Abstract
We studied 31 clinical isolates of Salmonella enterica serotype Mbandaka resistant to broad-spectrum cephalosporins and recovered in Tunisia over a 5-year period. The transferability of this resistance was demonstrated by conjugation experiments. Thirty of the 31 isolates were positive in the double-disk synergy test. By isoelectric focusing analysis, all of the isolates were found to produce a band of β-lactamase activity with a pI of 5.9. Three of these isolates produced an additional band with a pI of 7.6. PCR and DNA sequencing identified these β-lactamases as TEM-4 and SHV-2a, respectively. The remaining isolate, highly resistant to ceftazidime but susceptible to cefepime, produced a β-lactamase that focused at pI 7.8. No synergy was detected by the double-disk synergy test. Sequence analysis of the bla gene amplified by PCR showed that the plasmid-mediated AmpC-type enzyme was ACC-1a. Fingerprinting analysis by repetitive-element PCR and enterobacterial repeat intergenic consensus-PCR suggested that 29 of the 31 Salmonella serotype Mbandaka isolates belonged to the same clonal population.
Salmonella enterica is a major endemic and epidemic pathogen in animals and humans worldwide. Animals and their products, particularly meat, chicken eggs, and milk, are major sources of human infection. The incidence of nontyphoid Salmonella infections has increased considerably in many countries, but with marked differences among countries (http:/www.who.int/emc/diseases/zoo/SALM-SURV). In Tunisia, studies done in the 1980s suggested that the most common S. enterica serotype isolated from human and animal sources was serotype Wien (25). In contrast, recent reports show that S. enterica serotype Enteritidis is becoming the predominant serotype in both humans and other animals in Tunisia. Other serotypes, such as Mbandaka, Braendrup, Typhimurium, Anatum, and Infantis, are also frequently encountered in this country (12-14). Antibiotics are necessary to treat most children and neonates with salmonellosis, broad-spectrum cephalosporins (e.g., ceftriaxone) being the primary drugs of choice. Over the past 2 decades and particularly in developing countries, Salmonella strains resistant to broad-spectrum cephalosporins have been reported (30). These strains produce extended-spectrum β-lactamases (ESBLs), such as TEM and SHV (TEM-3, TEM-4, TEM-25, TEM-27, SHV-2, and SHV-5) (1, 4, 7, 8, 16, 18, 22, 25, 33, 41, 42), CTX-M-2 (10, 23), CTX-M-3 (7), PER-1 (46), and PER-2 (11) and, more recently, plasmid-mediated AmpC-type enzymes, such as DHA-1 (21), CMY-2 (19, 20, 31, 35, 47-49), and ACC-1 (43). In North Africa, SHV-2 has been reported in Salmonella serotypes Wien (25), Typhimurium (22), and Mbandaka (27), TEM-3 has been reported in Salmonella serotype Typhimurium (1), CMY-2 has been reported in Salmonella serotype Senftenberg (31), and ACC-1 has been reported in Salmonella serotype Livingstone (43). Only TEM-25 and SHV-2 have been reported in Salmonella serotype Mbandaka (27, 41).
We studied 31 Salmonella serotype Mbandaka strains resistant to broad-spectrum cephalosporins and isolated from 1995 to 1999 at La Rabta University Hospital in Tunis, Tunisia. The β-lactamases were characterized by isoelectric focusing and molecular methods. Mechanisms involved in the transmission of bla genes among these Salmonella strains were investigated by conjugation experiments. We also used antimicrobial susceptibility patterns, plasmid profiles, and enterobacterial repeat intergenic consensus (ERIC)-PCR and repetitive-element PCR (rep-PCR) fingerprinting (36) for epidemiological characterization.
MATERIALS AND METHODS
Bacterial strains and media.
Thirty-one isolates of Salmonella serotype Mbandaka obtained from 1995 to 1999 at La Rabta University Hospital in Tunis, Tunisia, were studied (Table1). The strains were isolated from stools, blood cultures, the urinary tract, and pus (Table 1). All isolates were identified with the API 20E system (Biomerieux SA, Marcy l'Etoile, France). Isolates were serotyped on the basis of somatic O, phase 1 flagellar, and phase 2 flagellar antigen expression by agglutination tests with antisera (Bio-Rad SA, Marnes-la-Coquette, France). The following media, from Bio-Rad SA, were used for bacterial culturing: Mueller-Hinton (MH) agar, salmonella-shigella agar, Trypticase soy (TS) broth and agar, and Drigalski agar.
TABLE 1.
Date and site of isolation and β-lactamase and epidemiological characterizations of Tunisian clinical isolates of Salmonella serotype Mbandaka resistant to broad-spectrum cephalosporins
| Isolate | Yr of isolation | Site of isolation | pI(s) | β-Lactamase(s) | Resistance cotransferreda | Plasmid profile | rep-PCR profile | ERIC-PCR profile |
|---|---|---|---|---|---|---|---|---|
| MMAS1 | 1998 | Stools | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS2 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P2 | A | A2 |
| MMAS3 | 1997 | Pus | 5.9 | TEM-4 | G, TM, N | P3 | A | A2 |
| MMAS4 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P3 | A | A2 |
| MMAS5 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS6 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P4 | A | A2 |
| MMAS7 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P4 | A | A2 |
| MMAS8 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS10 | 1995 | Stools | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS13 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P5 | A | A2 |
| MMAS14 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P5 | A | A2 |
| MMAS15 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P2 | A | A2 |
| MMAS16 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P3 | A | A2 |
| MMAS17 | 1997 | Stools | 5.9 | TEM-4 | G, TM, N | P3 | A | A2 |
| MMAS19 | 1998 | Stools | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS21 | 1998 | Stools | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS22 | 1998 | Stools | 5.9 | TEM-4 | None | P7 | B | B |
| MMAS23 | 1998 | Stools | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS24 | 1998 | Stools | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS26 | 1998 | Blood | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS27 | 1998 | Blood | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS28 | 1998 | Stools | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS29 | 1998 | Blood | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS30 | 1999 | Stools | 5.9 | TEM-4 | N, G, TM, N | P6 | A | A1 |
| MMAS31 | 1999 | Stools | 5.9, 7.6 | TEM-4, SHV-2a | G, TM, N, TP, S | P6 | A | A1 |
| MMAS32 | 1999 | Stools | 5.9 | TEM-4 | G, TM, N | P6 | A | A1 |
| MMAS33 | 1999 | Stools | 5.9 | TEM-4 | G, TM, N | P6 | A | A1 |
| MMAS35 | 1999 | Stools | 5.9, 7.6 | TEM-4, SHV-2a | G, TM, N, TP, S | P6 | A | A1 |
| MMAS37 | 1999 | Stools | 5.9, 7.6 | TEM-4, SHV-2a | G, TM, N, TP, S | P6 | A | A1 |
| MMAS39 | 1999 | Urinary tract | 5.9 | TEM-4 | G, TM, N | P1 | A | A1 |
| MMAS40 | 1999 | Pus | 7.8 | ACC-1a | G, TM, N, TP, S | P8 | C | C |
G, gentamicin; TM, tobramycin; N, netilmicin; TP, trimethoprim; S, sulfonamides.
Antimicrobial susceptibility testing.
We used the disk diffusion assay with MH agar in accordance with French National Antibiogram Committee guidelines (Communiqué 2002; www.sfm.asso.fr/Sect4/atbfr.html). The isolates were tested for ESBL production by the double-disk synergy (DDS) method. The disks were placed 25 mm from center to center.
Conjugation experiments.
Escherichia coli strain K-12 (resistant to nalidixic acid) and E. coli J53-2 (resistant to rifampin) were used as recipients in transfer experiments. Overnight cultures of donor and recipient strains in TS broth at 37°C were mixed on MH agar at a ratio of 1:1 (200 μl of donor culture and 200 μl of recipient culture). The mixture was spread with Pasteur pipettes and incubated at 37°C for 18 h. Colonies from the mixed culture were suspended in 5 ml of sterile water, and 200 μl of the suspension was plated on Drigalski agar containing rifampin at 250 μg/ml or nalidixic acid at 50 μg/ml and cefotaxime at 2.5 μg/ml. Growing colonies were subjected to the DDS test to confirm the presence of ESBL transconjugants. All of the transconjugants were tested for their susceptibility to all of the antibiotics used for the donors.
Plasmid DNA analysis.
Plasmid DNA was extracted from Salmonella serotype Mbandaka isolates and their transconjugants by the method of Kado and Liu (29). The resulting DNA preparation was submitted to electrophoresis on horizontal slab gels containing 0.8% agarose (Bio-Rad SA). Plasmids with known molecular sizes—pIP112 (100 kb), pIP173 (128 kb), pCFF04 (85 kb), and pBK-CMV (4.4 kb) (Stratagene, Amsterdam, The Netherlands)—were used for size estimation. Electrophoresis was carried out at 100 V for 3 h. The slab gels were stained in ethidium bromide solution (10 μg/ml) for 10 min, followed by immersion in distilled water for 45 min. The slab gels were placed under UV light, and the bands were analyzed by using a computer-based program (Gel Doc 1000; Bio-Rad SA).
PCR fingerprinting.
Template DNA was extracted from each isolate by using a commercial genomic DNA purification kit (QIAampDNA mini kit; Qiagen, Courtaboeuf, France). For rep-PCR (final volume, 100 μl), the primers were REP1R-Dt (5′-IIINCGNCGNCATCNGGC-3′) and REP2-Dt (5′-NCGNCTTATCNGGCCTAC-3′), used as a pair. We used 50 pM each of primers REP1R-Dt and REP2-Dt, 5 U of Taq DNA polymerase (Amersham Pharmacia Biotech, Saclay, France), and 10 μl of PCR buffer. The rep-PCR parameters were as follows: initial denaturation at 95°C for 3 min; 40 cycles of denaturation at 92°C for 30 min, annealing at 40°C for 1 min, and extension at 65°C for 8 min; and a final extension at 65°C for 15 min. For ERIC-PCR, we used only one primer, ERIC2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′). The PCR parameters were identical to those used for rep-PCR, except for the annealing temperature (52°C).
PCR amplicons were resolved on 1% (wt/vol) agarose (Bio-Rad SA) containing ethidium bromide (0.5 μg/ml) by horizontal electrophoresis in Tris-borate-EDTA buffer. Gels were visualized under UV light and photographed with a computer-controlled image analyzer (Gel Doc 1000).
IEF of β-lactamases.
Crude β-lactamase preparations were obtained by sonication. Briefly, 50 ml of overnight cultures at 37°C in TS broth were centrifuged at 5,000 rpm for 5 min. The supernatants were eliminated, and the cells were resuspended in sterile distilled water (weight/weight). The bacterial suspensions were disrupted by sonication, twice for 30 s each at 40 Hz (Vibracell 300; Bioblock, Ilkirch, France). Enzyme activity was detected before electrophoresis by applying crude extracts to nitrocefin disks (Biomerieux SA). The extracts were subjected to isoelectric focusing (IEF) on pH 3.5 to 10 Ampholine (Amersham) polyacrylamide gels (32). TEM-25 and TEM-2 (Salmonella serotype Mbandaka CF1509; pIs, 5.3 and 5.6, respectively), TEM-3 (pCFF04; pI, 6.3), ACC-1 and TEM-1 (Klebsiella pneumoniae SLK54; pIs, 7.8 and 5.4, respectively), and CTX-M-9 and TEM-1 (E. coli RAJ; pIs, 8.2 and 5.4, respectively) were used as pI markers. Electrophoresis was performed at 14°C, 400 V, 6 W, and 15 mA for 18 h. β-Lactamase activity was detected by the chromogenic nitrocefin test by overlaying the gels with filter paper soaked with nitrocefin solution (0.5 mg/ml).
Molecular characterization of β-lactamase-encoding genes.
Genomic DNA was extracted (QIAampDNA mini kit). Specific primers were used for PCR. Primers OT3 and OT4 were used to amplify the putative blaTEM genes (5). The putative blaSHV genes were amplified with the corresponding primers, namely, OS5 and OS6 (6). The internal primers ACC upper and ACC lower were used to amplify the internal fragments of the blaACC gene (34). PCR assays were performed with a final volume of 100 μl containing 4 U of Taq DNA polymerase and 50 pM each primer. The PCR parameters were as follows: initial denaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 30 s, and extension at 72°C for 1 min; and a final extension at 72°C for 7 min.
The amplified bla genes were sequenced by using two independent PCR products along both strands (44) with PCR primers as sequencing primers on an ABI 373A DNA sequencer (Applied Biosystems, Foster City, Calif.). The nucleotide sequences and deduced protein sequences were analyzed with the BLAST and ClustalW programs (2, 45).
RESULTS AND DISCUSSION
Identification of Salmonella isolates.
The 31 isolates were identified as Salmonella by biochemical characterization and as Salmonella serotype Mbandaka by serotyping (O 6,7, Z10, e,n,z15).
Antimicrobial susceptibility and transferability of resistance.
All of the isolates except for MMAS40 were resistant to amoxicillin, cephalothin, ticarcillin, piperacillin, cefuroxime, cefotaxime, ceftazidime, and ceftriaxone. They were susceptible to β-lactam- β-lactamase inhibitor combinations and to cefoxitin and imipenem. The DDS test was positive for all of these isolates. MMAS40 was highly resistant to the amoxicillin-clavulanic acid and tazobactam-piperacillin combinations. MMAS40 was also highly resistant to ceftazidime and fully susceptible to cefepime (almost all of the other isolates showed intermediate susceptibility), cefoxitin, and imipenem. These β-lactam resistance phenotypes were always associated with gentamicin, tobramycin, and netilmicin resistance. Six isolates (MMAS30, MMAS31, MMAS32, MMAS35, MMAS37, and MMAS40) were resistant to the trimethoprim-sulfonamide combination. All of the isolates were fully susceptible to nalidixic acid, ofloxacin, ciprofloxacin, rifampin, and chloramphenicol. Our findings and those reported over the last 10 years in Tunisia and other countries worldwide show the emergence of resistance to broad-spectrum cephalosporins in Salmonella isolates (1, 4, 7-11, 16, 18-23, 25, 27, 31, 33, 35, 41-43, 46-49). Such isolates are also often resistant to chloramphenicol and co-trimoxazole. Fluoroquinolones are the last family of antibiotics to remain active against these isolates, but they are not yet approved for use in children.
The transferability of broad-spectrum cephalosporin resistance by conjugation has been frequently observed among the Enterobacteriaceae. This phenomenon is incriminated in the diffusion of this resistance, which is generally encoded by large transferable plasmids. Resistance to β-lactams, aminoglycosides, and co-trimoxazole is readily transferable by conjugation to E. coli. A single large plasmid (molecular size of between 110 and 140 kb) was detected in each transconjugant (data not shown).
Characterization of broad-spectrum cephalosporin resistance.
In IEF studies, all but one of the isolates (MMAS40) produced a β-lactamase with a pI of 5.9. Three isolates (MMAS31, MMAS35, and MMAS37) produced an additional β-lactamase with a pI of 7.6. MMAS40 differed from the other isolates, producing a β-lactamase with a pI of 7.8 (data not shown) (Table 1). Similar results were obtained for the transconjugants.
blaTEM amplification was obtained with primers OT3 and OT4 for all of the isolates and their transconjugants that produced a β-lactamase with a pI of 5.9 in IEF (i.e., except for MMAS40). With primers OS5 and OS6, the blaSHV gene was amplified only for MMAS31, MMAS35, and MMAS37 and their transconjugants that produced an additional band of β-lactamase activity at pI 7.6. DNA sequencing of PCR products was performed only for clinical isolates. Analysis of deduced amino acid sequences showed that blaTEM encoded a TEM-4 β-lactamase (pI 5.9) and that blaSHV encoded an SHV-2a β-lactamase (pI 7.6). The β-lactam resistance patterns, together with the positive DDS test results for 30 of 31 isolates, suggested the presence of class A ESBLs (15). SHV-2, one of the most frequently described ESBLs worldwide (28, 38, 40), was previously described for S. enterica and notably for a nosocomial outbreak in Tunisia (25). On the other hand, this was the first reported identification of the TEM-4 ESBL in Tunisian Salmonella isolates. TEM-4 was first reported for E. coli in France (37) and subsequently for E. coli and K. pneumoniae in Spain (17). This enzyme was recently described for an isolate of Salmonella collected during a French national survey in 1998 (18); unfortunately, no information on the origin of this isolate was available. This report was also the second description of two different ESBLs in salmonellae (7). The last isolate, MMAS40, had a particular β-lactam resistance phenotype—resistance to β-lactam-β-lactamase inhibitor combinations and susceptibility to cefepime—suggesting the presence of an AmpC-type enzyme (39). Susceptibility to cefoxitin and a pI of 7.8 suggested an ACC-type β-lactamase (34). This suggestion was confirmed by sequencing of the PCR product obtained with the ACC upper and ACC lower primers. Analysis of the deduced amino acid sequence showed the presence of the ACC-1a enzyme in this isolate. ACC-1 was first described in Germany and then in France during a multiresistant K. pneumoniae outbreak in an intensive care unit following the admission of a patient transferred from Sfax in Tunisia (34). A subsequent investigation in a Sfax hospital revealed the presence of the ACC-1 enzyme in many isolates of the Enterobacteriaceae, including K. pneumoniae, Proteus mirabilis, and S. enterica serotype Livingstone (43). All of these results suggest that the ACC-1 β-lactamase is probably widespread in Tunisia.
Epidemiological characteristics.
We used PCR fingerprinting to evaluate the relatedness of the Salmonella serotype Mbandaka isolates. Primer ERIC2 was used to amplify the enterobacterial repeat intergenic consensus sequence, yielding three patterns, A, B, and C. One pattern (A) contained two subtypes, A1 and A2 (lanes C and I versus lanes A, B, D, F, G, and H in Fig. 1), which differed by only one band of about 1 kb. Fingerprinting with combined primers REP1R-Dt and REP2-Dt also generated three patterns (data not shown). These observations suggested that the isolates belonged to three clonal Salmonella populations (Table 1). One population predominated, comprising 26 isolates producing TEM-4 and 3 isolates producing both TEM-4 and SHV-2. The last two fingerprints each corresponded to one isolate (MMAS22 and MMAS40, producing TEM-4 and ACC-1, respectively).
FIG. 1.
PCR fingerprint patterns obtained with primer ERIC2 for selected Tunisian clinical isolates of Salmonella serotype Mbandaka resistant to broad-spectrum cephalosporins. Lane A, isolate MMAS1 (pattern A1); lane B, isolate MMAS2 (pattern A1); lane C, isolate MMAS5 (pattern A2); lane D, isolate MMAS21 (pattern A1); lane E, isolate MMAS22 (pattern B); lane F, isolate MMAS23 (pattern A1); lane G, isolate MMAS29 (pattern A1); lane H, isolate MMAS31 (pattern A1); lane I, isolate MMAS32 (pattern A2).
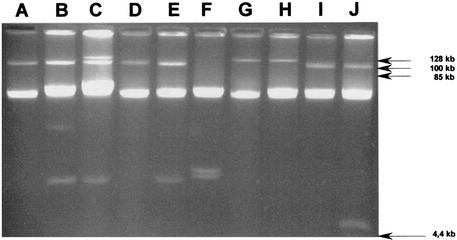
The 31 Salmonella isolates yielded eight plasmid profiles (Table 1) comprising between one and three bands (Fig. 2). The estimated molecular sizes of the plasmids were between 5 and 140 kb. The most predominant profile (P1) was shared by 13 isolates and corresponded to a single large plasmid of about 110 kb. The second profile (P6) (only one plasmid of about 130 kb) was shared by six isolates. The third profile, P3, shared by four isolates, was characterized by two large plasmids of about 110 and 140 kb. Plasmid profiles P2, P4, and P5 were each represented by two isolates and differed from P1 by the additional presence of one or two small plasmids (less than 20 kb). Profile P7, which corresponded only to isolate MMAS22, was unusual, being characterized by the absence of a large plasmid and the presence of two small plasmids. Profile P8 (MMAS40) corresponded to the presence of a large plasmid of about 110 kb and a small plasmid of about 5 kb.
FIG. 2.
Plasmid patterns of selected Tunisian clinical isolates of Salmonella serotype Mbandaka resistant to broad-spectrum cephalosporins. Lane A, isolate MMAS1 (pattern P1); lane B, isolate MMAS2 (pattern P2); lane C, isolate MMAS3 (pattern P3); lane D, isolate MMAS5 (pattern P1); lane E, isolate MMAS7 (pattern P4); lane F, isolate MMAS22 (pattern P7); lane G, isolate MMAS30 (pattern P6); lane H, isolate MMAS32 (pattern P6); lane I, isolate MMAS39 (pattern P1); lane J, isolate MMAS40 (pattern P8).
Epidemiological results based on PCR fingerprinting and plasmid analyses showed that 29 of the 31 isolates (with the exception of MMAS22 and MMAS40) belonged to the same clonal population, despite their different plasmid contents. The plasmid profiles of this clonal population distinguished at least three subclonal populations. The first corresponded to the predominant P1-like profiles (P1, P2, P4, and P5), characterized by a large conjugative plasmid (110 kb); the isolates yielding these profiles were recovered throughout the 5-year study period. Profile P3 corresponded to two isolates recovered in 1997. Profile P6 corresponded to six isolates recovered in 1999, three of which produced SHV-2. The last two isolates, MMAS22 and MMAS40, were unusual: no conjugative transfer was obtained with MMAS22, which harbored only two small plasmids, and MMAS40 was the only isolate to produce ACC-1.
In conclusion, our study confirms that S. enterica isolates are an important reservoir of genes encoding resistance to broad-spectrum cephalosporins in Tunisia. Most such genes encode ESBLs derived from TEM- or SHV-type β-lactamases (28, 38, 40). More recently, other enzymes were described; these belonged to Ambler class A (3), such as CTX-M-2 (10, 23, 26) (derived from the chromosome-encoded β-lactamase of Kluyvera ascorbata), CTX-M-3 (7), and PER-type enzymes (11, 46), or were derived from chromosomal class C β-lactamases, such as CMY-2 (19, 20, 31, 35, 47-49), DHA-1 (21), and ACC-1 (43), originating from Citrobacter freundii, Morganella morganii, and Hafnia alvei, respectively (9, 34, 39). These multiresistant isolates of Salmonella are often responsible for nosocomial outbreaks (23, 25, 42, 46) or for animal-to-human transmission (20, 48).
Salmonellae are ubiquitous, being found in aquatic environments, animals, and humans. Acquisition of new resistance genes by this genus could be facilitated by the simultaneous presence in the environment of naturally resistant Enterobacteriaceae or gram-negative bacilli and by traces of antimicrobial agents used in human and veterinary medicine. There is an increasingly urgent need to restrict the use of antimicrobial agents in animals (24) and humans alike and to control the disposal of these drugs in the environment.
Acknowledgments
This work was partially financed by grants from the Ministère de la Recherche (Réseau Beta-Lactamase) and from the Faculté de Médecine Saint-Antoine.
We thank D. Sirot for the gift of S. enterica serotype Mbandaka CF1509.
REFERENCES
- 1.AitMhand, R., A. Soukri, N. Moustaoui, H. Amarouch, N. ElMdaghri, D. Sirot, and M. Benbachir. 2002. Plasmid-mediated TEM-3 extended-spectrum β-lactamase production in Salmonella typhimurium in Casablanca. J. Antimicrob. Chemother. 49:169-172. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein data-base search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambler, R. P. 1980. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B 289:321-331. [DOI] [PubMed] [Google Scholar]
- 4.Archambaud, M., G. Gerbaud, E. Labau, N. Marty, and P. Courvalin. 1991. Possible in-vivo transfer of β-lactamase TEM-3 from Klebsiella pneumoniae to Salmonella kedougou. J. Antimicrob. Chemother. 27:427-436. [DOI] [PubMed] [Google Scholar]
- 5.Arlet, G., G. Brami, D. Decre, A. Flippo, O. Gaillot, P. H. Lagrange, and A. Philippon. 1995. Molecular characterisation by PCR-restriction fragment length polymorphism of TEM beta-lactamases. FEMS Microbiol. Lett. 134:203-208. [DOI] [PubMed] [Google Scholar]
- 6.Arlet, G., M. Rouveau, and A. Philippon. 1997. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum beta-lactamase. FEMS Microbiol. Lett. 152:163-167. [DOI] [PubMed] [Google Scholar]
- 7.Baraniak, A., E. Sadowy, W. Hryniewicz, and M. Gniadkowski. 2002. Two different extended-spectrum beta-lactamases (ESBLs) in one of the first ESBL-producing Salmonella isolates in Poland. J. Clin. Microbiol. 40:1095-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barguelli, F., C. Burucoa, A. Amor, J. L. Fauchere, and C. Fendri. 1995. In vivo acquisition of extended-spectrum β-lactamase in Salmonella enteritidis during antimicrobial therapy. Eur. J. Clin. Microbiol. Infect. Dis. 14:703-706. [DOI] [PubMed] [Google Scholar]
- 9.Barnaud, G., G. Arlet, C. Verdet, O. Gaillot, P. H. Lagrange, and A. Philippon. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob. Agents Chemother. 42:2352-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauernfeind, A., J. M. Casellas, M. Goldberg, M. Holley, R. Jungwirth, P. Mangold, T. Rohnisch, S. Schweighart, and R. Wilhelm. 1992. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20:158-163. [DOI] [PubMed] [Google Scholar]
- 11.Bauernfeind, A., I. Stemplinger, R. Jungwirth, P. Mangold, S. Amann, E. Akalin, O. Ang, C. Bal, and J. M. Casellas. 1996. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob. Agents Chemother. 40:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben Aissa, R., A. Bel Haj, N. Ben Haj, A. Chourabi, S. Ben Rhouma, F. Moknassi, and H. Troudi. 1996. Rapport d'activité du Centre National de Référence des Salmonella, Shigella et Vibrio cholerae—1995. Arch. Inst. Pasteur Tunis 73:35-40. [Google Scholar]
- 13.Ben Aissa, R., A. Bel Haj, N. Ben Haj, A. Chourabi, S. Ben Rhouma, F. Moknassi, and H. Troudi. 1997. Rapport d'activité du Centre National de Référence des Salmonella, Shigella et Vibrio cholerae—1996. Arch. Inst. Pasteur Tunis 74:33-40.15945175 [Google Scholar]
- 14.Ben Aissa, R., A. Bel Haj, N. Ben Haj, A. Chourabi, I. Hammami, F. Moknassi, and H. Troudi. 1998. Rapport d'activité du Centre National de Référence des Salmonella, Shigella et Vibrio cholerae—1997. Arch. Inst. Pasteur Tunis 75:35-43. [Google Scholar]
- 15.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabie, A., J. Jouanelle, and C. Saintaime. 1989. β-Lactamase à spectre élargi (CTX-1) chez Salmonella panama à Fort-de-France (Martinique). Med. Mal. Infect. 19:418-420. [Google Scholar]
- 17.Coque, T. M., A. Olivier, J. C. Pérez-Díaz, F. Baquero, and R. Canton. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum β-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46:500-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, et al. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunne, E. F., P. D. Fey, P. Kludt, R. reporter, F. Mostashari, P. Shillam, J. Wicklund, C. Miller, B. Holland, K. Stamey, T. J. Barrett, J. K. Rasheed, F. C. Tenover, E. M. Ribot, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC β-lactamase. JAMA 284:3151-3156. [DOI] [PubMed] [Google Scholar]
- 20.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone resistant Salmonella infections acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 21.Gaillot, O., C. Clement, M. Simonet, and A. Philippon. 1997. Novel transferable β-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J. Antimicrob. Chemother. 39:85-87. [DOI] [PubMed] [Google Scholar]
- 22.Garbarg-Chenon, A., H. Vu Thien, R. Labia, H. Ben Yaglane, V. Godart, P. Deny, F. Bricout, and J. C. Nicolas. 1989. Characterization of a plasmid coding for resistance to broad-spectrum cephalosporins in Salmonella typhimurium. Drugs Exp. Clin. Res. 15:145-150. [PubMed] [Google Scholar]
- 23.Gazouli, M., S. V. Sidorenko, E. Tzelepi, N. S. Kozlova, D. P. Gladin, and L. S. Tzouvelekis. 1998. A plasmid-mediated β-lactamase conferring resistance to cefotaxime in a Salmonella typhimurium clone found in St. Petersburg, Russia. J. Antimicrob. Chemother. 41:119-121. [DOI] [PubMed] [Google Scholar]
- 24.Gorbach, S. L. 2001. Antimicrobial use in animal feed—time to stop. N. Engl. J. Med. 345:1202-1203. [DOI] [PubMed] [Google Scholar]
- 25.Hammami, A., G. Arlet, S. Ben Redjeb, F. Grimont, A. Ben Hassen, A. Rekik, and A. Philippon. 1991. Nosocomial outbreak of acute gastroenteritis in a neonatal intensive care unit in Tunisia caused by multiply drug resistant Salmonella wien producing SHV-2 β-Lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 10:641-646. [DOI] [PubMed] [Google Scholar]
- 26.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Issak, M. I., K. P. Shannon, S. A. Qureshi, and G. L. French. 1995. Extended-spectrum β-lactamase in Salmonella. J. Hosp. Infect. 30:319-321. [DOI] [PubMed] [Google Scholar]
- 28.Jacoby, G. A., and A. A. Medeiros. 1991. More extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 35:1697-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kariuki, S., and C. A. Hart. 2001. Global aspects of antimicrobial-resistant enteric bacteria. Curr. Opin. Infect. Dis. 14:579-586. [DOI] [PubMed] [Google Scholar]
- 31.Koeck, J. L., G. Arlet, A. Philippon, S. Basmaciogullari, H. Vu Thien, Y. Buisson, and J.-D. Cavallo. 1997. A plasmid-mediated CMY-2 β-lactamase from an Algerian clinical isolate of Salmonella senftenberg. FEMS Microbiol. Lett. 152:255-260. [DOI] [PubMed] [Google Scholar]
- 32.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 33.Morosini, M. A., R. Canton, J. M. Martinez-Beltran, M. C. Negri, J. C. Perez-Diaz, F. Baquero, and J. Blazquez. 1995. New extended-spectrum TEM-type β-lactamase from Salmonella enterica subsp. enterica isolated in a nosocomial outbreak. Antimicrob. Agents Chemother. 39:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadjar, D., M. Rouveau, C. Verdet, J.-L. Donay, J.-L. Herrmann, P. H. Lagrange, A. Philippon, and G. Arlet. 2000. Outbreak of Klebsiella pneumoniae producing transferable AmpC-type β-lactamase (ACC-1) originating from Hafnia alvei. FEMS Microbiol. Lett. 187:35-40. [DOI] [PubMed] [Google Scholar]
- 35.Navarro, F., E. Perez-Trallero, J. M. Marimon, R. Aliaga, M. Gomariz, and B. Mirelis. 2001. CMY-2-producing Salmonella enterica, Klebsiella pneumoniae, Klebsiella oxytoca, Proteus mirabilis and Escherichia coli strains isolated in Spain (October 1999-December 2000). J. Antimicrob. Chemother. 48:383-389. [DOI] [PubMed] [Google Scholar]
- 36.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul, G. C., G. Gerbaud, A. Bure, A. M. Philippon, B. Pangon, and P. Courvalin. 1989. TEM-4, a new plasmid-mediated beta-lactamase that hydrolyzes broad-spectrum cephalosporins in a clinical isolate of Escherichia coli. Antimicrob. Agents Chemother. 33:1958-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philippon, A., G. Arlet, and P. H. Lagrange. 1994. Origin and impact of plasmid-mediated extended-spectrum β-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 13(Suppl. 1):2-11. [DOI] [PubMed] [Google Scholar]
- 39.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Philippon, A., R. Labia, and G. Jacoby. 1989. Extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 33:1131-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poupart, M. C., C. Chanal, D. Sirot, R. Labia, and J. Sirot. 1991. Identification of CTX-2, a novel cefotaximase from a Salmonella mbandaka isolate. Antimicrob. Agents Chemother. 35:1498-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Revathi, G., K. P. Shannon, P. D. Stapleton, B. K. Jain, and G. L. French. 1998. An outbreak of extended-spectrum β-lactamase-producing Salmonella senftenberg in a burns ward. J. Hosp. Infect. 40:295-302. [DOI] [PubMed] [Google Scholar]
- 43.Rhimi-Mahjoubi, F., M. Bernier, G. Arlet, Z. Ben Jemaa, P. Jouve, A. Hammami, and A. Philippon. 2002. Mise en évidence de la céphalosporinase plasmidique ACC-1 dans différentes entérobactéries (Klebsiella pneumoniae, Proteus mirabilis, Salmonella) isolées dans un Hôpital Tunisien (Sfax 1997-2000). Pathol. Biol. 50:7-11. [DOI] [PubMed] [Google Scholar]
- 44.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vahaboglu, H., S. Dodanli, C. Eroglu, R. Ozturk, G. Soyletir, I. Yildirim, and V. Avkan. 1996. Characterization of multiple-antibiotic-resistant Salmonella typhimurium strains: molecular epidemiology of PER-1-producing isolates and evidence for nosocomial plasmid exchange by a clone. J. Clin. Microbiol. 34:2942-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffman, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, S., D. G. White, P. F. McDermott, S. Friedman, L. English, S. Ayers, J. Meng, J. J. Maurer, R. Holland, and R. D. Walker. 2001. Identification and expression of cephamycinase blaCMY genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45:3647-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]