. Newsl. 10:11. [Google Scholar]
- 3.Calcagno, A. M., G. Niño-Vega, F. San-Blás, and G. San-Blás. 1998. Geographic discrimination of Paracoccidioides brasiliensis strains by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 36:1733-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Efron, B., and G. Gong. 1983. A leisurely look at the bootstrap, the jacknife, and croos validation. Am. Stat. 37:36-48. [Google Scholar]
- 5.Fava-Netto, C. 1995. Estudos quantitativos sobre a fixação do complemento na blastomicose Sul Americana com antígeno polissacarídico. Arq. Cir. Exp. 18:197-254. [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 7.Garcia, N. M., G. M. B. Del Negro, E. M. H. Vaccari, N. T. Melo, C. M. Assis, and C. S. Lacaz. 1993. Paracoccidioides brasiliensis, nova amostra isolada de fezes de um pinguim (Pygoscelis adeliae). Rev. Inst. Med. Trop. Sao Paulo 35:227-235. [DOI] [PubMed] [Google Scholar]
- 8.Hahn, R. C., and J. S. Hamdan. 2000. In vitro susceptibilities of Paracoccidioides brasiliensis yeast form to antifungal drug. Mycoses 43:403-407. [PubMed] [Google Scholar]
- 9.Hahn, R. C., A. M. Macedo, N. L. Santos, J. C. P. Resende, and J. S. Hamdan. 2002. Characterization of Paracoccidioides brasiliensis atypical isolates by random amplified polymorphic DNA analysis. Rev. Iberoam. Micol. 19:49-51. [PubMed] [Google Scholar]
- 10.Hahn, R. C., Y. T. M. Conceição, J. F. Ferreira, N. L. Santos, and J. S. Hamdan. Disseminated paracoccidioidomycosis: correlation between clinical and in vitro resistance to ketoconazole and trimethoprim/sulfamethoxazole. Mycoses, in press. [DOI] [PubMed]
- 11.Manocha, M. S. 1980. Lipid composition of Paracoccidioides brasiliensis: comparison between the yeast and mycelial forms. Sabouraudia 18:281-286. [DOI] [PubMed] [Google Scholar]
- 12.Molinari-Madlum, E. E. W. I., M. S. S. Felipe, and C. M. A. Soares. 1998. Virulence of Paracoccidioides brasiliensis isolates can be correlate to groups defined by random amplified polymorphic DNA analysis. Med. Mycol. 37:269-276. [PubMed] [Google Scholar]
- 13.Motta, T. R., C. A. Moreira-Filho, R. P. Mendes, et al. 2002. Evaluation of DNA polymorphisms amplified by arbitrary primers (RAPD) as genetically associated elements to differentiate virulent and non-virulent Paracoccidioides brasiliensis isolates. FEMS Immunol. Med. Microbiol. 1407:1-7. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 1998. Reference method for broth dilution antifungal susceptibility testing of conidium forming filamentous fungi. Proposed standard M38-P. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Nei, M., and W. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaller, M. A., M. G. Rinaldi, J. N. Galgiani, M. S. Bartlett, B. A. Body, et al. 1990. Colaborative investigation of variables in susceptibility testing of yeasts. Antimicrob. Agents Chemother. 34:1648-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Restrepo, A. 1990. Actualización sobre la paracoccidioidomicosis y su agente etiológico. Interciencia 15:193-199. [Google Scholar]
- 18.Restrepo, A. M., and B. E. Jimenez. 1980. Growth of Paracoccidioides brasiliensis yeast phase on a chemically defined culture medium. J. Clin. Microbiol. 12:279-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruma, P., S. C. A. Chen, T. C. Sorrel, and A. G. Brownlee. 1996. Characterization of Cryptococcus neoformans by random DNA amplification. Lett. Appl. Microbiol. 23:312-316. [DOI] [PubMed] [Google Scholar]
- 20.San-Blás, G. 1993. Paracoccidioidomycosis and its etiologic agent Paracoccidioides brasiliensis. J. Med. Vet. Mycol. 31:99-113. [PubMed] [Google Scholar]
- 21.Sano, A., R. Tanaka, K. Yokoyama, M. Franco, E. Bagagli, et al. 1999. Comparison between human and armadillo Paracoccidioides brasiliensis isolates by random amplified polymorphic DNA analysis. Mycopathologia 143:165-169. [DOI] [PubMed] [Google Scholar]
- 22.Santos, F. R., S. D. Pena, and J. T. Epplen. 1993. Genetic and population study of a y-linked tetranucleotide repeat DNA polymorphism with a simple non-isotopic technique. Hum. Genet. 90:655-656. [DOI] [PubMed] [Google Scholar]
- 23.Soares, C. M. A., E. E. W. I. Molinari-Madlum, S. P. da Silva, M. Pereira, and M. S. S. Felipe. 1995. Characterization of Paracoccidioides brasiliensis isolates by random amplified polymorphic DNA analysis. J. Clin. Microbiol. 33:505-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swofford, D. L., and G. J. Olsen. 1990. Phylogeny reconstruction, p. 411-501. In D. M. Hills and C. Moritz (ed.), Molecular systematics. Sinauer Associates, Sunderland, Mass.
- 25.Van de Peer, Y., and R. De Wacher. 1994. Treecom for windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 26.Williams, J. G. K., A. R. Kubelik, K. J. Livak, J. A. Rafalski, and S. Y. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods, J. P. D., W. Kersulyte, E. Goldman, and D. E. Berg. 1993. Fast DNA isolation from Histoplasma capsulatum: methodology for arbitrary primer polymerase chain reaction-based epidemiological and clinical studies. J. Clin. Microbiol. 31:463-464. [DOI] [PMC free article] [PubMed] [Google Scholar]