Abstract
The viral load assays AMPLICOR HIV-1 Monitor Test 1.5, Nuclisens HIV-1 QT, and Quantiplex HIV RNA 3.0 (bDNA) were evaluated for their abilities to quantify human immunodeficiency virus type 1 (HIV-1) RNA in 64 plasma samples from 21 children infected in Portugal. The children were infected with HIV-1 subtypes A1, B, F1, G, and BG recombinant virus. AMPLICOR v1.5 and Quantiplex v3.0 detected all samples, and there was a good correlation of results between the two kits. Thirty-eight specimens containing HIV-1 subtype B, G, or recombinant BG, could not be detected by Nuclisens HIV-1 QT. We also evaluated the new Retina HIV-1 assay on 21 samples that were HIV-1 positive; Retina HIV-1 failed to detect 5 of 11 subtype G specimens. AMPLICOR v1.5 and Quantiplex v3.0 assays may be used for HIV-1 RNA quantification in Portugal, whereas an improvement in sensitivity for subtype G and recombinant BG is required for Nuclisens HIV-1 QT and Retina HIV-1.
Quantification of the human immunodeficiency virus type 1 (HIV-1) RNA levels (viral load) in plasma is currently used to assess patient prognosis and manage antiretroviral therapy (11, 14, 18). The reduction in viral load induced by antiretroviral treatment indicates treatment efficacy and correlates with improved clinical outcome (12, 26). Detection and quantification of HIV RNA may also help to diagnose HIV-1 infection in children borne by infected mothers (20) and to screen organ (15) and blood (28) donations.
Phylogenetic analyses of viral sequences collected worldwide have revealed the existence of three distinct HIV-1 groups: M, O, and N. Nine subtypes and two subsubtypes have been described within the most prevalent M group: A1, A2, B, C, D, F1, F2, G, H, J, and K (19). Fourteen circulating recombinant forms (CRF), or intersubtype recombinant HIV-1 with epidemic significance, have been identified and named CRF01_AE to CRF14_BG (25). The HIV-1 epidemics in Western Europe and North America are caused mainly by subtype B. However, non-B subtypes and recombinant forms have been identified in most countries, an indication that the epidemics may change with time (25). In Lisbon, Portugal, subtype G is already the second most prevalent HIV-1 subtype, accounting for approximately 21% of all infections (10). To maintain the clinical and diagnostic usefulness, viral load assays should accurately detect and quantify all HIV-1 group M subtypes and subsubtypes, as well as group O HIV-1.
Among the commercially available medical devices used to quantify HIV-1 RNA in plasma, the most widely used are AMPLICOR HIV-1 Monitor Test 1.5 (Roche Molecular Systems), Nuclisens HIV-1 QT (Organon Tecknika), and Quantiplex HIV RNA 3.0 (bDNA) (Bayer Diagnostics) (2, 8, 16, 22, 23, 27). AMPLICOR v1.5 and Quantiplex v3.0 detect and quantify accurately HIV-1 subtypes A to H, CRF01_AE, and CRF02_AG. Nuclisens tends to underestimate HIV-1 subtype A levels and detects poorly subtype G and CRF02_AG (4, 5, 16, 17, 22). Few studies have directly compared the clinical performances of these three test versions with different subtypes of HIV-1 (16). In all studies evaluating clinical sensitivity, only a small number of subtype G clinical samples were tested and no BG recombinant viruses were tested (3, 5, 23). Retina HIV-1 assay is a nucleic acid sequence-based amplification assay that uses primers targeting the long terminal repeat region and real-time detection with molecular beacons (5). Compared with Nuclisens, Retina HIV-1 displays improved quantification of subtypes A, G, CRF01_AE, and CRF02_AG (5).
As a reference laboratory, we perform HIV-1 viral load measurements for the diagnostic, follow-up, and therapy management of children infected by vertical transmission in Portugal. To determine which viral load assay would be more efficient in our population, we compared the abilities of AMPLICOR HIV-1 monitor test 1.5, Nuclisens HIV-1 QT, and Quantiplex HIV RNA 3.0 (bDNA) to detect and quantify HIV-1 RNA in 64 plasma samples drawn from 21 HIV-1-infected children being cared for at three hospitals in Lisbon and one hospital in Faro. Between two and four consecutive plasma samples were obtained from each child, with a mean interval between collections of samples of 3.6 months (range, 1 to 8 months). The overall mean (standard deviation [SD]) age of the subjects at the beginning of the study was 5.8 (3.4) years. Seven (33%) children were born to African mothers. Eleven (52%) were female. All patients were receiving antiretroviral therapy.
Quantification of HIV-1 RNA and genetic subtypes.
The viral load was determined as described by the manufacturers. For AMPLICOR v1.5 and Nuclisens the standard protocols were used (22, 27); the limit of quantification of these two assays is 400 copies of HIV-1 RNA per ml of plasma. The limit of quantification of Quantiplex v3.0 is 50 copies of HIV-1 RNA per ml of plasma (9). The lower quantification level of the Retina HIV-1 assay is approximately 100 copies per reaction (5).
HIV-1 genetic subtypes were determined for 18 subjects. HIV-1-specific reverse-transcription PCR was done on RNA recovered from 200 μl of plasma. Nested PCR was performed to obtain a 409-bp fragment from the C2-V3 env region by using outer primer pairs JA167 and JA170 and inner primers JA168 and JA169 and to obtain a 582-bp fragment from the p17 gag region by using outer primer pairs JA152 and JA155 and inner primers JA153 and JA154. The thermal cycling conditions for PCR and primer numbers and positions have been described previously (13). Amplified DNA fragments were sequenced with an automated DNA sequencer.
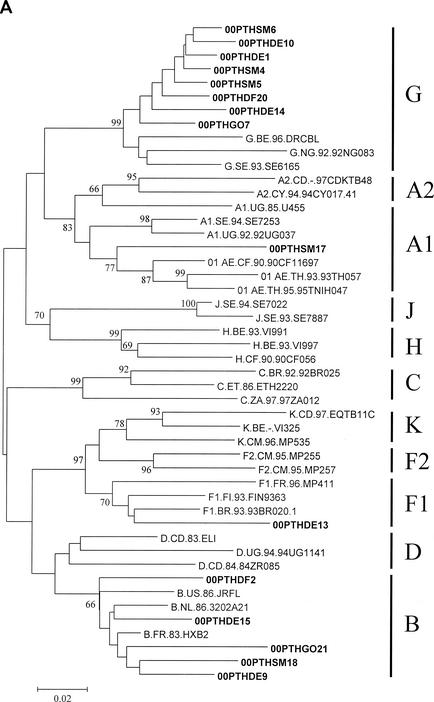
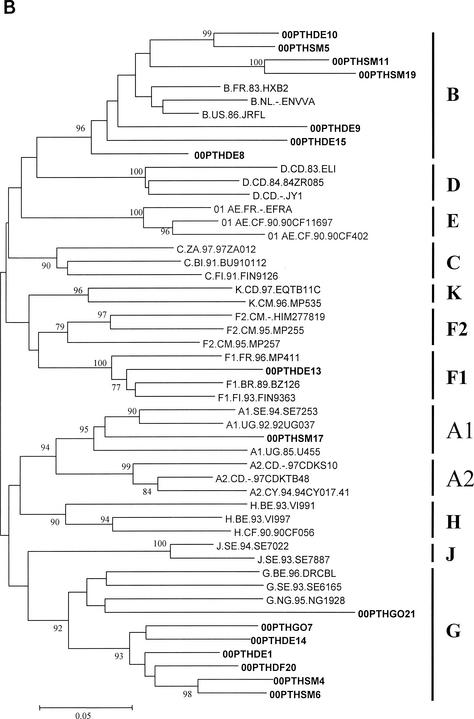
To determine the genetic subtype, the sequences were aligned with the 2001 HIV-1 reference sequence set obtained from the Los Alamos AIDS Database, the genetic distances between sequences were calculated by using the two-parameter substitution model of Kimura, and phylogenetic analysis was performed by using the neighbor-joining method with bootstrapping (S. Kumar, K. Tamura, I. B. Jakobsen, and M. Nei, MEGA2: Molecular Evolutionary Genetics Analysis software, Arizona State University, Tempe, Ariz., 2001). The distribution of the genetic subtypes was as follows (gag/env): 1 subtype A1/A1, 2 subtype B/B, 2 subtype B/−, 3 subtype −/B, 1 subtype F1/F1, and 6 subtype G/G (Fig. 1). Discordant subtype classifications were found in the gag and env gene sequences of the remaining three patients: patients 00PTHSM5 and 00PTHDE10 were subtype G in gag and subtype B in env (G/B), whereas patient 00PTHGO21 was subtype B in gag and subtype G in env (B/G). We were unable to genotype HIV-1 RNA from three patients due to insufficient amounts of plasma. These genotyping data are consistent with a complex HIV-1 epidemic in Portugal, with cocirculation of multiple subtypes and recombinant viruses (10). The predominance of HIV-1 subtype G and BG recombinant forms over other subtypes in Portugal resembles the pattern seen in HIV-1 epidemics in Galicia, Spain (7, 24). Preliminary full-length genomic sequencing studies indicate that two of our BG recombinant viruses (00PTHSM5 and 00PTHDE10) are representative of CRF14_BG (N. Taveira and F. McCutchan, unpublished data). The G/B recombinant virus composed of a subtype B gag and subtype G env identified for patient 00PTHGO21 has not been described previously.
FIG. 1.
Genetic subtypes and evolutionary relationships of the virus sequenced in this study based on neighbor-joining phylogenetic trees of partial gag (A) sequences (p17) and partial env (B) sequences (C2-V3). The phylogenetic trees were constructed with reference sequences from all HIV-1 subtypes and subsubtypes as well as with the Portuguese (PT) sequences (shown in boldface type). The bootstrap values supporting each of the internal branches defining a subtype or subsubtype are shown. Bootstrap values of 70% or greater provide reasonable confidence for assignment of an individual segment to one or the other genotype. The scale represents genetic distance.
Overall, AMPLICOR v1.5, Quantiplex v3.0, and Nuclisens detected viral RNA in 64 (100%), 64 (100%), and 33 (52%) of the 64 patient samples, respectively (Table 1). Retina HIV-1 detected viral RNA in 15 of 21 (71%) samples from 10 patients. There was a direct association between the genetic subtypes of HIV-1 and the low sensitivity of Nuclisens, as it detected all A1 and F1 samples but only 67% subtype B samples and 15.4% subtype G and BG recombinant samples (Table 1). This accounted for overall false-negative results for three patients infected with subtype G/G and one patient infected with G/B recombinant. The poor performance of this test with HIV-1 subtype G had already been demonstrated in other studies with a more restricted number of clinical samples (4-6, 16, 17, 22). Importantly, two of three samples from one subtype B-infected individual (00PTHDE9) and all samples from one individual (00PTHGO21) infected with a gag B and env G recombinant virus were not detected with Nuclisens but were detected with AMPLICOR v1.5 and Quantiplex v3.0. To our knowledge, this is the first demonstration that Nuclisens may fail to detect subtype B samples. To investigate whether these negative results were due to recombination within the gag gene, the p17 sequence from these patients was analyzed by bootscanning using SimPlot version 2.5 (21; S. C. Ray, SimPlot for Windows [version 2.5], 1999, Baltimore, Md.; http://www.welch.jhu.edu/∼sray/download). No evidence for recombination was obtained for patient 00PTHDE9. In contrast, there was a B/G recombination breakpoint located within the gag gene of patient 00PTHGO21 (data not shown). This may explain the negative results obtained with this patient, since Nuclisens primers hybridize within the gag gene region (5, 22).
TABLE 1.
Numbers and percentages of patient samples in which HIV-1 RNA was detected with AMPLICOR HIV-1 Monitor Test version 1.5, Nuclisens HIV-1 QT, and Quantiplex HIV RNA 3.0 (bDNA) tests
| Genetic subtype (gag/env)a | No. of samples tested | No. (%) of samples in which HIV-1 RNA was detected by:
|
||
|---|---|---|---|---|
| AMPLICOR HIV-1 Monitor Test version 1.5 | Quantiplex 3.0 HIV RNA (bDNA) | Nuclisens HIV-1 QT | ||
| A1/A1 | 4 | 4 (100) | 4 (100) | 4 (100) |
| B/B | 6 | 6 (100) | 6 (100) | 4 (67) |
| −/B | 9 | 9 (100) | 9 (100) | 5 (56) |
| B/− | 5 | 5 (100) | 5 (100) | 5 (100) |
| F1/F1 | 4 | 4 (100) | 4 (100) | 4 (100) |
| G/G | 17 | 17 (100) | 17 (100) | 3 (18) |
| B/G | 3 | 3 (100) | 3 (100) | 0 (0) |
| G/B | 6 | 6 (100) | 6 (100) | 1 (17) |
| U | 10 | 10 (100) | 10 (100) | 7 (70) |
| Total | 64 | 64 (100) | 64 (100) | 33 (52) |
−/B, genotyped only in the env gene; B/−, genotyped only in the gag gene; U, unknown genotype.
In the small subset of samples that were tested, the Retina HIV-1 assay performed better than Nuclisens. HIV-1 RNA was not detected in 5 of 11 (45%) subtype G/G samples with the Retina HIV-1 assay (data not shown). There was one overall false-negative result for patient 00PTHDE14, who was infected with a subtype G/G virus. Of 21 plasma samples tested by Nuclisens and Retina HIV-1 assay, 5 samples from 3 subtype G-infected patients had no viral RNA that was detectable by either assay. Nuclisens detected one sample that was not detected with Retina HIV-1. Retina HIV1 detected HIV-1 RNA in 11 samples, 5 G/G, 2 −/B, 2B/B, and 2 G/B, that were not detected with Nuclisens. In contrast to what was observed for the Nuclisens assay, HIV-1 RNA was detected in all subtype B/B samples that were tested with Retina HIV-1.
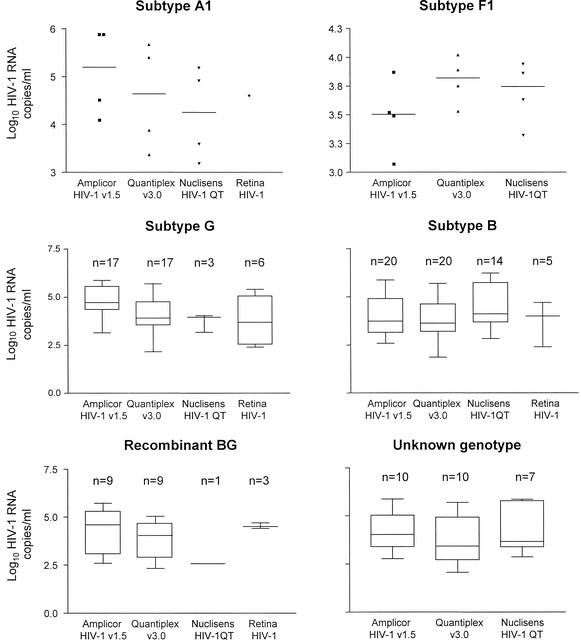
To determine the abilities of the four tests to quantify HIV-1 RNA levels of various subtypes, we compared the viral loads obtained from the patient's samples with each test (Fig. 2). The overall mean (SD) viral loads measured in the clinical samples were 4.33 (1.04; n = 64), 4.16 (1.01; n = 33), 3.94 (0.98; n = 64), and 3.70 (1.30; n = 15) log10 copies per ml for AMPLICOR v1.5, Nuclisens, Quantiplex v3.0, and Retina HIV-1 tests, respectively. Excluding the Retina HIV-1 results, the highest mean (SD) viral loads for subtype A1, 5.090 (0.928) log10 HIV-1 RNA copies/ml; subtype G, 4.828 (0.721); and recombinant BG, 4.282 (1.161), samples were obtained with AMPLICOR v1.5. The highest mean (SD) viral load measurements for samples of unknown genotype and subtype B samples were obtained with Nuclisens: 4.161 (1.187) and 4.476 (1.083) log10 HIV-1 RNA copies/ml, respectively. Quantiplex v3.0 yielded the highest mean viral load with subtype F1, 3.798 (0.210). Nuclisens produced the lowest mean viral load for subtype A1, G, and BG; AMPLICOR v1.5 produced the lowest mean viral load for subtype F1; and Quantiplex v3.0 produced the lowest mean viral loads for subtype B and samples not genotyped. Except for subtype F1 samples and samples of unknown genotype, the differences between the higher and lower mean viral load values obtained with the three assays for the different subtypes or recombinant samples were >0.5 log10 copies per ml (range, 0.616 to 1.712). This restates the importance of using the same test for patient follow-up.
FIG. 2.
HIV-1 RNA levels (log10 copies/ml) for subtypes A1, B, G, F1, and BG recombinant forms. The box extends from the 25th percentile to the 75th percentile, with a line at the median (50th percentile). The tabs extend above and below the boxes to show the highest and lowest values. Only data for samples that were positive were included in the figure. Due to the low number of cases, individual data points are shown for subtypes A1 and F1. V, version. Subtype B graphic includes B/B, −/B, and B/− samples. Recombinant B/G and G/B samples are included in the recombinant BG graphic.
The frequencies of discordant results (i.e., when the difference in log10 copies per ml was >0.5) were 34% between Nuclisens and Quantiplex v3.0, 47% between AMPLICOR v1.5 and Nuclisens, and 45% between Quantiplex v3.0 and AMPLICOR v1.5 (Table 2). The poor agreement obtained between Quantiplex v3.0 and AMPLICOR v1.5 in our population is in contrast to the 92.7% concordant results obtained in a recent study involving mostly subtype B samples (1). The frequency of discordant results obtained in our study was higher (76%) for the subtype G samples. In addition, a higher mean viral load, reaching clinical significance (0.697 log10 copies per ml), was obtained for subtype G with AMPLICOR v1.5. These results, therefore, highlight a significant difference in performance between AMPLICOR and Quantiplex with subtype G virus that may have an impact in patient management.
TABLE 2.
Numbers and percentages of discordant results between AMPLICOR HIV-1 Monitor Test version 1.5, Nuclisens HIV-1 QT, and Quantiplex HIV RNA 3.0 (bDNA) tests
| Genetic subtype | No. (%) of discordant resultsa/total no. of results obtained with viral load tests:
|
||
|---|---|---|---|
| Quantiplex vs. Amplicor | Amplicor vs. Nuclisens | Nuclisens vs. Quantiplex | |
| A1 | 2/4 (50) | 4/4 (100) | 0/4 (0) |
| B | 6/20 (30) | 6/14 (43) | 5/14 (36) |
| F1 | 0/4 (0) | 0/4 (0) | 0/4 (0) |
| G | 13/17 (76) | 4/5 (80) | 4/5 (80) |
| U | 4/10 (40) | 2/7 (29) | 2/7 (29) |
| B/G | 4/9 (44) | 2/4 (50) | 2/4 (50) |
| Total | 29/64 (45) | 18/38 (47) | 13/38 (34) |
Results for which the difference in log10 HIV-1 RNA copies per ml was >0.5.
The Spearman rank test was used to analyze the correlation between assays. AMPLICOR v1.5 and Quantiplex v3.0 were strongly correlated (r = 0.8805, P < 0.0001, n = 64), and this was independent of subtype (subtype B, r = 0.7960, P = 0.0007; subtype G, r = 0.8505, P < 0.0001; recombinant BG, r = 0.8619, P = 0.0045), demonstrating that these assays are well suited to quantify subtype G and recombinant BG virus RNA in clinical samples. In contrast, a weaker but significant correlation was found between Nuclisens and Quantiplex v3.0 (r = 0.7123, P < 0.0001, n = 33) and between Nuclisens and AMPLICOR v1.5 (r = 0.5612, P = 0.0007, n = 33). The coefficient of correlation between Nuclisens and Quantiplex v3.0 increased to 0.9373 for subtype B samples (P < 0.0001) and 0.8929 (P = 0.0123) for samples of unknown subtype. Higher correlation coefficients were also found between Nuclisens and AMPLICOR v1.5 when only subtype B (r = 0.7960, P < 0.0007) and samples of unknown subtype (r = 0.9286, P = 0.0067) were analyzed.
In summary, in 38 plasma specimens from Portuguese children infected with HIV-1 subtype B, G, or recombinant BG the infecting virus was not detected by the licensed Nuclisens HIV-1 QT. In contrast, HIV-1 RNA was detected and quantified in all samples with the two other assays that were tested, AMPLICOR v1.5 and Quantiplex v 3.0. Nuclisens HIV-1 QT is, therefore, unsuited for viral load quantification in HIV-1-infected patients living in Portugal and, probably, other countries with a high prevalence of subtype G virus.
Nucleotide sequence accession numbers.
The sequences determined in this study have been assigned GenBank accession no. AY161864 through AY161894.
Acknowledgments
Critical review of the manuscript by Anne-Mieke Vandamme is gratefully acknowledged.
This work was partially supported by grants CRIA-CR3751 and CR3753 from Comissão Nacional de Luta Contra a SIDA (CNLS), Portugal.
REFERENCES
- 1.Anastassopoulos, C. G., G. Touloumi, A. Katsoulidou, H. Hatzitheodorou, M. Pappa, D. Paraskevis, M. Lazanas, P. Gargalianos, and A. Hatzakis. 2001. Comparative evaluation of the Quantiplex HIV-1 RNA 2.0 and 3.0 (bDNA) assays and the Amplicor HIV-1 Monitor v1.5 test for the quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Virol. Methods 91:67-74. [DOI] [PubMed] [Google Scholar]
- 2.Berndt, C., U. Muller, F. Bergmann, U. Schmitt, R. Kaiser, and C. Muller. 2000. Comparison between a nucleic acid sequence-based amplification and branched DNA test for quantifying HIV RNA load in blood plasma. J. Virol. Methods 89:177-181. [DOI] [PubMed] [Google Scholar]
- 3.Burgisser, P., P. Vernazza, M. Flepp, J. Boni, Z. Tomasik, U. Hummel, G. Pantaleo, and J. Schupbach, et al. 2000. Performance of five different assays for the quantification of viral load in persons infected with various subtypes of HIV-1. J. Acquir. Immune Defic. Syndr. 23:138-144. [DOI] [PubMed] [Google Scholar]
- 4.de Baar, M. P., A. M. Van Der Schoot, J. Goudsmit, F. Jacobs, R. Ehren, K. H. Van Der Horn, P. Oudshoorn, F. de Wolf, and A. de Ronde. 1999. Design and evaluation of a human immunodeficiency virus type 1 RNA assay using nucleic acid sequence-based amplification technology able to quantify both group M and O viruses by using the long terminal repeat as target. J. Clin. Microbiol. 37:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Baar, M. P., M. W. van Dooren, E. de Rooij, M. Bakker, B. van Gemen, J. Goudsmit, and A. de Ronde. 2001. Single rapid real-time monitored isothermal RNA amplification assay for quantification of human immunodeficiency virus type 1 isolates from groups M, N, and O. J. Clin. Microbiol. 39:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debyser, Z., E. Van Wijngaerden, K. Van Laethem, K. Beuselinck, M. Reynders, E. De Clercq, J. Desmyter, and A.-M. Vandamme. 1998. Failure to quantify viral load with two of the three commercial methods in a pregnant woman harbouring an HIV type 1 subtype G strain. AIDS Res. Hum. Retrovir. 14:453-459. [DOI] [PubMed] [Google Scholar]
- 7.Delgado, E., M. M. Thomson, M. L. Villahermosa, M. Sierra, A. Ocampo, C. Miralles, R. Rodriguez-Perez, J. Diz-Aren, R. Ojea-de Castro, E. Losada, M. T. Cuevas, E. Vazquez-de Parga, R. Carmona, L. Perez-Alvarez, L. Medrano, L. Cuevas, J. A. Taboada, and R. Najera. 2002. Identification of a newly characterized HIV-1 BG intersubtype circulating recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J. Acquir. Immune Defic. Syndr. 29:536-543. [DOI] [PubMed] [Google Scholar]
- 8.Elbeik, T., W. G. Alvord, R. Trichavaroj, M. de Souza, R. Dewar, A. Brown, D. Chernoff, N. L. Michael, P. Nassos, K. Hadley, and V. L. Ng. 2002. Comparative analysis of HIV-1 viral load assays on subtype quantification: Bayer Versant HIV-1 RNA 3.0 versus Roche Amplicor HIV-1 Monitor version 1.5. J. Acquir. Immune Defic. Syndr. 29:330-339. [DOI] [PubMed] [Google Scholar]
- 9.Erice, A., D. Brambilla, J. Bremer, J. B. Jackson, R. Kokka, B. Yen-Lieberman, and R. W. Coombs. 2000. Performance characteristics of the Quantiplex HIV-1 RNA 3.0 assay for detection and quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 38:2837-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteves, A., R. Parreira, T. Venenno, M. Franco, J. Piedade, J. Germano de Sousa, and W. F. Canas-Ferreira. 2002. Molecular epidemiology of HIV type 1 infection in Portugal: high prevalence of non-B subtypes. AIDS Res. Hum. Retrovir. 18:313-325. [DOI] [PubMed] [Google Scholar]
- 11.Grant, L. A., M. J. Silverberger, H. Palacio, H. Minkoff, F. Anastos, M. A. Young, M. Nowicki, A. Kovacs, M. Cohen, and A. Munoz. 2001. Discontinuation of potent antiretroviral therapy: predictive value of and impact on CD4 cells counts and HIV RNA levels. AIDS 15:2101-2108. [DOI] [PubMed] [Google Scholar]
- 12.Haubrich, R. H., J. S. Currier, D. N. Forthal, G. Beall, C. A. Kemper, D. Johnson, M. P. Dube, J. Hwang, J. M. Leedom, J. Tilles, and J. A. McCutchan. 2001. A randomized study of the utility of human immunodeficiency virus RNA measurement for the management of antiretroviral therapy. Clin. Infect. Dis. 33:1060-1068. [DOI] [PubMed] [Google Scholar]
- 13.Leitner, T., D. Escanilla, S. Marquina, J. Wahlberg, C. Brostrom, H. B. Hansson, M. Uhlen, and J. Albert. 1995. Biological and molecular characterisation of subtype D, G, and A/D recombinant HIV-1 transmissions in Sweden. Virology 209:136-146. [DOI] [PubMed] [Google Scholar]
- 14.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 15.Miedouge, M., M. Chatelut, J. M. Mansuy, L. Rostaing, F. Malecaze, K. Sandres-Saune, F. Boudet, J. Puel, M. Abbal, and J. Izopet. 2002. Screening of blood from potential organ and cornea donors for viruses. J. Med. Virol. 2002. 66:571-575. [DOI] [PubMed] [Google Scholar]
- 16.Murphy, D. G., L. Cote, M. Fauvel, P. Rene, and J. Vincelette. 2000. Multicenter comparison of Roche COBAS AMPLICOR MONITOR version 1.5, Organon Teknika NucliSens QT with Extractor, and Bayer Quantiplex version 3.0 for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 38:4034-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nkengasong, J. N., M. Kalou, C. Maurice, C. Bile, M.-Y. Borget, S. Koblavi, E. Boateng, M. Sassan-Morokro, E. Anatole-Ehounou, P. Ghys, A. E. Greenberg, and S. Z. Wiktor. 1998. Comparison of Nuclisens and Amplicor monitor assays for quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma of persons with HIV-1 subtype A infection in Abidjan, Côte d'Ivoire. J. Clin. Microbiol. 36:2495-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Brien, W. A., P. M. Hartigan, D. Martin, J. Esinhart, A. Hill, S. Benoit, M. Rubin, M. S. Simberkoff, J. D. Hamilton, et al. 1996. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N. Engl. J. Med. 334:426-431. [DOI] [PubMed] [Google Scholar]
- 19.Peeters, M. 2000. Recombinant HIV sequences: their role in the global epidemics, p. 54-72. In C. Kuiken, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, J. Mullins, S. Wolinsky, and B. Korber (ed.), HIV Sequence Compendium 2000. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 20.Prud'homme, I. T., J. E. Kim, R. G. Pilon, T. Minkus, N. Hawley-Foss, W. Cameron, and E. W. Rud. 1998. Amplicor HIV Monitor, Nasba HIV-1 RNA QT and Quantiplex HIV RNA version 2.0 viral load assays: a Canadian evaluation. J. Clin. Virol. 11:189-202. [DOI] [PubMed] [Google Scholar]
- 21.Salminen, M. O., J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 11:1423-1425. [DOI] [PubMed] [Google Scholar]
- 22.Segondy, M., T. D. Ly, M. Lapeyre, and B. Montes. 1998. Evaluation of the Nuclisens HIV-1 QT assay for quantitation of human immunodeficiency virus type 1 RNA levels in plasma. J. Clin. Microbiol. 36:3372-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson, P., V. Soriano, S. G. Devare, and J. Hackett, Jr. 2001. Comparative performance of three viral load assays on human immunodeficiency virus type 1 (HIV-1) isolates representing group M (subtypes A to G) and group O: LCx HIV RNA Quantitative, AMPLICOR HIV-1 Monitor version 1.5, and Quantiplex HIV-1 RNA version 3.0. J. Clin. Microbiol. 39:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson, M. M., E. Delgado, N. Manjon, A. Ocampo, M. L. Villahermosa, A. Marino, I. Herrero, M. T. Cuevas, E. Vazquez-de Parga, L. Perez-Alvarez, L. Medrano, J. A. Taboada, and R. Najera. 2001. HIV-1 genetic diversity in Galicia, Spain: BG intersubtype recombinant viruses circulating among injecting drug users. AIDS 15:509-516. [DOI] [PubMed] [Google Scholar]
- 25.Thomson, M. M., L. Perez-Alvarez, and R. Najera. 2002. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect. Dis. 2:461-471. [DOI] [PubMed] [Google Scholar]
- 26.Tilling, R., S. Kinloch, L. E. Goh, D. Cooper, L. Perrin, F. Lampe, J. Zaunders, B. Hoen, C. Tsoukas, J. Andersson, and G. Janossy. 2002. Parallel decline of CD8+/CD38++ T cells and viraemia in response to quadruple highly active antiretroviral therapy in primary HIV infection. AIDS 16:589-596. [DOI] [PubMed] [Google Scholar]
- 27.Triques, K., J. Coste, J. L., Perret, C. Segarra, E. Mpoudi, J. Reynes, E. Delaporte, A. Butcher, K. Dreyer, S. Herman, J. Spadoro, and M. Peeters. 1999. Efficiencies of four versions of the AMPLICOR HIV-1 MONITOR test for quantification of different subtypes of human immunodeficiency virus type 1. J. Clin. Microbiol. 37:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, Y., M. H. Lamendola, M. Mendoza, D. Xu, M. Nguyen, S. Yeh, Y. Wu, J. Ku, M. Rosenstraus, and R. Sun. 2001. Performance characteristics of the COBAS AmpliScreen HIV-1 test, version 1.5, an assay designed for screening plasma mini-pools. Transfusion 41:643-651. [DOI] [PubMed] [Google Scholar]