Abstract
The recovery of Ralstonia and Pandoraea species from respiratory tract cultures of patients with cystic fibrosis has recently been reported. These species are difficult to identify, and especially to differentiate from Burkholderia cepacia complex organisms, with classical methods. The discriminatory power of amplified ribosomal DNA restriction analysis (ARDRA) within the two genera was assessed by comparing the restriction profiles of reference strains of each species by using a panel of six enzymes already proven suitable for the identification of Burkholderia species. ARDRA provided differentiation of all the Ralstonia species tested and of Pandoraea norimbergensis. Pandoraea species P. pnomenusa, P. sputorum, P. pulmonicola, and P. apista were not discriminated to the species level. This method allowed the identification of five clinical isolates recovered from French cystic fibrosis patients as Ralstonia mannitolilytica.
The recovery of various gram-negative bacilli, mainly Ralstonia and Pandoraea species, from the sputa of patients with cystic fibrosis (CF) on Burkholderia cepacia-selective media has been recently pointed out (4, 5). The genus Ralstonia, described in 1995 by Yabuuchi et al.(16), contains environmental gram-negative bacilli including major plant pathogen R. solanacearum (12); potential bioremediation agents R. eutropha and the metal-resistant R. campinensis, R. metallidurans, and R. basilensis (11); and opportunistic human pathogens. Among strains of clinical importance, four Ralstonia species have been isolated from CF patients: R. pickettii (formerly Pseudomonas pickettii and Burkholderia pickettii) (2), R. mannitolilytica (formerly R. pickettii biovar 3/thomasii) (5, 10), and quite rarely R. gilardii (8) and R. taiwanensis (3, 8), whereas R. paucula (formerly CDC group IV c-2) has not been involved in CF to date. The novel genus Pandoraea (4) contains five species: P. pnomenusa, P. sputorum, P. pulmonicola, and P. apista, mainly associated with clinical isolates recovered from CF patients, and P. norimbergensis (formerly Burkholderia norimbergensis), rarely isolated from the environment and from human clinical specimens. Four Pandoraea genomospecies, containing one strain each, were also described (9). The misidentification of Ralstonia and Pandoraea species as organisms belonging to the B. cepacia complex (Bcc) is fraught with consequences since the recovery of a Bcc organism in a CF patient leads to strict infection control measures in order to reduce patient-to-patient spread, and possibly to exclusion from lung transplantation. At present, the prevalence, patient-to-patient transmissibility, and clinical impact of Ralstonia and Pandoraea organisms in CF patients are unknown and cannot be evaluated if they are badly recognized in the clinical microbiology laboratory. Thus, an accurate identification of these species is essential. Unfortunately, the usual manual and automated phenotypic identification methods are not satisfactory (1, 14, 15). The aim of this study was to determine whether amplified ribosomal DNA restriction analysis (ARDRA), already successfully applied to the identification of Burkholderia species (13), could be used for Ralstonia and Pandoraea species.
The type strains of the following Ralstonia and Pandoraea species were tested: R. solanacearum, R. eutropha, R. campinensis, R. metallidurans, R. basilensis, R. pickettii, R. mannitolilytica, R. gilardii, R. taiwanensis, and R. paucula and P. pnomenusa, P. sputorum, P. pulmonicola, P. apista, and P. norimbergensis. As R. solanacearum is known to be a heterogeneous species, several strains, representative of races 1, 2, and 3, kindly provided by Christian Boucher (Centre National de la Recherche Scientifique-Institut National de la Recherche Agronomique, Castanet-Tolosan, France) were included. The other reference strains were obtained from international culture collections. Five clinical isolates recovered from the sputum of CF patients and transmitted to the French Observatoire Cepacia/Vaincre la Mucoviscidose were also analyzed. These isolates had been grown on B. cepacia-selective media and were resistant to colistin, aminoglycosides, ticarcillin, and aztreonam but did not belong to the Burkholderia genus according to ARDRA. Two of these isolates, kindly provided by A. Ferroni, had been identified as R. mannitolilytica by use of 16S rRNA sequencing (10). The bacterial strains studied and their sources are listed in Table 1.
TABLE 1.
List of bacterial strains studied and ARDRA resultsc
| Strain or isolate | Source of isolation | ARDRA pattern for:
|
ARDRA group | |||||
|---|---|---|---|---|---|---|---|---|
| AluI | CfoI | DdeI | MspI | NciI | BssKI | |||
| Reference strains | ||||||||
| R. solanacearum | ||||||||
| Race 1 K 60T/UW 25a | Tomato | H | D | D | H | J | A | R1 |
| Race 1 GMI 1000a | Tomato | K | D | D | H | J | A | R2 |
| Race 2 S 210/UW 70a | Plantain | H | D | D | H | J | A | R1 |
| Race 3 S 206/UW 80a | Potato | I | D | D | H | J | A | R3 |
| Race 3 S 207/UW 81a | Potato | I | D | D | H | J | A | R3 |
| R. eutropha LMG 1199T | Soil | L | F | L | D | L | L | R4 |
| R. campinensis LMG 19282T | Zinc desert | H | F | D | I | K | I | R5 |
| R. metallidurans LMG 1195T | Wastewater zinc factory | H | H | D | I | J | A | R6 |
| R. basilensis LMG 18990T | Freshwater pond | L | F | L | D | D | N | R7 |
| R. pickettii ATCC 27511T | Human (tracheotomy) | D | D | D | D | D | D | R8 |
| R. mannitolilytica LMG 6866T | Outbreak of hospital infection | K | D | D | H | K | A | R9 |
| R. gilardii LMG 5886T | Whirlpool | H | G | D | I | J | Ib | R10 |
| R. taiwanensis LMG 19424T | Mimosa | H | K | L | I | K | Ib | R11 |
| R. paucula LMG 3244T | Human (respiratory tract) | H | K | D | I | J | A | R12 |
| P. pnomenusa LMG 18087T | Human (CF patient, United Kingdom) | H | L | K | B | J | M | P1 |
| P. sputorum LMG 18819T | Human (CF patient, United States) | H | L | K | B | J | M | P1 |
| P. pulmonicola LMG 18106T | Human (CF patient, Canada) | H | J | K | B | J | M | P2 |
| P. apista LMG 16407T | Human (CF patient, Denmark) | H | J | K | B | J | M | P2 |
| P. norimbergensis LMG 18379T | Water layer, Germany | J | I | M | B | J | M | P3 |
| B. cepacia genomovar III LMG 12615 | Human (CF patient, United Kingdom) | A | A | A | A | A | A | |
| B. multivorans LMG 13010T | Human (CF patient, Belgium) | A | A | I | A | A | AI | |
| Clinical isolates | ||||||||
| 5-6 | Human (CF patient, France) | K | D | D | H | K | A | R9 |
| 5-51 | Human (CF patient, France) | K | D | D | H | K | A | R9 |
| 5-52 | Human (CF patient, France) | K | D | D | H | K | A | R9 |
| 6-81 | Human (CF patient, France) | K | D | D | H | K | A | R9 |
| 9-26 | Human (CF patient, France) | K | D | D | H | K | A | R9 |
Strain provided by C. Boucher, INRA, Castanet-Tolosan, France.
Subtle difference in the size of one fragment compared to the I pattern.
Results for the Bcc species were previously published (13).
The restriction fragment length polymorphism (RFLP) analysis of amplified 16S rDNA was performed as previously described (13). Briefly, the primers fD1 (5′-CCGAATTCGTCGACAACAGAGTTTGATCCTGGCTCAG-3′) and rD1 (5′-CCCGGGATCCAAGCTTAAGGAGGTGATCCAGCC-3′) were used to amplify approximately 1,500 bp within the 16S rRNA gene. The PCR products were digested with the following endonucleases: AluI, CfoI, DdeI, MspI, NciI, and BssKI.
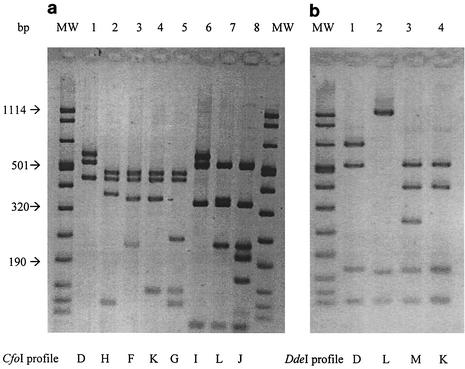
Some of the restriction profiles obtained are shown in Fig. 1, and ARDRA results are summarized in Table 1. The previously published results (13) obtained for the two main Bcc species involved in cystic fibrosis, i.e., B. cepacia genomovar III and B. multivorans, are included in Table 1 for comparison. The 14 Ralstonia type and reference strains tested, representing 10 species, were classified in 12 ARDRA groups; the 5 R. solanacearum strains were classified in 3 ARDRA groups, and one of the strains belonging to race 1 was not discriminated from the representative strain of race 2 tested. The type strains of all the Ralstonia species included in this study harbored specific ARDRA patterns. The five Pandoraea type strains tested were classified in three ARDRA groups; thus, P. pnomenusa and P. sputorum on one hand and P. pulmonicola and P. apista on the other hand harbor the same RFLP profiles. The five clinical strains tested presented the same RFLP profiles as the R. mannitolilytica type strain. Finally, the ARDRA patterns clearly differentiate Ralstonia and Pandoraea species from Burkholderia species, as well as from other nonfermenting gram-negative bacilli frequently recovered from patients with CF, such as Pseudomonas aeruginosa, Stenotrophomonas maltophilia, and Achromobacter xylosoxidans (data not shown). The prevalence of Ralstonia species in patients with CF is probably low but may be underestimated due to misidentification. Coenye et al. recently reported that 42 of about 4,000 bacterial isolates collected from American CF patients and sent to the Burkholderia cepacia Research Laboratory and Repository (University of Michigan) belonged to the genus Ralstonia (8), with R. mannitolilytica being the most prevalent species (25 out of 42 isolates). In a French pediatric CF center, R. mannitolilytica was recovered from 2 out of 259 patients (10). Finally, 5 clinical isolates out of 247 nonredundant isolates recovered from 243 French CF patients and collected by the Observatoire Cepacia/Vaincre la Mucoviscidose, including the two isolates mentioned above, were identified as R. mannitolilytica in the present study. The pathogenic potential of this species is not clear; however, chronic colonization is possible. Pandoraea species have been recovered from CF patients in Canada, Denmark, Brazil, the United States, and the United Kingdom, but to our knowledge there are no available epidemiological data concerning the prevalence of these organisms in CF patients.
FIG. 1.
ARDRA patterns. MW, molecular weight marker VIII (Roche). (a) Different Cfo I restriction profiles obtained within Ralstonia (lanes 1 to 5) and Pandoraea species (lanes 6 to 8). (b) Different Dde I restriction profiles obtained within Ralstonia (lanes 1 and 2) and Pandoraea species (lanes 3 and 4).
Phenotypic tests for Ralstonia and Pandoraea species can be misleading: confusion of Ralstonia sp. with Burkholderia sp., Pseudomonas sp., and S. maltophilia (8, 10) and of Pandoraea sp. with Burkholderia sp. and Ralstonia sp. has been reported (4). Thus, molecular methods are necessary for an accurate diagnosis. 16S rRNA gene sequencing (10) is widely applicable to any problematic isolate but requires a sequencer; 16S ribosomal DNA-based genus- and/or species-specific PCR has been developed for R. mannitolilytica and R. pickettii (8) and for Pandoraea species (7). Analysis of the gyrB gene seems promising for the identification of Pandoraea species (6). The ARDRA method tested in this study, though unable to separate all Pandoraea species, proves to be a useful identification tool; it requires the constitution of a data bank of profiles but has the advantage of being equally applicable to the main organisms growing on B. cepacia-selective media, i.e., Burkholderia, Ralstonia, and Pandoraea.
Acknowledgments
This study was supported by the Université Paul-Sabatier, Toulouse III, France and by the French CF Association Vaincre La Mucoviscidose.
We thank A. Ferroni (Hôpital Necker, Paris), H. Monteil (Institut de Bactériologie, Strasbourg), P. Plessis (Centre Hospitalier des Pays-de-Morlaix), and J. Thubert (Centre de Perharidy, Roscoff) for providing clinical strains.
REFERENCES
- 1.Brisse, S., S. Stefani, J. Verhoef, A. Van Belkum, P. Vandamme, and W. Goessens. 2002. Comparative evaluation of the BD Phoenix and VITEK 2 automated instruments for identification of isolates of the Burkholderia cepacia complex. J. Clin. Microbiol. 40:1743-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burns, J. L., J. Emerson, J. R. Stapp, D. L. Yim, J. Krzewinski, L. Louden, B. W. Ramsey, and C. R. Clausen. 1998. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin. Infect. Dis. 27:158-163. [DOI] [PubMed] [Google Scholar]
- 3.Chen, W., S. Laevens, T. Lee, T. Coenye, P. De Vos, M. Mergeay, and P. Vandamme. 2001. Ralstonia taiwanensis sp. nov., isolated from root nodules of mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. E vol. Microbiol. 51:1729-1735. [DOI] [PubMed] [Google Scholar]
- 4.Coenye, T., E. Falsen, B. Hoste, M. Ohlen, J. Goris, J. Govan, M. Gillis, and P. Vandamme. 2000. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pnomenusa sp. nov., and Pandoraea norimbergensis comb. nov. Int. J. Syst. E vol. Microbiol. 50:887-899. [DOI] [PubMed] [Google Scholar]
- 5.Coenye, T., J. Goris, T. Spilker, P. Vandamme, and J. LiPuma. 2002. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J. Clin. Microbiol. 40:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coenye, T., and J. LiPuma. 2002. Use of the gyrB gene for the identification of Pandoraea species. FEMS Microbiol. Lett. 208:15-19. [DOI] [PubMed] [Google Scholar]
- 7.Coenye, T., L. Liu, P. Vandamme, and J. LiPuma. 2001. Identification of Pandoraea species by 16S ribosomal DNA-based assays. J. Clin. Microbiol. 39:4452-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coenye, T., P. Vandamme, and J. LiPuma. 2002. Infection by Ralstonia species in cystic fibrosis patients: identification of R. pickettii and R. mannitolilytica by polymerase chain reaction. Emerg. Infect. Dis. 8:692-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daneshvar, M., D. G. Hollis, A. G. Steigerwalt, A. M. Whitney, L. Spangler, M. P. Douglas, J. G. Jordan, J. P. MacGregor, B. C. Hill, F. C. Tenover, D. J. Brenner, and R. S. Weyant. 2001. Assignment of CDC weak oxidizer group 2 (Wo-2) to the genus Pandoraea and characterization of three new Pandoraea genomospecies. J. Clin. Microbiol. 39:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferroni, A., I. Sermet-Gaudelus, E. Abachin, G. Quesne, G. Lenoir, P. Berche, and J.-L. Gaillard. 2002. Use of 16S rRNA gene sequencing for identification of nonfermenting gram-negative bacilli recovered from patients attending a single cystic fibrosis center. J. Clin. Microbiol. 40:3793-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goris, J., P. De Vos, T. Coenye, B. Hoste, D. Janssens, H. Brim, L. Diels, M. Mergeay, K. Kersters, and P. Vandamme. 2001. Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int. J. Syst. E vol. Microbiol. 51:1773-1782. [DOI] [PubMed] [Google Scholar]
- 12.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 13.Segonds, C., T. Heulin, N. Marty, and G. Chabanon. 1999. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J. Clin. Microbiol. 37:2201-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaneechoutte, M., T. De Baere, G. Wauters, S. Steyaert, G. Claeys, D. Vogelaers, and G. Verschraegen. 2001. One case each of recurrent meningitis and hemoperitoneum infection with Ralstonia mannitolilytica. J. Clin. Microbiol. 39:4588-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wauters, G., G. Claeys, G. Verschraegen, T. De Baere, E. Vandecruys, L. Van Simaey, C. De Ganck, and M. Vaneechoutte. 2001. Case of catheter sepsis with Ralstonia gilardii in a child with acute lymphoblastic leukemia. J. Clin. Microbiol. 39:4583-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yabuuchi, E., Y. Kosako, I. Yano, H. Hotta, and Y. Nishiuchi. 1995. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudouroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol. Immunol. 39:897-904. [DOI] [PubMed] [Google Scholar]