Abstract
Campylobacter species are the leading agents of bacterial gastroenteritis in developed countries. In this study 320 specimens of feces from patients with symptoms of acute gastroenteritis were cultured for Campylobacter species by direct plating on modified charcoal cefoperazone deoxycholate agar and by enrichment in modified Preston broth, with or without blood added, for 48 h at 37°C prior to plating. A 16S/23S PCR/DNA probe membrane-based colorimetric detection assay was evaluated on a subset of the feces (n = 127), including 18 culture-positive and 109 culture-negative specimens. DNA was extracted directly from the fecal specimens by using the QIAamp DNA stool Minikit for the DNA probe-based PCR assay (PCR/DNA probe assay). A second PCR/DNA probe assay based on the 16S rRNA gene in Campylobacter spp. was applied to all specimens that were culture negative, PCR/DNA positive on initial analysis. Campylobacter species were cultured in 20 of the 320 specimens. The 16S/23S PCR/DNA probe assay detected campylobacter DNA in 17 of 18 (94% sensitivity) culture-positive specimens and in 41 (38%) culture-negative specimens. The presence of campylobacter DNA in 35 of 41 culture-negative specimens was confirmed by the 16S PCR/DNA probe assay. DNA sequence analysis of seven 16S/23S PCR products and five 16S PCR products amplified from a selection of these specimens confirmed the presence of campylobacter DNA and more specifically Campylobacter jejuni, C. concisus, C. curvus, and C. gracilis DNA in these specimens. The molecular assays described in this study are rapid methods for the detection and identification of Campylobacter species in fecal specimens. The finding of Campylobacter spp. DNA in a large number of specimens of feces from patients with no other identified cause of diarrhea may suggest that Campylobacter spp. other than C. jejuni and C. coli may account for a proportion of cases of acute gastroenteritis in which no etiological agent is currently identified.
Campylobacter enteritis is a leading cause of acute bacterial gastrointestinal infection worldwide. The genus Campylobacter includes many species of which Campylobacter jejuni and C. coli are common pathogens and account for the majority of diagnosed human campylobacter infections (3, 19). A number of Campylobacter species are recognized as uncommon human pathogens, whereas the status of other species as pathogens or commensals is not fully defined at present.
Routine detection of Campylobacter species in clinical laboratories is based on culture on selective media and subsequent phenotypic identification. Culture methods are biased toward the detection of C. jejuni and C. coli. A number of the antimicrobial agents incorporated into the commonly used selective media (e.g., Preston agar, Skirrow agar, and Butzler agar) may inhibit growth of some Campylobacter species. Cephalothin, colistin, and polymyxin B can be inhibitory to some strains of C. jejuni and C. coli and to many of the other less commonly encountered Campylobacter species, e.g., C. upsaliensis, C. hyointestinalis, and C. fetus (5, 8). The incubation temperature of 42°C routinely used is inhibitory to nonthermophilic Campylobacter species that can also be associated with gastroenteritis. Furthermore, a number of Campylobacter species, e.g., C. concisus, C. rectus, C. curvus, C. gracilis, and C. showae require incubation in a hydrogen-enriched microaerophilic atmosphere for recovery (17). Therefore, the true incidence of campylobacter enteritis may be underestimated because of the limitations of routine culture methods. In addition to the limitations of current methods for detection of the full range of Campylobacter species that may be associated with acute gastroenteritis, conventional methods are relatively slow. Presumptive results may be available after 2 days. However, definitive species-level identification based on phenotypic methods may require a further 3 to 4 days. Phenotypic identification can be challenging because of the fastidious growth requirements, the asaccharolytic nature and possession of few distinguishing biochemical characteristics by Campylobacter species (8). Most clinical laboratories do not perform more than presumptive identification.
Molecular methods based on PCR amplification may provide an alternative to culture methods for the detection of Campylobacter in clinical specimens. The application of PCR-based assays to the detection of Campylobacter species in clinical and food samples has been previously reported by this group and by others (4, 7, 9, 11, 14, 16, 18, 22). These reports describe amplification of a number of DNA targets including the campylobacter flagellin gene (22), 16S rRNA (7, 11, 16), and the 16S/23S rRNA intergenic spacer region (4, 18). Many of these assays have been applied to specimens of feces, with numbers of specimens analyzed varying from 50 to 3,736 specimens.
The first objective of the present study was to compare preenrichment, followed by plating with routine direct plating for the isolation of Campylobacter from specimens of feces (n = 320) submitted from hospital and community for the investigation of acute gastroenteritis. The second objective of the study was the evaluation of a previously described 16S/23S PCR/DNA probe membrane-based colorimetric assay (4, 18) for detection of C. jejuni, C. coli, and other Campylobacter spp. DNA on a subset (n = 127) of these specimens.
MATERIALS AND METHODS
Clinical samples.
A series of 320 consecutive liquid and semisolid specimens of feces submitted for routine investigation of acute infectious gastroenteritis were included in the present study. Specimens were obtained from the community practices (n = 201) and from hospital (n = 119) accident and emergency and pediatric departments.
Bacteriological investigation of clinical samples.
The fecal samples were examined for Campylobacter species by direct inoculation of feces on to modified charcoal cefoperazone desoxycholate agar (CCDA; Oxoid, Basingstoke, United Kingdom). The CCDA comprised of campylobacter blood-free selective agar base (Oxoid CM739) with campylobacter selective supplement (Oxoid SR155) and campylobacter growth supplement (Oxoid SR084). The plates were incubated for 48 h at 42°C under microaerophilic conditions (5% O2, 5% CO2, 2% H2, and 88% N2 by volume) generated by oxoid microaerophilic gas pack. C. jejuni NCTC 11322 was incubated with each lot of plates as a positive control. Gray, flat, and spreading colonies resembling those of Campylobacter were selected for further identification. Oxidase- and catalase-positive colonies exhibiting a characteristic Gram stain appearance (gram-negative S-shaped rods) were reported as Campylobacter species. The ability to hydrolyze hippurate was evaluated by the British Standards Method (BS 5763 part 17, 1996:9.5.5.5), and susceptibility to nalidixic acid was determined by disk diffusion with a 30-μg nalidixic acid disk applied to a blood agar plate inoculated with a suspension of the test isolate. Nalidixic acid-susceptible isolates capable of hippurate hydrolysis were reported as C. jejuni, while nalidixic acid-susceptible, hippurate hydrolysis-negative isolates were reported as C. coli.
Enrichment broths were used to examine the role of enrichment for the isolation of Campylobacter species from feces. The enrichment broth was a modified Preston enrichment broth (comprised of nutrient broth no. 2 [Oxoid CM67]), with campylobacter selective supplement [Oxoid SR084] and campylobacter growth supplement [Oxoid SR155]).The broth was evaluated with 5% lysed horse blood (broth A) and without lysed blood (broth B). Enrichment broth, in screw-cap tubes were inoculated with feces, and then additional broth was added to bring the level of fluid almost to the brim and loosely closed. This establishes a microaerophilic atmosphere. Enrichment cultures were incubated for 48 h at 37°C, followed by subculturing onto CCDA plates. Subsequent incubation and reading of plates were as described above. A suspension of C. jejuni NCTC 11322 (ca. 15 CFU/ml) was added to each lot of enrichment broth and processed as a positive control.
All specimens were also examined for the presence of Shigella spp., Salmonella enterica, Yersinia spp., and Escherichia coli O157 by culture on xylose lysine deoxycholate agar, Cefsulodin Irgasan Novobiocin agar, and Sorbitol MacConkey agar, respectively. Suspected colonies were biochemically identified (API 20 E strip; bioMérieux, Marcy l'Étoile, France), and the species was confirmed by serologic testing where appropriate. Specimens from children under 12 years were also examined for Cryptosporidium parvum by microscopy with a modified auramine stain. Specimens from children under 3 years were examined for adenovirus and rotavirus by using the Novamed combistick test (Novamed, Ltd., Jerusalem, Israel).
DNA extraction.
DNA was extracted from a total of 127 fecal specimens representing 18 culture-positive and 109 culture-negative specimens from the original cohort of 320 specimens. After culture, specimens were stored at −80°C until DNA extraction with QIAamp DNA stool Minikit (Qiagen GmbH, Hilden, Germany) was performed in accordance with the manufacturer's instructions. The option to incubate the specimen in lysis buffer at 95°C instead of 70°C was applied.
Oligonucleotides.
Biotinylated PCR primers and oligonucleotide probes modified with a 5′ amine listed in Table 1 were synthesized by MWG-Biotech, Milton Keynes, United Kingdom.
TABLE 1.
Sequences and descriptions of the oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′)a | Tm (°C) | Modification |
|---|---|---|---|
| A1-B | BIO-AGTCGTAACAAGGTAGCCG- | 51.0 | Biotinylated |
| B1-B | BIO-CYRYTGCCAAGGCATCCACC- | 58.0 | Biotinylated |
| CAMP1F-B | BIO-GTTAAGAGTCACAAGCAAGT- | 48.0 | Biotinylated |
| C412F | BIO-GGATGACACTTTTCGGAGC- | 51.0 | Biotinylated |
| C1288R | BIO-CATTGTAGCACGTGTGTC- | 48.0 | Biotinylated |
| CAMP4 | AMI-GGTAAGCTACTAAGAGCG- | 49.0 | Aminated |
| CJEJ7 | AMI-GCTTAGTTGAGACTAAATCA- | 46.0 | Aminated |
| CCOL2 | AMI-GACTTAGTTTAGATATTTTTAG- | 44.0 | Aminated |
| CLAR4 | AMI-GTATTATCTTTGCTTTAGTCTTTAAAGATTAAA- | 53.0 | Aminated |
| CUPS4 | AMI-GCTCTTTTATCATTAAACTTTAGC- | 49.0 | Aminated |
| CAMPM4 | AMI-GGTAAGCTACAAAGAGCA- | 46.0 | Aminated |
| 16S1 | AMI-TCTGCCTCTCCCYCACTC- | 53.0 | Aminated |
| POSDET | AMI-NNNNNNNNNNNN-BIO | 32.0 | Aminated and biotinylated |
BIO, biotinylation; AMI, amination; Y, C + T; R, A + G.
PCR amplification.
PCRs were performed in a final volume of 100 μl. Each reaction contained 0.25 mM concentrations of deoxynucleotide triphosphates, 2 mM MgCl2 (Promega, Madison, Wis.), 1× reaction buffer (Promega), 100 ng (12.5 pmol) of forward and reverse primers (either the universal primers A1-B and B1-B, the Campylobacter genus-specific primers CAMP1F-B/B1-B, or C412F/C1228R [13] as appropriate), 1.5 U of Taq polymerase (Promega, Madison, Wisconsin), and 5 μl of template DNA and made up to a final volume of 100 μl with nuclease-free water (Sigma-Aldrich, Dorset, United Kingdom). Thermocycling in either an MJ Research PTC 200 (MJ Research, Inc., Boston, Mass.) or Perkin-Elmer 9600 (Perkin-Elmer, Cambridge, United Kingdom) consisted of 35 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s for the primers sets A1-B/B1-B and CAMP1F-B/B1-B and annealing at 60°C for 60 s for the primer set C412F/C1228R, extension at 72°C for 30 s for the primer sets A1-B/B1-B and CAMP1F-B/B1-B and for 60 s for the primer set C412F/C1228R, and a final extension at 72°C for 10 min. Samples were held at 4°C until analysis. A no-template negative control and a positive control of purified campylobacter DNA (C. jejuni NCTC 11168) were included in each PCR run. PCR setup was performed in a biological cabinet by using dedicated pipettes and aerosol barrier tips to minimize the risk of PCR contamination. Two independent PCR amplifications (run 1 and run 2) with campylobacter genus primers CAMP1F-B/BI-B were performed on 119 samples in run 1 and 117 samples in run 2 (insufficient material remaining for 2 samples) from which PCR quality DNA had been successfully extracted. A total of 10 to 20 μl of each PCR product was run on a 1% agarose gel in 1× Tris borate-EDTA buffer.
Reverse hybridization colorimetric assay.
The reverse blot hybridization method was performed as previously reported (4, 18). PCR products generated in the PCR amplification run 1 were hybridized to membrane strips containing the following probes: POS-DET, CAMP4, CJEJ7, CCOL2, CLAR4, and CUPS4. PCR products generated in the PCR amplification run 2 were hybridized to membrane strips containing the same probes plus the additional probe CAMPM4. The probes were immobilized onto membranes by using a BioDot X-Y 3000 dispensing platform (BioDot, Inc., Irvine, Calif.). For the 16S DNA probe assay, DNA probe 16S1 was immobilized onto a membrane. A positive control probe (POS-DET) for the colorimetric component of the assay and a negative control probe were included on each membrane or on a separate membrane included in each assay run. Briefly, a 45-μl aliquot of the biotinylated PCR product was denatured by heating to 95°C for 7 min. The denatured PCR product was mixed with 1 ml of Hyb/Wash solution (5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA; pH 7.7] and 0.1% sodium dodecyl sulfate) and incubated with the membrane at 55°C for the 16S/23S assay and at 42°C for the 16S probe assay for 1 h. The membrane was washed twice for 10 min in Hyb/Wash solution and once for 5 min in Tris-buffered saline (TBS; 50 mM Tris-Cl [pH 7.5], 0.15 M NaCl). The membrane was incubated for 20 min in 10 ml of TBS-1% Tween 20 (Sigma-Aldrich, Steinheim, Germany) with a 1/2,000 dilution of streptavidin-alkaline phosphate conjugate (Calbiochem-Novobiochem, La Jolla, Calif.). The membrane was washed twice for 10 min in TBS and once for 5 min in color development solution (0.1 M Tris-Cl [pH 9.5], 50 mM MgCl2, 0.1 M NaCl). The substrates nitroblue tetrazolium (50 μl of a 100-mg/ml solution) and BCIP (5-bromo-4-chloro-3-indolylphosphate; 37.5 μl of a 50-mg/ml solution) were added to 10 ml of color development solution, and the membrane was incubated in this solution until clearly visible positive signals were obtained from the POS-DET-positive control probe. Soaking the membrane in distilled water stopped color development.
DNA sequencing.
DNA sequencing was performed on CAMP1F-B/BI-B and C412F/C1288R PCR products in the forward and reverse directions by using the sequencing services available at MWG-Biotech, Lark Technologies (Essex, United Kingdom), and Sequiserve (Vaterstetten, Germany). All sequence analysis was carried out by using the tools of the European Bioinformatics Institute (EMBL.EBI website [www.ebi.ac.uk]). Database searches were run by using the BLAST program against the EMBL and GenBank DNA databases. Pairwise comparisons were made by using the Bestfit programs. Multiple sequence alignments were carried out by using the CLUSTALW program (20).
RESULTS
Culture and enrichment.
The pathogenic organisms detected in 320 specimens of feces by standard methods for pathogens included adenovirus and rotavirus (3.6%), Cryptosporidium parvum (4.4%), S. enterica (0.3%), and Yersinia enterocolitica (0.3%). Campylobacter species was the most frequently identified pathogen. Culture and enrichment methods yielded Campylobacter species from 20 of the 320 samples (6.4% detection rate). Campylobacter species were detected only by direct culture on CCDA agar from five specimens, by both direct plating and enrichment in nine cases, and only after broth enrichment in six cases. Inclusion of the enrichment protocol resulted in a 30% increase in detection beyond that achieved with direct plating on CCDA alone. Of the 15 specimens yielding positive results from the enrichment broth, 10 were positive in both enrichment broths (with or without blood), 4 were positive in blood-free broth only, and 1 was positive in blood-containing broth only.
Evaluation of the application of the 16S/23S PCR/DNA probe assay to the detection of Campylobacter species directly in specimens of feces.
DNA extracted from 127 fecal specimens, including 18 culture-positive and 109 culture-negative samples was PCR amplified with universal primers A1-B/B1-B for the 16S/23S rRNA target region to determine whether PCR quality DNA had been successfully extracted from the feces. In total 119 of the 127 specimens (93.7%) yielded DNA that was amenable to PCR amplification. A PCR product was obtained from 93 specimens by using undiluted template DNA and from an additional 26 (20.5%) specimens following a 1 in 10 dilution of the template DNA. Eight specimens (6.3%) did not yield DNA suitable for PCR; one of these samples was culture positive for campylobacter.
Of the 17 culture-positive specimens from which PCR quality DNA was extracted, 100% were PCR/DNA probe positive for Campylobacter spp. in both assay runs. Of these 17 PCR/DNA probe-positive and culture-positive samples, all gave a positive signal with the CAMP4 genus-specific probe, whereas 15 and 13 were positive, respectively, in runs 1 and 2 with the C. jejuni-specific probe (CJEJ7). Of the 17 specimens, 16 were confirmed as C. jejuni positive by culture, with one specimen containing C. coli. This C. coli culture-positive specimen was PCR/DNA probe positive with CAMP4 and CJEJ7, the C. jejuni-specific probe, but not with CCOL2, the C. coli-specific probe.
In amplification run 1, CAMP1F-B/BI-B, PCR products were obtained from DNA extracted from 41 of 109 culture-negative specimens. Of the 41 PCR products, 27 hybridized with the CAMP4 (genus-specific) probe, and 3 of these 27 also hybridized with the CJEJ7, C. jejuni-specific probe (Table 2 and Fig. 1). On repeat testing (run 2), two of these three specimens were again positive with the C. jejuni-specific probe.
TABLE 2.
Summary of the results obtained for culture-negative specimensa
| No. of specimens in group | No. positive (run 1)
|
No. positive (run 2)
|
|||
|---|---|---|---|---|---|
| PCR | CAMP4 | PCR | CAMP4 | CAMPM4 | |
| 27 | 27 | 27 | 25b | 13c | 25 |
| 14 | 14 | 0 | 10 | 2 | 10 |
| 12 | 0 | 0 | 12 | 1 | 6 |
| 10 | 4 | 0 | 6 | 0 | 0 |
PCR results from run 1 and run 2 and DNA probe hybridization results with probes CAMP4 and CAMPM4.
Insufficient template DNA remaining for PCR from 2 of the 27 specimens.
Note that the competition for available PCR product from CAMPM4 may have reduced the amount of binding of amplified product to CAMP4 in run 2.
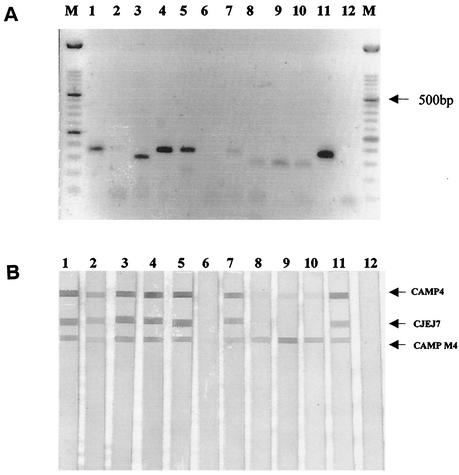
FIG. 1.
16S/23S PCR/DNA probe assay for the detection of Campylobacter. (A) PCR amplification of 5 μl of DNA with primer set CAMPIF-B/BI-B after extraction from stool specimens by using the QIAamp DNA stool kit. Lanes 1 to 5, culture-positive samples 203, 251, 276, 352, and 360, respectively; lanes 6 to 10, culture-negative samples 172, 356, 55, 89, and 128, respectively; lane 11, PCR-positive control C. jejuni 11168; lane 12, PCR-negative control. (B) DNA probe colorimetric assay. Strips 1 to 5, PCR products hybridized from culture-positive samples 203, 251, 276, 352, and 360, respectively; strips 6 to 10, PCR products hybridized from culture-negative samples 172, 356, 55, 89, and 128, respectively; strip 11, PCR-positive C. jejuni 11168; strip 12, PCR-negative control.
Four CAMP1F-B/BI-B PCR products amplified from culture-negative specimens in run 1 were selected for sequencing. Two of these amplicons yielded a hybridization signal with the CAMP4 probe, and two did not give a hybridization signal with the CAMP4 probe. One sequencing reaction was performed on each strand, and a consensus sequence was obtained and compared by using CLUSTALW. All four sequences showed ca. 98% sequence homology. BLAST searches performed with these sequences indicated that they produced significant alignments but with low homology to 16S/23S rRNA gene sequences available for Campylobacter species. All four sequences had 89% homology to the CAMP4 probe with two differences in base pairs: one at the 3′ end of the CAMP4 probe and one in the center of the probe. A new oligonucleotide probe CAMPM4 was designed incorporating these base differences and applied to the membrane strips in addition to the CAMP4 probe and species-specific oligonucleotides for run 2 of the DNA probe colorimetric membrane hybridization analysis.
In amplification run 2, CAMP1F-B/BI-B PCR products were obtained from DNA extracted from 47 of 107 culture-negative specimens. Of the 27 specimens that yielded CAMP1F-B/BI-B products and were CAMP4 probe positive in run 1, 25 yielded CAMP1F-B/BI-B products that hybridized with either or both the CAMP4 probe and the new probe CAMPM4 in run 2 (Table 2). There was insufficient DNA available in run 2 to allow testing for 2 of these 27 specimens. Twelve PCR products did not yield a signal with CAMP4 in run 2, possibly due to competition for the available product by the CAMPM4 probe. Ten samples that yielded PCR products but no probe signal in run 1 yielded PCR products and a DNA probe signal with CAMPM4 in run 2 with two of the samples, also yielding a signal with the CAMP4 probe. In run 2, PCR products were obtained from 12 specimens negative on run 1. Of these 12 PCR products, 6 yielded hybridization signals with the CAMPM4, and 1 of these PCR products also yielded a signal with the CAMP4 probe (Table 2). Four samples that yielded a CAMP1F-B/BI-B PCR product in run 1 but no probe signal with the CAMP4 probe did not yield a PCR amplification product in run 2.
After assay run 2, DNA sequencing of PCR products from an additional three culture-negative DNA probe-positive samples was performed, one from a culture-negative sample (sample 1) that yielded a PCR/DNA probe hybridization signal with CAMP4 and CJEJ7 probes in both assay runs and two from culture-negative samples (sample 2 and sample 3) that had yielded a signal with CAMP4 in run 1 and with CAMP4 and CAMPM4 in run 2. A CAMP1F-B/BI-B PCR product from C. jejuni 11168 was also submitted for sequencing. Pairwise CLUSTALW analysis of the sequence obtained for the PCR product from sample 1 with the sequence obtained for C. jejuni 11168 revealed a homology of 99% between the sequences. Pairwise CLUSTALW analysis of the PCR product from samples 2 and 3 with the sequence obtained for C. jejuni 11168 revealed a homology of 51%. CLUSTALW analysis of samples 2 and 3 with the four sequences previously obtained for culture-negative 16S/23S rRNA PCR products (run 1) indicated a homology of 97% between the sequences.
Application of a 16S PCR/DNA probe assay to culture-negative 16S/23S PCR DNA probe-positive samples.
To further evaluate the 41 culture-negative 16S/23S PCR/DNA probe-positive samples, the specimens were subject to PCR amplification targeting a distinct region of the genome, the 16S gene. PCR amplification was performed with campylobacter-specific primers (C412F/C1288R) as previously described (13) but modified with 5′ biotin molecules. The PCR products were hybridized to membrane strips containing the DNA probe 16S1 designed for the detection of the 16S gene in Campylobacter. Of 41 samples assayed, 35 yielded positive probe signals with this probe. Six 16S PCR products were sequenced, including a 16S PCR product obtained from DNA extracted from a culture-positive sample. BLAST analysis and application of the “sequence match” function of the Ribosome Database Project (RDP) were performed on each sequence. The sequence obtained for the 16S PCR product amplified from the culture-positive sample showed 100% homology with C. jejuni as evaluated by BLAST and had a score of 1.00 for C. jejuni by using the “sequence match” in the RDP. The 16S PCR products amplified and sequenced from the culture-negative samples yielded the following results: one sample showed a 100% homology with C. jejuni in BLAST and had a score of 0.982 with C. jejuni by using the RDP sequence match function. Two 16S PCR products showed 99% homology as determined by BLAST to C. concisus and had a score of 0.94 in the RDP sequence match function. One 16S PCR product showed a homology of 96% to C. concisus and C. gracilis and, respectively, scores of 0.861 and 0.85 as determined from the sequence match function of the RDP. The remaining 16S PCR product showed 98% homology to C. curvus and C. concisus according to BLAST and, respectively, scores of 0.93 and 0.88 according to the sequence match function of the RDP.
DISCUSSION
Acute gastrointestinal infection represents a major disease burden in all societies, particularly in developing countries. Campylobacter spp. are now recognized as the most common bacterial agent of acute gastroenteritis in most countries (2). Although the list of bacterial, protozoan, and viral pathogens recognized as contributing to this burden is now very long, it remains the case that an etiological agent is not identified in the majority of specimens submitted from patients with acute diarrhea. Failure to diagnose acute diarrheal disease may be attributable to etiological agents (infectious or noninfectious) not yet recognized or to recognized pathogens that go undetected due to the insensitivity of existing routine methods. Our results suggest that the use of direct plating on CCDA agar only for detection of Campylobacter spp. may result in failure to detect approximately one-third of specimens from which viable Campylobacter spp. could be recovered with routine use of enrichment broth in addition to direct plating. This effect may simply be due to the enhanced probability of finding pathogens present in low number by culturing a larger volume of specimen. Two recent studies (10, 15) evaluated the performance of different selective media and the use of filtration onto nonselective media for the isolation of Campylobacter spp. from feces. One study (10) found that selective media was superior to the use of a filtration method for the isolation of Campylobacter spp., whereas the other study (15) concluded that the combined use of selective culture and a filtration method onto nonselective agar increased the overall rates of isolation of campylobacter from feces. The reported increased isolation rate by use of a second method is consistent with our experience. However, the routine use of blood-free enrichment broth rather than filtration may be more practical for routine use and is analogous to the common practice of enrichment for isolation of S. enterica from feces. Our data suggest that blood free enrichment broth is as effective as enrichment broth with added blood. More extensive studies of the value of enrichment broth are required.
The second objective of the study was to evaluate the application of a 16S/23S rRNA PCR/DNA probe colorimetric membrane assay to the detection of Campylobacter spp. DNA directly in feces. This assay was previously evaluated for this application on a small number of samples (n = 50), which included 30 culture-positive specimens, 12 culture-negative specimens and 8 “aged” specimens that were originally culture positive but were culture negative for campylobacter when the 16S/23S PCR/DNA probe assay was applied (4). We reported a sensitivity of 90% with respect to the culture-positive specimens and 63% with respect to the “aged” specimens. We used the QIAamp Mini Stool Kit to extract the DNA and found that DNA suitable for PCR amplification was extracted from 96% of samples.
In the present study the QIAamp Mini Stool Kit yielded DNA that was suitable for amplification from 93.7% of the specimens, 20% of which required a 10-fold dilution to enable successful PCR amplification. The failure to obtain DNA amenable to PCR from 6.3% of specimens and the requirement to perform a dilution in 20% of DNA samples to obtain PCR products most probably reflects the presence of PCR inhibitors in the DNA samples. PCR amplification was achieved in samples when a dilution of the PCR template (20%) was performed, suggesting that dilution of the PCR inhibitors allowed successful PCR amplification. Because of the heterogeneous nature of feces specimens it is very difficult to develop a DNA extraction method that will successfully remove inhibitors that may be present at various amounts in different samples to ensure that DNA of comparable quality is extracted from every sample. For runs 1 and 2, a sensitivity of 100% was achieved with the Campylobacter genus probe CAMP4 for samples that yielded DNA amenable to PCR, and sensitivities of 88 and 76% were achieved with the C. jejuni-specific probe CJEJ7 for runs 1 and 2, respectively. The CAMP4 probe has been previously shown to have a 10-fold-lower detection limit than the CJEJ7 probe (9).
PCR amplification of Campylobacter DNA from culture-negative specimens (n = 109 [run 1], n = 107 [run 2]) generated PCR products in run 1 and run 2 from 35 samples. Four samples amplified in run 1 were negative in run 2, and 12 samples that did not yield a PCR product in run 1 were PCR positive in run 2. We suggest that the inconsistencies in PCR amplification between the two runs may be a reflection of the concentration of the Campylobacter spp. DNA that is present in these specimens and the random variation in the amount of Campylobacter DNA in different aliquots of DNA selected for addition as a template in individual PCRs. Also, since the two runs were performed by independent operators with different thermocyclers, these variables may have contributed to the inconsistent results. In the highly heterogeneous matrix of DNA extracted from feces, it is possible that PCR amplification of noncampylobacter DNA may occur under certain circumstances and, consequently, the diagnostic endpoint of this assay is based on the hybridization of the PCR product to campylobacter-specific DNA probes.
DNA sequencing of a selection of CAMP1F/B1-B PCR products amplified from culture-negative samples in run 1 revealed sequences that had 89% homology to the CAMP4 probe. A new oligonucleotide probe CAMPM4 was designed incorporating these base differences and applied to the membrane strips in addition to the CAMP4 probe and species-specific oligonucleotides for run 2 of the DNA probe assay. In run 2, DNA probe signals were obtained for 41 samples that yielded CAMP1F/B1-B PCR products with more signals obtained from probe CAMPM4 than CAMP4. Of the 27 culture-negative specimens that generated CAMP1F-B/BI-B PCR products and yielded a DNA probe signal with CAMP4 genus probe in run 1, 12 yielded a probe signal with CAMPM4 but not with CAMP4 in run 2. This apparent discrepancy is most likely related to preferential hybridization of PCR product to the novel CAMPM4 probe that was designed with 100% homology to similar PCR products from run 1 compared to 89% homology for the original CAMP4 probe. DNA probe signals with CAMP4 and CAMPM4 were obtained only from those samples with the highest yield of CAMP1F-B/BI-B PCR products as judged by band intensity on agarose gel analysis. The use of CAMPM4 was as an additional probe to complement the original CAMP4 genus probe and to enable PCR products with sequence differences to CAMP4 to produce a clearly detectable hybridization signal in the DNA probe colorimetric assay. Subsequently, after run 2, three CAMP1F-B/B1-B PCR products were sequenced and were shown to have 97% homology with the sequences obtained for run 1 PCR products.
Application of an independent PCR/DNA probe assay based on the 16S gene confirmed the presence of campylobacter DNA in 35 of the 41 culture-negative and CAMP4- and/or CAMPM4-positive specimens. Seven of these culture-negative PCR/DNA probe-positive specimens were also positive for the presence of another common agent of gastroenteritis. However, no recognized pathogen was detected in the remaining 34 samples. DNA sequence results for the CAMP1F-B/B1-B (16S/23S rRNA) PCR products were available for seven culture-negative specimens. In one specimen the sequence data showed 99% homology to C. jejuni 11168, this product hybridized with the CJEJ7 probe. The six other 16S/23S rRNA sequences showed a 51% homology to C. jejuni 11168. 16S sequence data for four of these samples were most consistent with the presence of C. concisus (n = 2), C. gracilis (n = 1), and C. curvus (n = 1), with sequences showing ≥98% homology with 16S sequences in the ribosomal database for these species.
In the present study C. jejuni or C. coli were detected in 14 of 320 (4.4%) of specimens of feces with acute diarrhea by primary plating. By using enrichment broth, we identified a further six (1.9%) culture-positive specimens, and molecular methods applied to approximately one-third (109 of 300) of the culture-negative specimens identified a further three (2.6%) C. jejuni-positive specimens. These data suggest that estimates of the percentage of cases of acute infectious diarrhea attributable to C. jejuni based on routine direct plating (4.4% in our series) may represent a very significant underestimate of the actual proportion of acute gastroenteritis attributable to C. jejuni. The findings emphasize the limitations of routine culture methods currently used, which may inhibit the isolation of some strains of C. jejuni and are unsuitable for the isolation of species that require additional hydrogen for growth.
We have also detected DNA of Campylobacter spp. other than C. jejuni or C. coli in a very high proportion (>30%) of specimens of feces from patients with acute gastroenteritis. The species present appear to be C. curvus, C. concisus, and C. gracilis. Other researchers also have identified the presence of C. concisus (normally associated with the peridontal cavity) in feces from patients with gastroenteritis but also in healthy human subjects. Van Etterijck et al. (21) reported the isolation of C. concisus in 13.2% fecal samples from children with diarrhea and from 9% of healthy children, whereas another study identified the presence of C. concisus as determined by a PCR-based enzyme-linked immunosorbent assay assay in 15% of specimens of feces from healthy subjects (12). A recent study (6) identified the presence of C. curvus and C. concisus in 42 fecal specimens, 5 of which were also infected with an established bacterial enteric pathogen from 1,376 specimens being evaluated for the presence of Campylobacter. Of 214 healthy or control specimens, 8 were also positive for C. concisus. We suggest that although the role of C. concisus in gastroenteritis requires further investigation, it may have a role as an opportunistic pathogen in young children and immunocompromised individuals. This suggestion is supported by a study (1) that reports the finding based on protein profiling of two distinct types of C. concisus protein profiles occurring in patient samples, with one type infecting children and immunocompromised patients. Further studies to define the significance of DNA from these species in feces are required.
In the present study the application of a 16S/23S PCR/DNA probe assay identified the presence of C. jejuni in 2.6% of culture-negative specimens and the presence of Campylobacter spp. DNA in an additional 30% of culture-negative specimens. The routine application of the PCR/DNA probe assay described in the present study would, however, require some further development. In the present study the PCR quality of DNA extracted from feces was assessed by an independent PCR. This is not ideal for routine clinical use. For routine application, it would be preferable to redesign the assay to include an internal standard control that would be coamplified with the test sample, thereby indicating the presence of PCR inhibitors in the sample and the need to dilute the sample and repeat the PCR test.
Molecular methods may have practical advantages for studies to determine the frequency with which a range of Campylobacter spp. are present in feces since these methods can identify the presence of campylobacter DNA from a large number of species and can be applied retrospectively to stored specimens.
REFERENCES
- 1.Aabenhus, R., H. Permin, S. L. On, and L. P. Andersen. 2002. Prevalence of Campylobacter concisus in diarrhoea of immunocompromised patients. Scand. J. Infect. Dis. 34:248-252. [DOI] [PubMed] [Google Scholar]
- 2.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 3.Allos, B. M., and M. J. Blaser. 1995. Campylobacter jejuni and the expanding spectrum of related infections. Clin. Infect. Dis. 20:1092-1099. [DOI] [PubMed] [Google Scholar]
- 4.Collins, E., M. Glennon, S. Hanley, A. Murray, M. Cormican, T. Smith, and M. Maher. 2001. Evaluation of a PCR/DNA probe colorimetric membrane assay for identification of Campylobacter species in human stool specimens. J. Clin. Microbiol. 39:4163-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corry, J. E., D. E. Post, P. Colin, and M. J. Laisney. 1995. Culture media for the isolation of campylobacters. Int. J. Food Microbiol. 26:43-76. [DOI] [PubMed] [Google Scholar]
- 6.Engberg, J., S. L. On, C. S. Harrington, and P. Gerner-Smidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal specimens as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giesendorf, B. A., and W. G. Quint. 1995. Detection and identification of Campylobacter spp. using the polymerase chain reaction. Cell. Mol. Biol. 41:625-638. [PubMed] [Google Scholar]
- 8.Goossens, H., and J. P. Butzler. 1992. Isolation and identification of Campylobacter species, p. 93-109. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, D.C.
- 9.Grennan, B., N. A. O'Sullivan, R. Fallon, C. Carroll, T. Smith, M. Glennon, and M. Maher. PCR-ELISAs for the detection of Campylobacter jejuni and Campylobacter coli in poultry samples. BioTechniques 30:602-610. [DOI] [PubMed]
- 10.Kulkarni, S. P., S. Lever, J. M. J. Logan, A. J. Lawson, J. Stanley, and M. S. Shafi. 2002. Detection of Campylobacter species: a comparison of culture and polymerase chain reaction-based methods. J. Clin. Pathol. 55:749-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawson, A. J., J. M. Logan, G. L. O'Neill, M. Desai, and J. Stanley. 1999. Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37:3860-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawson, A. J., D. Linton, and J. Stanley. 1998. 16S rRNA gene of “Candidatus Campylobacter hominis” a novel uncultivated species, are found in the gastrointestinal tract of healthy humans. Microbiology 144:2063-2071. [DOI] [PubMed] [Google Scholar]
- 13.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of the five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 14.Linton, P., A. J. Lawson, R. J. Owen, and J. Stanley. 1997. PCR detection and identification to species level and fingerprinting of C. jejuni and C. coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClurg, K. R., R. B. McClurg, and J. E. Moore. 2002. Efficient isolation of campylobacters from stools: what are we missing? J. Clin. Pathol. 55:239-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metherell, L. A., J. M. Logan, and J. Stanley. 1999. PCR-enzyme linked immunosorbent assay for the detection and identification of Campylobacter species: application to isolates and stool samples. J. Clin. Microbiol. 37:433-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nachamkin, I. 1995. Campylobacter and Arcobacter, p. 483-491. In P. R. Murray, E. T. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology. ASM Press, Washington, D.C.
- 18.O'Sullivan, N. A., R. Fallon C. Carroll, T. Smith, and M. Maher. 2000. Detection and differentiation of C. jejuni and C. coli in broiler chickens samples using a PCR/DNA probe membrane based colorimetric detection assay. Mol. Cell Probes 14:7-16. [DOI] [PubMed] [Google Scholar]
- 19.Skirrow, M. B. 1994. Diseases due to Campylobacter, Helicobacter, and related bacteria. J. Comp. Pathol. 111:113-149. [DOI] [PubMed] [Google Scholar]
- 20.Thompson, J., D. Higgins, and T. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Etterijck, R., J. Breynaert, H. Revets, T. Devreker, Y. Vandenplas, P. Vandamme, and S. Lauwers. 1996. Isolation of Campylobacter concisus from feces of children with and without diarrhea. J. Clin. Microbiol. 34:2304-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waegal, A., and I. Nachamkin. 1996. Detection and molecular typing of C. jejuni in faecal samples by PCR. Mol. Cell. Probes 10:75-80. [DOI] [PubMed] [Google Scholar]