Abstract
Modern detection and identification tools can help to provide answers to urgent questions about the incidence, prevalence, and epidemiology of currently emerging diseases. We developed highly sensitive one-step TaqMan reverse transcription-PCR assays with sensitivities ranging from 104 to 101 molecules for 11 human pathogens of the orthobunyaviruses. We compared the performances of these assays on three currently available cyclers (ABI-PRISM 7700, LightCycler, and SmartCycler). The assay for Oropouche virus (OROV) was tested using sera collected from days 1 to 5 after onset of OROV disease and was found to be greatly superior to an established nested PCR system. A mean copy number of 1.31 × 107 OROV RNA/ml of serum was detected. Diagnostic RNA detection can be used as early as day 1 after onset of OROV disease. The use of a mobile SmartCycler and a hands-on time of less than 3 h could help to intensify outbreak surveillance and control, especially in field studies.
The Bunyaviridae family of viruses is subdivided into five genera. Except for the plant pathogenic genus Tospovirus, the Orthobunyavirus, Hantavirus, Nairovirus, and Phlebovirus genera are composed of enzootic viruses, some of which cause zoonotic disease in humans. As is the case with many zoonoses, humans act as accidental dead-end hosts of a zoonotic transmission cycle oscillating between mammals (mostly rodents) and arthropods (Orthobunyavirus, Nairovirus, and Phlebovirus) or among rodents via aerosols and bite wounds (Hantavirus). There are 330 known viruses in these four genera, and 174 are listed as belonging to the genus Orthobunyavirus, including half of the approximately 60 Bunyaviridae viruses causing disease in humans (10, 11).
The diseases elicited by Orthobunyavirus range from typical viral diseases with flu-like symptoms (e.g., caused by Tahyna virus [TAHV]) (34) to febrile arthralgia (e.g., caused by Oropouche virus [OROV]) (26), encephalitis (e.g., caused by La Crosse virus [LACV]) (35), and, as recently reported, even hemorrhagic fever (caused by Garissa virus) (4). Although most infections have a rather mild outcome, some can be deadly (15, 33).
Many orthobunyaviruses are underestimated with regard to their potential prevalence and distribution. Snowshoe Hare virus (SSHV), Inkoo virus (INKV), TAHV, and Batai virus (BATV), originally isolated in the United States (14), Finland (30), the Czech Republic (3), and Slovakia (2), respectively, have all been isolated in Siberia (20-22). TAHV and BATV have been isolated in several Western European countries (19), and the isolation of BATV has also been reported in Sudan (24) and India (12).
Some orthobunyaviruses have gradually become accepted as etiological agents of growing disease problems. LACV and related viruses from the California serogroup cause about 100 cases of encephalitis per annum in the United States (6). Oropouche fever caused by OROV has developed into the second most common arboviral disease, next to Dengue fever, in Brazil (27).
In recent decades, evidence has accumulated indicating that, especially in developing countries, the complex interaction of factors such as the growth of the human population, the accompanying demographic and rapid socioeconomic changes, urbanization, and ecological upheaval contribute to the emergence of new infectious diseases (36).
As an initial measure to counter the development of these diseases into major public health problems, it is essential to gather basic clinical and epidemiological data (8). Modern diagnostic detection and identification tools can help to provide answers to urgent questions about the incidence, prevalence, and epidemiology of currently emerging diseases and about pathogens that have the potential to emerge in the future. They can also help researchers pick up the trail of several viruses isolated 20 to 30 years ago (Bunyamwera virus [BUNV], Germiston virus [GERV], and Guaroa virus [GROV]) and used to study the molecular biology of the Bunyaviridae but for which essential information regarding public health issues are almost completely missing.
Our aim was to devise rapid diagnostic tools for human pathogens of the genus Orthobunyavirus. Therefore, we developed one-step TaqMan reverse transcription (RT)-PCR assays for all viruses for which sequence information was available and established quantification standards for each virus. A single set of reaction conditions and one temperature profile was used for all TaqMan RT-PCR assays. Here we describe the TaqMan RT-PCR protocols and present data from the testing of OROV serum samples collected during an outbreak in Brazil.
MATERIALS AND METHODS
Virus strains and patient material.
California encephalitis virus (CEV) strain BFS 283, Jamestown Canyon virus (JCV) strain 61V2235, SSHV, OROV strain TR 9760, GROV strain Co H 35211, GERV strain SA Ar 1050, and INKV were received as lyophilisates from R. E. Shope, University of Texas, Galveston, Tex.; LACV Thompson was obtained from Rayu Ramasamy, Meharry Medical College, Nashville, Tenn.; TAHV was obtained from J. Pilaski, Medizinisches Institut für Umwelthygiene, Düsseldorf, Germany; and BUNV and a pUC18 plasmid containing the S segment of BATV were provided by R. Elliot, University of Glasgow, Glasgow, Scotland. The patient material was collected from April to May 1996 during an outbreak of OROV disease in Oriximiná, Pará State, Brazil. Samples consisted of 30 sera from OROV-infected patients collected from days 1 to 5 after onset of disease and diagnosed by virus isolation at the Instituto Evandro Chagas (IEC), Belém, Brazil.
Virus culture and RNA preparation.
Viruses were grown in BHK-21 cells (INKV, TAHV, GROV, SSHV, and LACV), VeroE6 cells (JCV, California encephalitis virus [CALV], and OROV), or both (GERV) in 95% Dulbecco's modified Eagle medium-5% fetal calf serum in 175-cm2 flasks at 37°C in an atmosphere of 5% CO2. Each strain was passaged three times. RNA was prepared using RNeasy columns (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
RT-PCR and cloning of S segments.
One-step RT-PCR was performed using the RT enzyme RAV-2 (Amersham Pharmacia, Freiburg, Germany) and the polymerase Tth (Roche, Mannheim, Germany) as recommended by Kuno (17). Briefly, 1 μM concentrations of the S segment primers (Table 1) were used for RT-PCR amplification from viral RNA by using the following profile: RT at 53°C for 30 min and 30 cycles of PCR at 95°C for 60 s, 60°C for 60s, and 72°C for 60s. The annealing temperature differed for BUNV (55°C). Reaction mixtures contained 1 U of RAV-2, 1 U of Tth, and 500 μM concentrations of deoxynucleoside triphosphates (dNTPs) in 10 mM Tris-HCl (pH 8.9), 100 mM KCl, 3 to 5 mM MgCl2, 50 μg of bovine serum albumin (BSA)/ml, and 0.05% Tween 20. Betaine (1 μM) was added to amplify the S segments of CALV and OROV. Amplicons were ligated into pCRII, and ligations were transformed into Escherichia coli invα cells by using a TA cloning kit (Invitrogen, Breda, The Netherlands). Clones carrying a positively oriented DNA copy of the S segment were propagated in 50-ml overnight cultures, and plasmids were prepared by using a Maxiprep kit (Bio-Rad, München, Germany). The plasmids were sequenced using an ABI 377 sequencer to confirm the sequence identity.
TABLE 1.
Primers for S segments and species-specific RT-PCR
| Primer or probea | Sequence |
|---|---|
| S segment | |
| BUN+ | AGTAGTGTACTCCAC |
| BUN− | AGTAGTGTGCTCCAC |
| LACSFUP | AGTAGTGTACTCCACTTGAATACTTTGA |
| LACSFDP | AGTAGTGTGCTCCACTGAATACATT |
| TAHSFUP | AGTAGTGTACTCCACTTGAATACTTTGAA |
| TAHSFDP | AGTAGTGTGCTCCACTGAATACCTT |
| BUNSFUP | AGTAGTGTACTCCACACTACAAACTTG |
| BUNSFDP | AGTAGTGTGCTCCACCTAAAACTTA |
| GERSFUP | GTAGTGTACTCCACGCATAAAACTTT |
| GERSFDP | AGTAGTGTGCTCCACCTTAAACTTAA |
| OROSFUP | AGTAGTGTACTCCACTAT |
| OROSFDP | AGTAGTGTGGCTCCACAT |
| GROSFUP | AGTAGTGTACTCCACACTACATACCAAT |
| GROSFDP | AGTAGTGTGCTCCACCTAATACCTATA |
| Species specific | |
| LAC FP | CCAGATGGGTCCTTGATCA |
| LAC P | CGAGAATGATGATGAGTCTCAGCACG |
| LAC RP | CAATTGGGTTGATAATAGTTGTTCTG |
| CAL FP | GCGGAGTCAAATGGCAT |
| CAL P | AGAATGGTGCAGAAATTTATTTGGCATTC |
| CAL RP | GGTAAAATTTGAAAGTTTCCAAGAA |
| JC FP | GGATATCTAGCCAGATGGGTTCT |
| JC P | CTCAGATGATGACGAGTCTCAGAGAGAACTC |
| JC RP | ATTGGATTCTGCAATTGGATTT |
| SSH FP | GGATATTTAGCCAGATGGGTTC |
| SSH P | AGAGAATGAAGACGAGTCTCGGCG |
| SSH RP | TTGATGATTGTTGTCTTGATCAA |
| TAH FP | CAAAGCTGCTCTCGCTCG |
| TAH P | CCGGAGAGGAAGGCTAGTCCTAAATTTGGA |
| TAH RP | TTCCAGGAAAATGATWATTGACGA |
| INK FP | CATTGGAACAATGGCCC |
| INK P | TCCCAGGAACAGAAATGTTTCTAGAAGTTTTC |
| INK RP | AGGATCCATCATACCATGCTT |
| GER FP | TGTACTCAATACGAATTTCCCTGG |
| GER P | AGGAACAATGCAGTGCCTGACTACGGTC |
| GER RP | TCCACTGATACGGTGGAAGGTA |
| BUN FP | AATTTTCCTGGCAACCGGA |
| BUN P | AACCCAGTTCCTGACGATGGTCTTACCC |
| BUN RP | AAGGAATCCACTGAGGCGG |
| GRO FP | GAGGCTGGAGAATTCCTGT |
| GRO P | TAATACACATTTTCCTGGAAACCGGA |
| GRO RP | GAATCATCGAGGACTGGACT |
| ORO FP | CATTTGAAGCTAGATACGGACAA |
| ORO P | CAATGCTGGTGTTGTTAGAGTCTTCTTCCT |
| ORO RP | CCATGGGCCTCGATG |
| BAT FP | GACGCGAGATTAAAACTAGTCTCTC |
| BAT P | AAGTGAATGGGAGGTTACGCTTAACCTTG |
| BAT RP | AGGAAAATTTGTATTAAATACAGTAACCTTC |
| Nested | |
| ORO N5 | AAAGAGGATCCAATAATGTCAGAGTTCATTT |
| ORO N3 | GTGAATTCCCACTATATGCCAATTCCGAATT |
SFUP, S-segment upstream primer; SFDP, S-segment downstream primer; SFSP, S-segment single primer; FP, forward primer; RP, reverse primer; P, TaqMan probe. All sequences are given in the 5′-to-3′ orientation.
In vitro transcription and quantification of transcribed RNA.
Runoff transcription was performed from the T7 promoter downstream of the pCRII multiple cloning site to yield negative single-stranded RNA (−ssRNA). In preparation for T7 runoff transcription, 10 to 15 μg of plasmids was linearized and restriction enzymes were removed by phenol-chloroform extraction. The −ssRNA was transcribed in a total volume of 100 μl for 1 h at 37°C from 2 μg of linearized plasmid by using 40 U of T7 (Roche) in 40 mM Tris-HCl (pH 8.0), 6 mM MgCl2, 15 mM dithiothreitol, 2 mM spermidine, 0.05 mg of BSA/ml, 200 U of RNasin (Promega, Mannheim, Germany), and 100 μM concentrations of each ribonucleoside triphosphate. To remove the template DNA, the reaction mixtures were incubated twice with 10 U of DNase I (Roche) at 37°C for 10 min. After the first round of digestion, the RNAs were purified with the RNeasy clean-up kit (Qiagen). After the second round, a Trizol (Invitrogen) extraction was used to completely remove DNase. The efficiency of the DNase digestion improved markedly when, prior to incubation at 37°C, the reaction mixtures were sequentially incubated at 95°C for 1 min and in ice water for 1 min to melt down DNA-RNA hybrids. After two rounds of DNase treatment, template DNAs could no longer be detected in 1 μl of the undiluted RNA standards by species-specific PCR using a Fast Start DNA master kit (Roche), 300 nM concentrations of primers, 200 nM concentrations of probes, and 40 cycles of PCR at 95°C for 5 s and 60°C for 15 s on the LightCycler. Quantification of RNA was performed using the fluorescent dye RiboGreen (Molecular Probes, Eugene, Oreg.), which specifically binds to ssRNA. Briefly, RNA samples and standard RNA in the range of 20 to 1,000 ng/ml were mixed with RiboGreen in Tris-EDTA buffer. Fluorescent emission of samples and standards at 525 nm was measured using the plate read mode of the SDS1.6.3 software of ABI-PRISM 7700, and the quantities of the sample RNAs were calculated. ABI-PRISM 7700 has to be calibrated for the use of RiboGreen; alternatively, the emission spectrum of SybrGreen can be used for RiboGreen fluorescence detection. Copy numbers of the RNA transcripts were calculated from their molecular weight, and molecules, ranging from 107 to 101, were diluted in 100 μg of tRNA/ml (Sigma, München, Germany).
RT-PCR amplicon design.
Amplicons were placed into conserved regions of sequence alignments done with the Megalign software (DNAstar; Lasergene). Primers were designed for an annealing midpoint temperature (Tm) of 60°C by using the PCR document window of the Primer-Express software (Applied Biosystems), which operates by using the algorithm developed by Rychlik et al. (28). Species-specific amplicons were designed in conserved regions. Primer Tm ranged between 58 and 60°C, and the Tm of the 5′FAM- and 3′TAMRA-tagged probes ranged from 68 to 70°C. Species-specific primers were designed in reference to sequences with the following accession numbers: K00108 and K00610 (LACV); U12797 (CEV); U12796 and U12799 (JCV); J02390 and U12800 (SSHV); Z68497, U47142, and X73468 (TAHV); Z68496, U47137, and U47138 (INKV); D00353 (BUNV); M19420 (GERV); X37466 (GROV); and AF164531-58 (OROV). Nested primers were designed to hybridize to the S segment of OROV.
RT-PCR conditions.
The RT-PCR conditions for the ABI-PRISM 7700 (Applied Biosystems) were as follows: 53°C for 30 min and 40 cycles of 95°C for 15 s and 60°C for 60 s [reaction mixtures in a total volume of 25 μl contained 2.5 U of RAV-2-2.5 U of Tth, 500 μM dNTPs, 10 U of RNasin, and 2 μM Rox in 50 mM Bicine (pH 8.2), 115 mM potassium acetate (KOAc), 5 mM Mn(OAc)2, 8% glycerol, 1 μM concentrations of primers, and 600 nM concentrations of probes]. To increase sensitivity, 2 μg of the single-strand binding protein GP32 (Roche) was added per reaction (32). RT-PCR conditions for the LightCycler (Roche) were as follows: RT at 61°C for 20 min, activation at 95°C for 5 min, and 40 cycles of PCR at 95°C for 5 s and 60°C for 15 s. We used a RNA master hybridization probes kit with 500 nM concentrations of primers and 200 nM concentrations of probes. This kit includes an aptamer-blocked Tth which performs both RT and hotstart PCR amplification. RT-PCR conditions for the SmartCycler (Cepheid) were as follows: RT at 53°C for 5 min and 40 cycles of PCR at 95°C for 5 s and 60 to 63°C for 15 s [reaction mixtures in a 25-μl total volume contained 1 U of RAV-2-1 U of Tth, 500 μM dNTPs, 500 nM concentrations of primers, and 200 nM concentrations of probes in 50 mM Bicine (pH 8.2), 115 mM KOAc, 5 mM Mn(OAc)2, 8% glycerol, and SmartCycler additive reagent (200 mM Tris-HCl, pH 8.0, 200 ng of BSA/ml, 0.15 M trehalose, 0.2% Tween 20)]. To increase sensitivity, 2 μg of the single-strand binding protein GP32 was added per reaction. We had to adapt hybridization temperatures to 62°C for BUNV and JCV and 63°C for GERV, JCV, SSHV, INKV, and TAHV, but they remained at 60°C for GROV, CEV, and BATV. The nested PCR conditions were as described above for the amplification of S segments, with 25 cycles for each round and hybridization at 52°C. Outside primers OROSFUP and OROSFDP and nested primers ORO N5 and ORO N3 have been described previously (29) (Table 1).
RNA preparation of patient samples.
RNA was extracted from 125 μl of serum with Trizol LS (Invitrogen) according to the manufacturer's instructions and was resuspended in a volume of 20 μl.
RESULTS
Primer design.
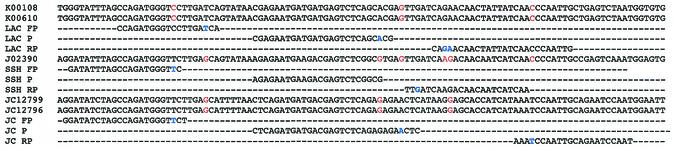
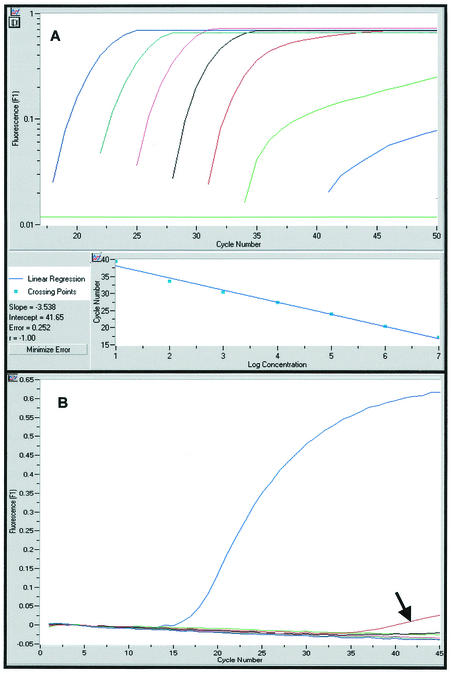
The majority of sequences published for Bunyaviridae species are S-segment sequences coding for the nucleocapsid. Primers and probes were placed into blocks of conserved sequences across sequence alignments of variant species isolates. The advantage of the TaqMan chemistry (two primers and one probe) is that three blocks of conserved sequences across the species variants are needed as opposed to four blocks for the hydprobe chemistry (two primers and two probes). We designed the primers by using the ARMS (amplification refractory mutation system) principle, which is based on the observation that at high stringency (60°C), the third nucleotide upstream of the 3-prime end of a primer is most decisive for the specificity of the overall 3-prime hybridization of a primer (25). This principle was adhered to wherever possible in order to design primers able to specifically distinguish between closely related sequences, as exemplified by the primers designed for some of the closely related California group Orthobunyavirus sequences (Fig. 1). Cross-reactivity was tested with 107 molecules of the respective standard RNA. Apart from the species-specific amplicon for LACV which showed a slight cross amplification with GROV (Fig. 2), we observed no cross amplification between any of the amplicons (primers and probes) and the RNA standards of all 11 viruses.
FIG. 1.
Primer design for California group Orthobunyavirus. According to the ARMS principle, the third nucleotide from the 3-prime end of a primer is the most decisive for specific binding. Shown is an alignment of the target area (sequence accession numbers) and the amplicons (FP, forward primer; P, probe; RP, reverse primer) of LACV, SSHV, and JCV. Variant nucleotides at the 3-prime ends of primers are highlighted in blue, and nonmatching nucleotides in nontarget sequences are printed in red. All sequences are given in sense orientation.
FIG. 2.
Sensitivity and specificity of the LACV amplicon. (A) Amplification blot of the LACV RNA standard from 107 to 101 molecules (from left to right: 107, blue; 106, green; 105, red; 104, black; 103, red; 102, green; and 102, blue). (B) The LACV species-specific assay was tested with 107 copies of the standard RNA of all of the other Orthobunyavirus species. The LACV amplicon showed minor cross amplification of 107 copies of the GROV standard RNA (red curve) but no cross amplification of the standard RNA of any other strain (all other colors).
Cell culture, cloning of S segments, and synthetic RNA standards.
We successfully cultured 10 orthobunyaviruses (BATV was the exception). They all showed marked cytopathic effects on BHK-21 or Vero E6 cells beginning from the second 3-day passage after inoculation. The presence of the viruses was confirmed by species-specific RT-PCR. We cultured all viruses sequentially and never in parallel to avoid laboratory cross-contamination of the strains. We performed the amplification of the S segments by using a one-step RT-PCR approach as suggested by Kuno (17) with a combination of the reverse transcriptase RAV-2 (Amersham) and the polymerase Tth. This worked remarkably well when we used S-segment primers designed with a Tm of about 60°C (Table 1). With the exception of JCV and CALV, for which primers BUN+ and BUN− were applied (9), species-specific S-segment primers were used to avoid false amplification. All cloned amplicons were confirmed by sequencing. The use of full-length negative-sense RNA from the cloned S segments as RNA standards enabled us to come as close as possible to the natural behavior of the viral RNA in the RT-PCR. We successfully produced synthetic S-segment RNA standards for all 11 viruses.
One-step TaqMan RT-PCR.
The performance of the amplicons was assessed on a standard range of 107 to 101 molecules of in vitro-transcribed −ssRNA. The sensitivities achieved with the amplicons on ABI-PRISM 7700 using RAV-2 and Tth are listed in Table 2. The addition of the single-strand binding protein GP32 of the T4 phage has been shown to increase the sensitivity of PCRs (32). The addition of GP32 increased the slope of the kinetic curves of the fluorescent signal produced in our TaqMan assays, especially in the lower standard range of 103 to 101 molecules. The increase in slope of the kinetic reaction curves allowed the threshold to be raised and placed into the elongated exponential phases of the low-copy-number standard curves of the range. This improved the correlation coefficient of the whole standard range, providing a solid basis for quantification at a higher sensitivity. We tested the standards on the LightCycler with an RNA master amplification hybridization probes kit (Roche). The results shown in Table 2 indicate that, in comparison with the results with the ABI-PRISM 7700, we found increased sensitivities of 1 to 2 logs for five species-specific TaqMan RT-PCR assays when using the Roche kit on the LightCycler. We could not generate comparable sensitivities using the Roche kit on the ABI PRISM 7700 (data not shown). Finally, we adapted all TaqMan RT-PCR assays to the SmartCycler. We again used the RAV-2-Tth enzyme mix we already had used on the ABI-PRISM 7700. We found that we had to increase the reaction temperatures to obtain good sensitivities, which on the whole were reduced by 1 to 0.5 logs compared with the performance on the LightCycler (Table 2). With our RNA standard, the nested assay for OROV showed a sensitivity of 105 RNA molecules detected in the first round of PCR and 103 RNA molecules detected in the second round of PCR in ethidium bromide stained gel analysis.
TABLE 2.
TaqMan RT-PCR sensitivities
| Species | Sensitivity (lowest no. of molecules of RNA standard detected)
|
||
|---|---|---|---|
| ABI-PRISM 7700 | LightCycler | SmartCycler | |
| BUNV | 102 | 101 | 102 |
| GERV | NDa | 102 | 103 |
| GROV | 104 | 102 | 103 |
| OROV | 102 | 102 | 102 |
| BATV | ND | 101 | 102 |
| CEV | ND | 102 | 102 |
| JCV | 102 | 102 | 103 |
| LACV | 103 | 101 | 101 |
| SSHV | ND | 104 | 104 |
| INKV | ND | 103 | 103 |
| TAHV | ND | 102 | 103 |
ND, not done.
Testing patient material.
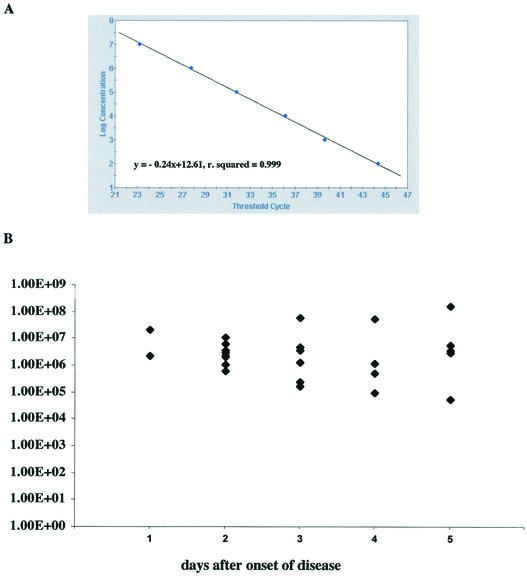
In a retrospective analysis, we prepared RNA from 30 serum samples from which OROV had been isolated at the IEC. A nested PCR assay established at the IEC laboratory detected OROV RNA in 8 of 30 samples (26.6%). The TaqMan RT-PCR assay detected viral RNA in 28 of the 30 samples (93.3%), and the number of RNA molecules per milliliter of serum ranged from 5.42 × 104 to 1.65 × 108, with a mean copy number of 1.31 × 107 (median, 2.36 × 106). Two samples were negative but also scored negative in the nested PCR assay. TaqMan RT-PCR positive samples had been drawn from patients at days 1 (two samples), 2 (nine samples), 3 (seven samples), 4 (five samples), and 5 (five samples), and negative samples had been drawn at days 2 and 3 after onset of disease (Fig. 3).
FIG. 3.
Detection of OROV in patient sera. (A) OROV RNA standard range from 107 to 101 molecules (as established on the SmartCycler). (B) Results are given as the number of copies of viral RNA detected per milliliter of serum obtained from each of 28 patients sampled on days 1 to 5 after onset of disease. Two patient samples drawn on days 2 and 3 after onset of disease tested negative. RNA was Trizol extracted from 125 μl of serum. OROV infection had been previously confirmed by virus isolation.
DISCUSSION
In recent decades, emerging infections have been a steady companion of the accelerated growth of the human population and its concomitant impact, including deforestation and urbanization, on inhabited ecosystems (36). The development of modern rapid diagnostics offers researchers the chance to collect clinical and epidemiological data on the etiological agents of these diseases (13, 16). We have developed a set of 11 fluorescence one-step TaqMan RT-PCR assays for human pathogens of the orthobunyaviruses.
Most fluorescence PCR approaches settle for cloning the target area of the amplicon, since the fluorescence PCR simply measures the ratio of the fluorescent signal to the copy numbers of target molecules. The nature of the target molecule is secondary. RNA target molecules, however, tend to form complex secondary structures that reduce accessibility of the target molecule to PCR and thus influence the sensitivity of RT-PCR assays (18). In recognition of this effect, we decided to use the complete viral S segment for our RNA standard. The T4 phage single-strand binding protein GP32 has been shown to increase the sensitivity of PCR due to breakdown of secondary structures in single-stranded DNA and RNA (1, 7). GP32 indeed increased the sensitivity of our TaqMan RT-PCR assays by up to 2 logs. The RNA standards were tested on the ABI-PRISM 7700 in a Bicine buffer using RAV-2, Tth as the polymerase (23), and the additive GP32. We obtained high sensitivities for most assays. The sensitivity of the SSHV assay, however, was very low (104 to 106 molecules). One reason for this may be the extremely stable secondary structures in the S segment of SSHV. We were able to increase the sensitivities of several assays when using a RNA master hybridization probes kit on the LightCycler. One ingredient of this mixture is an aptamer-blocked Tth which allows separation of RT activity from the polymerase activity of the enzyme. The sensitivity of the LACV assay improved markedly, i.e., down to 10 molecules, when this mixture was used on the LightCycler. Compared with the use of RAV-2-Tth, the Roche mixture improved the sensitivity by just 1 additional log on the ABI-PRISM 7700. We attribute the superior efficiency of the RT-PCR in the LightCycler to the minimal transition phases between the temperatures of the PCR steps due to the fast (20°C/s) heating and cooling rates of the LightCycler.
The TaqMan amplicons performed well on the SmartCycler, although slightly reduced sensitivities compared with those for the LightCycler were obtained. Again, this may be due to the slightly reduced performance in heating (10°C/s) and cooling (2.5°C/s) of the SmartCycler compared with that of the LightCycler.
Thanks to the ARMS principle, the specificity of the assays was very good, as no cross amplification was observed when we cross tested the 11 assays with very high copy numbers of the RNA standards (107 molecules) of each of the 11 viruses. Only the LACV assay showed a minor cross amplification of the GROV RNA. However, the crossing point on the threshold for this cross amplification was very late and the fluorescence intensity was very low, i.e., the cross amplification was not very efficient. It can be ruled out by raising the threshold above a fluorescence intensity value of 0.1.
In recent years, OROV has developed into an urban disease in Amazonia and it bears all the hallmarks of an emerging disease that could quickly spread beyond its present reach (27). The OROV TaqMan RT-PCR assay showed a higher sensitivity than the established nested RT-PCR assay. It detected viral RNA in 93.3% of the 30 patient samples collected from days 1 to 5 after onset of disease, compared with 26.6% for the nested RT-PCR assay. The most likely reason for the decreased sensitivity of the nested RT-PCR assay for OROV compared with the that of the TaqMan RT-PCR assay is the differences in the amplicon sizes produced by each assay (754 and 693 bp for the first and second rounds, respectively, of the nested RT-PCR assay versus 97 bp for the TaqMan RT-PCR assay). The high mean viral load of 1.31 × 107 molecules/ml of serum is typical for infections with Bunyaviridae. In sheep, Rift Valley fever virus, for example, produces a high viremia of up to 105 viruses/ml of serum in the early days of an infection (31). An early diagnostic window for this virus is therefore amenable to RT-PCR and has also been documented for Crimean-Congo hemorrhagic fever virus (5).
The higher sensitivity of our assay allowed detection and identification of OROV in two samples collected at day 1 and in nine samples collected at day 2 of the disease. This means that patients with typical symptoms can be confirmed to be infected with OROV at a very early stage of the disease. By using our assay, rapid detection of OROV infection should indeed be possible within a total hands-on time (sampling, preparation, and PCR) of less than 3 h. This method should therefore greatly facilitate the investigation of an OROV outbreak in time to differentiate it from a dengue outbreak for which it might be mistaken. To confirm this, we are now endeavoring to collect sequential serum samples from individual patients during the next outbreak.
The combination of easy RNA extraction, mobile cyclers such as the SmartCycler TD system, and the highly sensitive and rapid one-step RT-PCRs presented here could become an ideal tool for outbreak surveillance, epidemiological screening, or detection of released biological agents in the field.
Since detection with TaqMan probes is very specific, established amplicons can only be as good as the latest sequencing information available. Especially when trying to detect RNA viruses, one should always bear in mind that a negative result only means that the sequence the amplicon has been designed for was not detected in the sample. It does not formally exclude the possible presence of a variant of the particular virus. Virus isolation and sequencing is very important to keeping fluorescent nucleic acid detection up to date. Consequently, mobile surveillance work for pathogenic RNA viruses should not rest on mobile nucleic acid detection alone. Although mobile PCR can facilitate outbreak investigation and control, it does not obviate the necessity for serology assays.
Acknowledgments
We thank Robert E. Shope for providing the majority of strains from his collection. Without this generous support, the project would not have been possible. We are indebted to Melanie Riemer for perfect technical assistance.
This work was supported by grants InSanI 0598-V4301 and InSanI 030-V4304 from the Bundesministerium für Verteidigung, Germany.
REFERENCES
- 1.Abu Al-Soud, W., and P. Radstrom. 2000. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 38:4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardos, V., and C. E. Cupkova. 1962. The calovo virus—the second virus isolated from mosquitos in Czechoslovakia. J. Hyg. Epidemiol. Microbiol. Immunol. 6:186-192. [PubMed] [Google Scholar]
- 3.Bardos, V., and D. V. Danielova. 1959. The Tahyna virus—a virus isolated from mosquitos in Czechoslovakia. J. Hyg. Epidemiol. Microbiol. Immunol. 3:264-276. [PubMed]
- 4.Bowen, M. D., S. G. Trappier, A. J. Sanchez, R. F. Meyer, C. S. Goldsmith, S. R. Zaki, L. M. Dunster, C. J. Peters, T. G. Ksiazek, and S. T. Nichol. 2001. A reassortant bunyavirus isolated from acute hemorrhagic fever cases in Kenya and Somalia. Virology 291:185-190. [DOI] [PubMed] [Google Scholar]
- 5.Burt, F. J., P. A. Leman, J. F. Smith, and R. Swanepoel. 1998. The use of a reverse transcription-polymerase chain reaction for the detection of viral nucleic acid in the diagnosis of Crimean-Congo haemorrhagic fever. J. Virol. Methods 70:129-137. [DOI] [PubMed] [Google Scholar]
- 6.Calisher, C. H. 1994. Medically important arboviruses of the United States and Canada. Clin. Microbiol. Rev. 7:89-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler, D. P., C. A. Wagnon, and H. Bolton. 1998. Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Appl. Environ. Microbiol. 64:669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DaSilva, E., and M. Iaccarino. 1999. Emerging diseases: a global threat. Biotechnol. Adv. 17:363-384. [DOI] [PubMed] [Google Scholar]
- 9.Dunn, E. F., D. C. Pritlove, and R. M. Elliott. 1994. The S RNA genome segments of Batai, Cache Valley, Guaroa, Kairi, Lumbo, Main Drain and Northway bunyaviruses: sequence determination and analysis. J. Gen. Virol. 75:597-608. [DOI] [PubMed] [Google Scholar]
- 10.Elliot, R. M. (ed.). 1996. The Bunyaviridae. Plenum Press, New York. N.Y.
- 11.Elliott, R. M., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. Pettersson, A. Plyusnin, and C. S. Schmaljohn. 2000. Family Bunyaviridae. In M. H. V. van Regenmortel et al. (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 12.Geevarghese, G., N. Y. Prasanna, P. G. Jacob, Hanumaiah, and H. R. Bhat. 1994. Isolation of Batai virus from sentinel domestic pig from Kolar district in Karnataka State, India. Acta Virol. 38:239-240. [PubMed] [Google Scholar]
- 13.Heymann, D. L., and G. R. Rodier. 2001. Hot spots in a wired world: W. H. O. surveillance of emerging and re-emerging infectious diseases. Lancet Infect. Dis. 1:345-353. [DOI] [PubMed] [Google Scholar]
- 14.Hoff, G. L., R. O. Anslow, J. Spalatin, and R. P. Hanson. 1971. Isolation of Montana snowshoe hare serotype of California encephalitis virus group from a snowshoe hare and Aedes mosquitoes. J. Wildl. Dis. 7:28-34. [DOI] [PubMed] [Google Scholar]
- 15.Huang, C., W. H. Thompson, N. Karabatsos, L. Grady, and W. P. Campbell. 1997. Evidence that fatal human infections with La Crosse virus may be associated with a narrow range of genotypes. Virus Res. 48:143-148. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, J. M. 2001. Emerging infectious diseases: a CDC perspective. Emerg. Infect. Dis. 7:494-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuno, G. 1998. Universal diagnostic RT-PCR protocol for arboviruses. J. Virol. Methods 72:27-41. [DOI] [PubMed] [Google Scholar]
- 18.Kuo, K. W., M. F. Leung, and W. C. Leung. 1997. Intrinsic secondary structure of human TNFR-I mRNA influences the determination of gene expression by RT-PCR. Mol. Cell. Biochem. 177:1-6. [DOI] [PubMed] [Google Scholar]
- 19.Lundstrom, J. O. 1999. Mosquito-borne viruses in western Europe: a review. J. Vector Ecol. 24:1-39. [PubMed] [Google Scholar]
- 20.L'Vov, D. K., V. L. Gromashevskii, T. M. Skvortsova, V. A. Aristova, L. V. Kolobukhina, T. N. Morozova, I. V. Galkina, A. M. Butenko, M. S. Nedialkova, I. M. Selivanov, et al. 1998. Circulation of viruses of the California serocomplex (Bunyaviridae, Bunyavirus) in the central and southern parts of the Russian plain. Vopr. Virusol. 43:10-14. (In Russian.) [PubMed]
- 21.L'Vov, S. D., V. L. Gromashevskii, I. V. Voropanov, V. P. Andreev, and T. M. Skvortsova. 1989. Isolation of viruses of antigenic complexes of California encephalitis and Bunyamwera (Bunyaviridae, Bunuavirus) from mosquitoes in northeast Asia. Vopr. Virusol. 34:333-338. (In Russian.) [PubMed]
- 22.Mitchell, C. J., S. D. Lvov, H. M. Savage, C. H. Calisher, G. C. Smith, D. K. Lvov, and D. J. Gubler. 1993. Vector and host relationships of California serogroup viruses in western Siberia. Am. J. Trop. Med. Hyg. 49:53-62. [DOI] [PubMed] [Google Scholar]
- 23.Myers, T. W., and D. H. Gelfand. 1991. Reverse transcription and DNA amplification by a Thermus thermophilus DNA polymerase. Biochemistry 30:7661-7666. [DOI] [PubMed] [Google Scholar]
- 24.Nashed, N. W., J. G. Olson, and A. el-Tigani. 1993. Isolation of Batai virus (Bunyaviridae:Bunyavirus) from the blood of suspected malaria patients in Sudan. Am. J. Trop. Med. Hyg. 48:676-681. [DOI] [PubMed] [Google Scholar]
- 25.Newton, C. R., A. Graham, L. E. Heptinstall, S. J. Powell, C. Summers, N. Kalsheker, J. C. Smith, and A. F. Markham. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17:2503-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinheiro, F. P., A. P. Travassos da Rosa, J. F. Travassos da Rosa, R. Ishak, R. B. Freitas, M. L. Gomes, J. W. LeDuc, and O. F. Oliva. 1981. Oropouche virus. I. A review of clinical, epidemiological, and ecological findings. Am. J. Trop. Med. Hyg. 30:149-160. [PubMed] [Google Scholar]
- 27.Pinheiro, F. P., A. P. A. Travassos da Rosa, and P. F. C. Vasconcelos (ed.). 1998. An overview of Oropouche fever epidemics in Brazil and neighbouring countries. Instituto Evandro Chagas, Belem, Brazil.
- 28.Rychlik, W., W. J. Spencer, and R. E. Rhoads. 1990. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 18:6409-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeed, M. F., H. Wang, M. Nunes, P. F. Vasconcelos, S. C. Weaver, R. E. Shope, D. M. Watts, R. B. Tesh, and A. D. Barrett. 2000. Nucleotide sequences and phylogeny of the nucleocapsid gene of Oropouche virus. J. Gen. Virol. 81:743-748. [DOI] [PubMed] [Google Scholar]
- 30.Saikku, P., C. H. von Bonsdorff, M. Brummer-Korvenkontio, and A. Vaheri. 1971. Isolation of non-cubical ribonucleoprotein from Inkoo virus, a Bunyamwera supergroup arbovirus. J. Gen. Virol. 13:335-337. [DOI] [PubMed] [Google Scholar]
- 31.Sall, A. A., J. Thonnon, O. K. Sene, A. Fall, M. Ndiaye, B. Baudez, C. Mathiot, and M. Bouloy. 2001. Single-tube and nested reverse transcriptase-polymerase chain reaction for detection of Rift Valley fever virus in human and animal sera. J. Virol. Methods 91:85-92. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz, K., T. Hansen-Hagge, and C. Bartram. 1990. Improved yields of long PCR products using gene 32 protein. Nucleic Acids Res. 18:1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sexton, D. J., P. E. Rollin, E. B. Breitschwerdt, G. R. Corey, S. A. Myers, M. R. Dumais, M. D. Bowen, C. S. Goldsmith, S. R. Zaki, S. T. Nichol, C. J. Peters, and T. G. Ksiazek. 1997. Life-threatening Cache Valley virus infection. N. Engl. J. Med. 336:547-549. [DOI] [PubMed] [Google Scholar]
- 34.Simkova, A., and F. Sluka. 1973. Isolation of Tahyna virus from the blood of a case of influenza-like disease. Acta Virol. 17:94. [PubMed] [Google Scholar]
- 35.Sokol, D. K., M. B. Kleiman, and B. P. Garg. 2001. LaCrosse viral encephalitis mimics herpes simplex viral encephalitis. Pediatr. Neurol. 25:413-415. [DOI] [PubMed] [Google Scholar]
- 36.Vasconcelos, P. F., A. P. Travassos da Rosa, S. G. Rodrigues, E. S. Travassos da Rosa, N. Dégallier, and J. F. Travassos da Rosa. 2001. Inadequate management of natural ecosystem in the Brazilian Amazon region results in the emergence and reemergence of arboviruses. Cad. Saúde Pública 17:155-164. [DOI] [PubMed] [Google Scholar]