Abstract
The transduction of type I interferon signals to the nucleus relies on activation of a protein complex, ISGF3, involving two signal transducers and activators of transcription (STAT) proteins, STAT1 and STAT2, and the interferon (IFN) regulatory factor (IRF) protein, p48/ISGF3γ. The STAT subunits are cytoplasmically localized in unstimulated cells and rapidly translocate to the nucleus of IFN-stimulated cells, but the p48/ISGF3γ protein is found in both the nucleus and the cytoplasm, regardless of IFN stimulation. Here, we demonstrate that p48 is efficiently and constitutively targeted to the nucleus. Analysis of the subcellular distribution of green fluorescent protein-p48 fragments indicates that p48 contains a bipartite nuclear retention signal within its amino-terminal DNA-binding domain. This signal is preserved in two other IRF proteins involved in immune responses, ICSBP and IRF4. Mutations to clustered basic residues within amino acids 50–100 of p48 or IRF4 disrupt their nuclear accumulation, and DNA-binding ability is not required for nuclear targeting. This is the only example of a nuclear localization signal for any ISGF3 component and assigns a second function to the IRF DNA-binding domain. We also demonstrate that the nuclear distribution of p48 is dramatically altered by coexpression of the STAT2 protein, indicating that STAT2 forms a cytoplasmic complex with p48, overriding the intrinsic p48 nuclear targeting. Retention by STAT2 may serve to regulate the activity of free p48 and/or guarantee that cytoplasmic pools of preassociated STAT2:p48 are available for rapid activation of the IFN response. These findings suggest that analogous mechanisms may exist for regulating the distribution of other IRF proteins.
The innate antiviral mechanism for most cells involves the actions of type I interferons (IFNα and IFNβ), leading to induction of IFN responsive gene expression. Two families of transcriptional regulators, members of the signal transducers and activators of transcription (STAT) and IFN regulatory factor (IRF) families, work in conjunction to establish a cascade of gene regulation and signal transduction events, producing the antiviral response. Recently, the IRF family members IRF3 and IRF7 have been shown to play an important role in the regulation of IFNβ synthesis in response to virus infection in the primary infected cells (1). Infection causes their mobilization from the cytoplasm to the nucleus, where they bind to and induce the expression of the IFNβ gene. In the adjacent cells, most of the immediate actions of IFN have been linked to activation of latent cytoplasmic STAT proteins to produce a multiprotein complex, ISGF3, which induces transcription from target promoter interferon-stimulated response elements (ISRE). ISGF3 is comprised of three protein subunits: STAT1, STAT2, and p48/ISGF3γ. The p48 protein belongs to the IRF family, which includes at least nine mammalian members. The IRF proteins are most homologous in their NH2- terminal DNA-binding domain, which is characterized by a unique helix–turn–helix structure with the DNA recognition helix positioned by a cluster of five conserved tryptophan residues (2). The COOH terminus of the p48 protein has been demonstrated to mediate ISGF3 formation (3) by binding directly to the STAT1 and STAT2 subunits (4, 5) (see Fig. 2A).
Figure 2.
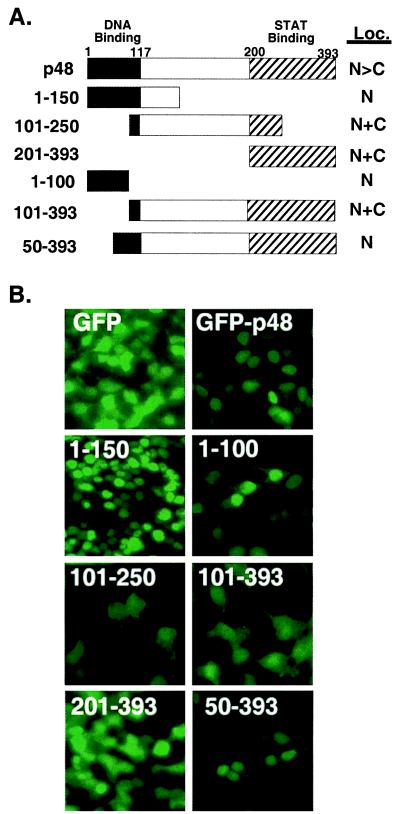
The DNA-binding region of p48 specifies nuclear localization. (A) Diagram of the p48 fragments fused to GFP and their subcellular localization (Loc.) when transiently expressed in 293T cells. Functional domains of p48 are labeled. N, nuclear; C, cytoplasmic. (B) Fluorescence patterns of the GFP and GFP fusions after transient expression in 293T cells. Expressed protein and fragment boundaries are indicated.
The p48 protein has long been recognized as the DNA sequence recognition subunit of ISGF3 and is required for IFNα responses (6, 7). The p48 protein has also been demonstrated to play a role in IFN-γ responses. After IFN-γ treatment, the p48 level can be increased 10-fold because of new protein synthesis (8, 9). In addition, fibroblasts from mice deficient in p48 fail to respond to IFN-γ, presumably because of a defect in the IFN-γ dependent p48 expression (10). Transfection-based studies of p48-mediated ISRE transcription have demonstrated that increased p48 levels are required to achieve maximal induction of some IFN-stimulated genes (ISGs) (11), and that these genes are inducible simply by treatment with IFN-γ, which increases the p48 protein level. Recently, p48 has been linked to the IFN-γ induction of the IP-10 chemokine gene (12). These findings suggest an ISGF3-independent role for p48 in gene expression.
The subcellular distribution of several IRF family proteins has been investigated, and amino acids important for their localization have been determined. The IRF1 and IRF2 proteins contain positive-acting nuclear localization signals (NLS) located immediately C-terminal to the DNA-binding domain, involving amino acids ≈120–140 (13). The IRF3 protein has a nuclear export signal (NES) specifying its cytoplasmic retention (14). Examination of the p48 protein primary amino acid sequence does not reveal any obvious regions significantly homologous to these signals or to any other known NLS or NES. The IRF factors most homologous to p48, ICSBP and IRF4, similarly lack identifiable NLS or NES regions, but recent reports indicate that they can undergo regulated nuclear translocation (15, 16). The mechanisms underlying this translocation remain uncharacterized.
An important unresolved issue in the activation of the ISGF3 complex by IFN treatment concerns the location and dynamics of complex assembly and the signals used for its nuclear transport. The STAT1:2 heterodimer forms at the cytoplasmic face of the cell membrane while bound to the intracellular domain of the IFN receptor (17). The cellular location where STAT proteins first encounter p48 remains unclear, but previous studies of ISGF3 assembly determined that the p48 protein is distributed in both the nucleus and cytoplasm of cells and the protein from either fraction can form ISGF3 in vitro (8, 9, 18).
In this report, the subcellular distribution of the p48 protein has been investigated and, in agreement with previously reported data, a significant portion of the protein is found in the cytoplasm. In contrast, analysis of green fluorescent protein (GFP)-p48 fusion proteins shows that the p48 protein contains within its DNA-binding domain the necessary sequences to specify its nuclear accumulation. Mutations to amino acids important for p48 DNA-binding ability do not alter the distribution of this protein, indicating that the two functions of this region are distinct, but overlapping. Nonetheless, mutations to basic amino acids within the DNA-binding region can abolish nuclear accumulation of the protein, defining two essential elements of the p48 NLS. Similar results are obtained with the IRF4 protein, implying that this conserved functional region defines a bipartite NLS motif for a subset of IRF proteins.
We also report that coexpression of the STAT2 protein can dramatically alter the localization of p48, causing its retention in the cytoplasm. The ability of STAT2 to retain p48 in the cytoplasm correlates with protein:protein interaction in the p48 C terminus. Together, these results indicate a previously unrecognized bipartite nuclear localization signal for the p48 protein that is conserved in other IRF family proteins, and also suggest a role for STAT2 as a cytoplasmic chaperone for the p48 protein.
Experimental Procedures
Cell Culture and Transfection.
Human 2fTGH, U3A, U6A, 293T, and murine NIH 3T3 cells were maintained in DMEM supplemented with 10% cosmic calf serum (HyClone). Transfection of cells with cDNAs was carried out by standard CaPO2 procedures (19, 20) and directly observed 24 h later (see below).
Fluorescence Microscopy.
For indirect immunofluorescence, cells grown on chamber slides were fixed in 1:1 methanol:acetone at −20°C for 15 min, washed with PBS, then blocked with 1% BSA in PBS for 15 min. Samples were stained with p48 antiserum (diluted 1:50 in 1% BSA/PBS solution) and FITC-conjugated goat anti-rabbit for 1 h each at 37°C. GFP fluorescence of transfected cells was observed at 24 h posttransfection with a fluorescence microscope (Olympus BX60) with the fluorescein filter set. Images were captured with a charge-coupled device camera (Optronics International, Chelmsford, MA) and processed with adobe photoshop software on a Power MacIntosh G3 computer.
Plasmid Construction.
Inserts for the GFP-p48 and GFP-IRF4 fusions were generated by PCR amplification of p48 or IRF4 (gift of Alex Grossman, Amgen Institute, Ontario, Canada) segments with in-frame EcoRI and BamHI restriction sites by using Vent Polymerase (NEB, Beverly, MA). The resulting fragments were cloned into the pEGFPC1 vector (CLONTECH). His-p48 was constructed similarly, by subcloning the PCR-generated p48 ORF into a modified pCDNA3 vector containing a linker encoding a methionine, arginine, serine, and six histidines before the initiating methionine of p48. Site-directed mutagenesis was performed with a four-primer PCR method described previously (20). All constructs were verified by DNA sequencing. The FLAG tagged STAT2 cDNA was described previously (21) and was the gift of Markus Heim (University Hospital, Basel).
Antibodies, Extracts, Immunoprecipitation, and Immunoblotting Assays.
Antibodies to p48, His-tag, FLAG-tag, and GFP were obtained from Santa Cruz Biotechnology, Qiagen (Chatsworth, CA), Kodak, and CLONTECH, respectively, and used according to manufacturers' instructions. For immunoprecipitations, whole cell extracts were prepared and precipitated as described (4). The immune complexes collected on protein A/G agarose (Calbiochem) were washed four times with cell extraction buffer, the proteins separated by SDS/PAGE on 12% gels and transferred to nitrocellulose filters and immunoblotted by standard procedures (4).
Electrophoretic Mobility Shift Assay.
Electrophoretic gel mobility shift assays were carried out essentially as described previously (20). Double-stranded oligonucleotides representing the ISG15 ISRE element were radiolabeled by filling in protruding ends with 32P by using Klenow fragment of DNA polymerase. Cell extracts were mixed with 1 × 105 cpm of probe for 15 min before separation on a 5% polyacrylamide gel. For antibody supershifts, 0.1 μl of antibody was added to the reaction during incubation. Gels were dried and subjected to autoradiography.
Results
Subcellular Distribution of GFP-p48 Fusion Proteins.
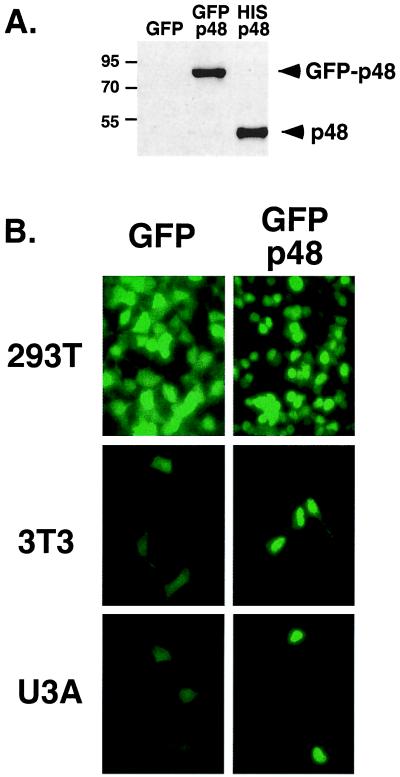
The p48 protein was found to be localized in both nuclear and cytoplasmic HeLa cell extracts (8, 18). Our own investigations of endogenous p48 distribution by cell fractionation (data not shown) agree with these prior results, and the distribution of p48 can be further demonstrated by indirect immunofluorescence of endogenous p48 in intact cells (see Fig. 4C). The lack of identifiable nuclear targeting sequences in the p48 primary amino acid sequence and the fact that the apparent molecular weight of the p48 protein is near the theoretical diffusion limit for the nuclear pore complex (22) suggested a hypothesis that the p48 protein might freely diffuse between the nucleus and the cytoplasm of cells. To test the diffusion theory, we constructed a GFP fusion with p48 (GFP-p48). The larger fusion protein exhibited a substantially slower gel migration than a hexahistidine-tagged p48 (His-p48; Fig. 1A) and is well beyond the theoretical diffusion limit for the nuclear pore (22). The distribution of GFP fluorescence revealed that the GFP-p48 protein clearly accumulates in the cell nucleus. As shown in Fig. 1B, the accumulation of GFP-p48 in the nucleus is a general property of the protein observed in transiently transfected 293T cells, NIH 3T3 cells, and U3A cells. Thus, the p48 protein is actively transported to and can accumulate within the nucleus, implying that the protein (i) contains a nuclear import signal, and (ii) is normally held in the cytoplasm of cells by either protein:protein interaction or posttranslational modification. To demonstrate that the GFP tag did not influence nuclear localization, the localization of His-p48 was examined by indirect immunofluorescence. His-p48 was also found to accumulate in the nucleus (see Fig. 4A, square E).
Figure 4.
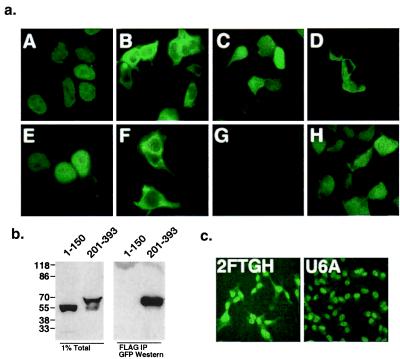
Cytoplasmic retention of p48 by STAT2. (a) Fluorescent patterns of GFP fusions in the presence and absence of STAT2 expression. (A–D) GFP-p48 fusions were expressed in 293T cells alone (A and C) or with STAT2 (B and D) and were examined by fluorescence microscopy 24 h later. (A) Full-length GFP-p48 (residues 1–393). (B) Full-length GFP-p48 expressed with STAT2. (C) GFP-p48201–393. (D) GFP-p48201–393 expressed with STAT2. (E–G) GFP does not influence p48 localization. Anti-p48 indirect immunofluorescence: (E) Expression of His-p48. (F) His-p48 expressed with STAT2. (G) Mock-transfected control. (H) Control GFP vector expressed with STAT2. (×40 objective magnification.) (b) Cytoplasmic retention of p48 correlates with STAT2 interaction region. GFP-p481–150 and GFP-p48201–393 were expressed in 293T cells with FLAG-tagged STAT2, and whole cell lysates were prepared. (Left) Direct application of 1% of the lysates to the gel. (Right) The remaining lysate was immunoprecipitated with anti-FLAG epitope antibody before SDS/PAGE and immunoblotting with GFP polyclonal antiserum. Positions of prestained molecular weight markers are indicated to the left. (c) Abundance of cytoplasmic p48 depends on STAT2 expression. Localization of endogenous p48 in the presence (2FTGH) or absence (U6A) of STAT2 was observed by indirect immunofluorescence using p48 specific antiserum.
Figure 1.
Subcellular localization of GFP-p48. (A) Identification of the GFP-p48 fusion protein expressed in 293T cells by immunoblot of whole cell lysates with p48-specific antiserum. Control lysates with GFP alone and His-p48 are included. (B) Fluorescence patterns of cells transiently transfected with GFP vector (Left) or GFP-p48 vector (Right). The p48 sequence specifies efficient transport to and accumulation in the nucleus of 293T cells, NIH 3T3 cells, and U3A cells as indicated on the left margin.
The DNA-binding Domain of p48 Directs Accumulation in the Nucleus.
Two major functional domains have been defined in the p48 protein: the amino-terminal DNA-binding domain (23), and the carboxyl-terminal STAT binding domain (4) (Fig. 2A). To determine whether the NLS of p48 is contained within either of these two domains, GFP-p48 cDNAs encoding p48 amino acids 1–150, 101–250, and 201–393 were constructed and expressed in 293T cells (Fig. 2B). The fluorescence pattern of the GFP protein alone reveals that this protein localizes to both the nucleus and cytoplasm, presumably by free diffusion of the ≈27-kDa protein through the nuclear pore complex. Examination of the GFP-p48 fusion proteins indicates that only the full-length GFP-p48 and the GFP fusion containing the amino-terminal 150 amino acids of p48 accumulate in the nucleus. Fragments containing amino acids 101–250 or 201–393 appeared in both nuclear and cytoplasmic compartments similar to the GFP control. The accumulation of GFP-p48 and GFP-p481–150 in the nucleus and the fact that GFP-p48101–250 and GFP-p48201–393 do not exhibit nuclear accumulation even though all of the fusions are similar in size (≈50 kDa, data not shown), suggests that the NLS of p48 resides in the amino terminus, specifically within amino acids 1–100.
To further localize the p48 nuclear-targeting region, GFP fusions with p48 amino acids 1–100, 101–393, and 50–393 were constructed and expressed. Both the GFP-p481–100 and GFP-p4850–393 were targeted to and accumulated in the nucleus, but the GFP-p48101–393 was not. This result further localizes the p48 NLS to amino acids 50–100, which contains the DNA recognition helix.
Mutational Analysis of p48 Nuclear Localization.
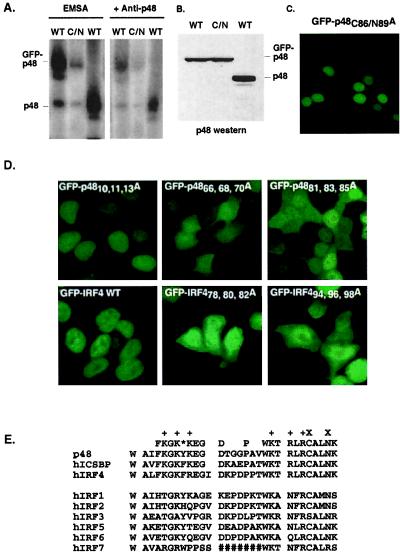
The accumulation of the GFP-p48 in the nucleus might be explained by nuclear diffusion followed by retention in the nucleus because of the protein's inherent DNA-binding ability. To test this model, site-directed mutagenesis of the GFP-p48 protein was used to target amino acids involved in DNA binding by p48. The rationale for mutagenesis of the DNA-binding domain was based on the co-crystal structure of IRF1 bound to DNA (2). The defined region of p48, amino acids 50–100, encompasses the helix–turn–helix region of the DNA-binding domain. In this region, two α helices, α2 and α3, are separated by a large flexible loop, L2. Residues involved in the overall folding of the DNA-binding domain were avoided to preserve key structural elements, and basic amino acids that might be involved in nuclear import were also maintained. Instead, two residues of helix α3, which make specific nucleotide base contacts, were targeted for mutagenesis (see Fig. 3E). A cysteine at position 86 of p48 and an asparagine at position 89 of p48 were both changed to alanine to create GFP-p48C86/N89A. The mutated GFP-p48 protein was expressed in 293T cells and extracts prepared for electrophoretic mobility shift assays. The mutant protein has a greatly reduced DNA-binding capacity compared with p48 or GFP-p48 (Figs. 3 A and B). Analysis of the subcellular distribution of GFP-p48C86/N89A revealed that it was capable of accumulating in the nucleus indistinguishably from wild-type GFP-p48 (Fig. 3C). This result demonstrates that the two functions of the amino-terminal region, DNA binding and nuclear accumulation, are separable.
Figure 3.
Mutational analysis of the p48 and IRF-4 NLS. DNA binding is not required for p48 nuclear accumulation. (A) Electrophoretic mobility shift assay illustrating that mutation GFP-p48C86/N89A (C/N) disrupts p48 DNA-binding ability. Antibody-mediated supershift reveals the specificity of the protein:DNA complexes. (B) Anti-p48 immunoblot illustrating equal amounts of p48 proteins in the cell extracts used for EMSA. (C) Nuclear fluorescence of GFP-p48C86/N89A mutant expressed in 293T cells. (D) Effects of mutations to basic amino acids on GFP-p48 and GFP-IRF4 nuclear accumulation. Mutants GFP-p4810,11,13A, GFP-p4866, 68, 70A, and GFP-p4881, 83, 85A were expressed in 293T cells (Upper). GFP-IRF4, and mutants GFP-IRF478,80,82A and GFP-IRF494,96,98A were expressed in 293T cells (Lower). (E) Alignment of nuclear localization regions of IRF family proteins representing p48 amino acids 62–90. The consensus derived from p48, ICSBP, and IRF4 sequences is written on top, with basic residues marked (+). *, Position of a conserved aromatic residue in the consensus. X, Residues C86 and N89. #, A long extension in IRF7, which is deleted for the purpose of this alignment.
Further mutagenesis was used to determine specific amino acids that contribute to p48 nuclear accumulation. Because most of the known NLS domains contain clusters of basic amino acids, three clusters of basic amino acids in the amino terminus of p48 were separately targeted by alanine substitution. R10, K11, and R13 were changed to alanine to create GFP-p4810,11,13A, K66, K68, and K70 were changed to alanine to create GFP-p4866, 68, 70A, and K81, R83, and R85 were changed to alanine to create GFP-p4881, 83, 85A. On expression in 293T cells, only the GFP-p4810, 11, 13A mutant (which lies outside the defined NLS region) accumulated in the nucleus similarly to GFP-p48 (Fig. 3D). The other two proteins (GFP-p4866, 68, 70A and GFP-p4881, 83, 85A) exhibited a much greater cytoplasmic distribution pattern and did not accumulate in the nucleus (Fig. 3D). These results indicate the importance of both basic amino acid clusters in p48 nuclear import and retention.
Comparison of the IRF protein amino acid sequences in this region reveals two other family members that have identically positioned basic amino acid clusters, IRF4 and ICSBP (Fig. 3E). To determine the general role of these amino acids in IRF nuclear accumulation, GFP-IRF4 fusion proteins were constructed and expressed in 293T cells (Fig. 3D). Like p48, wild-type GFP-IRF4 efficiently accumulates in the nucleus, but mutations to either cluster of basic amino acids (residues 78, 80, 82 or 94, 96, 98 were changed to alanine) result in the loss of nuclear retention. Thus, the use of these two basic clusters is not restricted to the p48 protein, but represents a common signal for this branch of the IRF family.
Retention of p48 in the Cytoplasm by STAT2.
The GFP-p48 protein accumulates in the nucleus when overexpressed in transient transfection assays. In contrast, significant cytoplasmic p48 protein is observed in normal cells (Fig. 4C, and refs. 8, 9, and 18), and in stable cell lines expressing modest levels of His-p48 (data not shown). One explanation for these differences is that, in the transient GFP-p48 transfections, a cytoplasmic retention factor becomes saturated, allowing the GFP-p48 to freely enter the nucleus. Because p48 is known to associate with STAT1 and STAT2 in the cytoplasm in the absence of IFN signaling (4), the ability of these proteins to influence the distribution of p48 was tested. Expression of STAT1 had little effect on GFP-p48 localization (not shown), but expression of STAT2 had a profound effect on the subcellular distribution of GFP-p48, causing the majority of the fluorescence to be retained in the cytoplasm and excluded from the nucleus (Fig. 4A, square B). This relocalization phenomenon is not influenced by the GFP tag, as His-p48 is also retained in the cytoplasm by STAT2 (Fig. 4A, square F).
To determine the regions of p48 required for the STAT2-dependent effect and test the specificity of relocalization in this assay, the GFP-p48 fragments described above were coexpressed with STAT2. Only GFP-p48201–393, which contains the known STAT2 binding region of p48 (4), was retained in the cytoplasm by STAT2 expression (Fig. 4A, square D). Thus, the ability of highly abundant STAT2 to override the p48 protein's intrinsic NLS relies only on the C-terminal domain.
To verify that the STAT2 protein is binding the GFP-p48 protein fragments used in this experiment, a coimmunoprecipitation assay was carried out. GFP-p48 fusions were coexpressed with a FLAG epitope-tagged STAT2 protein. Cell extracts were prepared, immunoprecipitated with FLAG antibodies, and immunoblotted with GFP-specific antibodies. STAT2 was able to precipitate the carboxyl-terminal GFP fusion (Fig. 4B, 201–393), but not the amino terminal fusion (Fig. 4B, 1–150). Thus, protein interactions between STAT2 and the p48 carboxy terminus correlate with the cytoplasmic retention phenotype.
To examine this cytoplasmic retention phenomenon in a physiological context, the steady state distribution of p48 was analyzed by indirect immunofluorescent staining of both 2fTGH cells and a STAT2-deficient derivative, U6A (24). In both cell lines, p48 was found in the nucleus, but in the 2fTGH cells, p48 was also detected in the cytoplasm (Fig. 4C). This result confirms that the presence of STAT2 plays a role in determining the p48 subcellular distribution.
Discussion
The results presented here define a nuclear localization signal within the DNA-binding domain of a set of IRF family proteins. We have mapped the signal of p48/ISGF3γ to amino acids 50–100 within the DNA recognition region. Basic amino acids at positions 66–70 and at positions 81–85 are required for efficient nuclear accumulation of the p48 protein. Significantly, we demonstrate that this NLS arrangement is also functional in a second IRF family member, IRF4.
The IRF family contains at least nine mammalian members (p48, IRF1, IRF2, IRF3, IRF4, IRF5, IRF6, IRF7, and ICSBP), which are most homologous in their DNA-binding domain. Only two of these, IRF1 and IRF2, have identifiable nuclear localization signals, which are not conserved among the family members and have been demonstrated experimentally to fall outside of the regions defined here for p48 targeting to the nucleus (13). Sequence alignment (Fig. 3E) reveals that other members of the IRF family share elements of the p48 NLS within their amino termini. Despite the high overall homology among the DNA-binding regions of the IRFs, only IRF4 and ICSBP exhibit conservation of the basic amino acids shown here to be critical for p48 nuclear accumulation. We demonstrate that these residues function analogously in IRF4, affirming the generality of this signal for IRF proteins. The arrangement of basic amino acids found in these proteins is consistent with a variation of the bipartite basic nuclear localization signal (25). Bipartite basic signals typically consist of two interdependent clusters of basic amino acids separated by a flexible linker of at least 10 amino acids (25, 26). In agreement with this definition, the determined IRF DNA-binding domain crystal structure illustrates that residues 66–70 and 80–85 are separated by 10 residues including the flexible loop, L2.
It is notable that the IRF3 protein, which is a subunit of the double-stranded RNA activated transcription factor, DRAF (27), contains a nuclear export signal in its carboxy terminus (14). Thus, regulated subcellular distribution emerges as a common theme for controlling IRF functions. The results indicate the principal NLS for these IRFs is the bipartite signal; nevertheless, it remains possible that other residues, protein interactions, or structural modifications make additional contributions to the subcellular distribution of these proteins. The lack of complete nuclear exclusion for any of the GFP-p48 fusions or the NLS mutants suggests that neither p48 nor IRF4 have an exposed nuclear export signal.
A unique means of regulating the distribution of the p48 protein has evolved. As we demonstrate, p48 exhibits constitutive nuclear localization when overexpressed and is primarily nuclear in the absence of STAT2. Further, coexpression of the STAT2 protein can override the intrinsic localization of p48 by interaction with a distinct region of the protein (amino acids 201–393), resulting in retention of p48 in the cytoplasm. Interestingly, expression of STAT1 had little effect on the localization of the GFP-p48 protein (not shown). This result suggests that the contact between p48 and STAT1 is either of too low affinity to retain p48, or is important only for the mature trimeric ISGF3 complex, which forms following IFN activation of ISGF3.
The physiological role of the observed chaperone function of STAT2 and the mechanistic details of ISGF3 assembly and translocation will require further investigation. Nonetheless, it is tempting to speculate that the preassociation of p48 with STAT2 in the cytoplasm might regulate IFN signaling in several ways. For example, STAT2 might regulate ISGF3-independent activities of p48 by retaining it in a cytoplasmic location. This function would be similar to the role of IκB in regulating the nuclear import of the NFκB transcription factor (28), except that, in this analogy, STAT2 is not degraded in response to stimulus like IκB, but is instead activated for dimerization with STAT1, and the entire trimeric ISGF3 complex is transported to the nucleus. As p48 was first identified as an essential part of the ISGF3 complex, little is known about its ability to independently regulate transcription, but p48 is known to be involved in IFNγ responses. For example, p48 has recently been implicated in the regulation of a protein called IP-10, a chemokine known to be involved in inflammatory and neoplastic disorders (12). The IP-10 gene is inducible by IFNγ, supporting a role for p48 in the actions of this cytokine independent of ISGF3. As p48 levels are increased by IFNγ treatment, the available STAT2 might become saturated, allowing free p48 to translocate to the nucleus.
A different role for STAT2:p48 cytoplasmic retention might be to maintain the cell's ability to rapidly generate the ISGF3 complex in response to type I IFN signals. In this scenario, in response to IFNα stimulation, the preexisting STAT2:p48 complex in the cytoplasm would bind as a unit to the IFN receptor-kinase complex, where STAT2 becomes phosphorylated on tyrosine and oligomerizes with STAT1. The trimeric complex is now competent for transfer to the nucleus together by using the currently undetermined STAT nuclear import signals. This mechanism would ensure a readily available pool of STAT2:p48 for the rapid assembly of ISGF3. These models are not necessarily mutually exclusive, and the precise role of STAT2:p48 complexes in ISGF3 assembly and transport in response to IFN treatment is under investigation.
Our results demonstrate that IRF4 uses a homologous signal to direct its accumulation in the nucleus. IRF4, a regulator of lymphocyte development and functions, has been found to undergo LPS-regulated nuclear translocation in macrophage cell lines (16). Like the interaction of p48 with STATs to form ISGF3, IRF4 combines with the ETS family factor, PU.1, to recognize a hybrid DNA element (29). IRF4 also interacts with the immunophilin, FKBP52, a peptidyl-prolyl isomerase that can inhibit IRF4 DNA-binding activity (30). It will be interesting to determine whether association with these or other proteins regulates the distribution of IRF4. The regulated nuclear transport of IRF proteins now appears to be a common theme by which the functions of these proteins can be controlled and will provide many interesting questions for future study.
Acknowledgments
We thank Abigail Greiner and Wei Zhang for assistance with early experiments on p48 localization, Aurelian Radu for assistance with fluorescence microscopy, and Markus Heim for the gift of the FLAG-tagged STAT2 expression vector. We thank Aneel Aggarwal, Scott Plevy, Serafin Piñol-Roma, Günther Blobel, and Aurelian Radu for helpful discussions and for reading this manuscript. This work was supported in part by the New York City Council Speaker's Fund for Biomedical Research Toward the Science of Patient Care and a Leukemia Research Foundation New Investigator Award (to C.M.H.).
Abbreviations
- IFN
interferon
- STAT
signal transducer and activator of transcription
- IRF
interferon regulatory factor
- ISRE
IFN-stimulated response element
- ISG
IFN-stimulated gene
- NLS
nuclear localization signal
- NES
nuclear export signal
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 2.Escalante C R, Yie J, Thanos D, Aggarwal A K. Nature (London) 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 3.Veals S A, Schindler C, Leonard D, Fu X-Y, Aebersold R, Darnell J E, Jr, Levy D E. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horvath C M, Stark G R, Kerr I M, Darnell J E., Jr Mol Cell Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Moczygemba M, Gutch M J, French D L, Reich N C. J Biol Chem. 1997;272:20070–20076. doi: 10.1074/jbc.272.32.20070. [DOI] [PubMed] [Google Scholar]
- 6.McKendry R, John J, Flavell D, Muller M, Kerr I M, Stark G R. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.John J, McKendry R, Pellegrini S, Flavell D, Kerr I M, Stark G R. Mol Cell Biol. 1991;11:4189–4195. doi: 10.1128/mcb.11.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy D E, Kessler D S, Pine R I, Darnell J E., Jr Genes Dev. 1989;3:1362–1372. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 9.Levy D E, Lew D J, Decker T, Kessler D S, Darnell J E., Jr EMBO J. 1990;9:1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan R S, Takasugi T, Matsuyama T, et al. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 11.Bluyssen H A R, Muzaffar R, Vlieststra R J, Made A C J V d, Leung S, Stark G R, Kerr I M, Trapman J, Levy D E. Proc Natl Acad Sci USA. 1995;92:5645–5649. doi: 10.1073/pnas.92.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majumder S, Zhou L Z, Chaturvedi P, Babcock G, Aras S, Ransohoff R M. J Immunol. 1998;161:4736–4744. [PubMed] [Google Scholar]
- 13.Schaper F, Kirchhoff S, Posern G, Koster M, Oumard A, Sharf R, Levi B Z, Hauser H. Biochem J. 1998;335:147–157. doi: 10.1042/bj3350147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Nagineni C N, Ge H, Efiok B, Chepelinsky A B, Egwuagu C E. J Biol Chem. 1999;274:9686–9691. doi: 10.1074/jbc.274.14.9686. [DOI] [PubMed] [Google Scholar]
- 16.Marecki S, Atchison M L, Fenton M J. J Immunol. 1999;163:2713–2722. [PubMed] [Google Scholar]
- 17.Li X, Leung S, Kerr I M, Stark G R. Mol Cell Biol. 1997;17:2048–2056. doi: 10.1128/mcb.17.4.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kessler D S, Veals S A, Fu X-Y, Levy D E. Genes Dev. 1990;4:1753–1765. doi: 10.1101/gad.4.10.1753. [DOI] [PubMed] [Google Scholar]
- 19.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 20.Horvath C M, Wen Z, Darnell J E., Jr Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 21.Heim M H, Kerr I M, Stark G R, Darnell J E., Jr Science. 1995;267:1347–1349. doi: 10.1126/science.7871432. [DOI] [PubMed] [Google Scholar]
- 22.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 23.Veals S, Maria T S, Levy D E. Mol Cell Biol. 1993;13:196–206. doi: 10.1128/mcb.13.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung S, Qureshi S A, Kerr I M, Darnell J E, Jr, Stark G R. Mol Cell Biol. 1995;15:1312–1317. doi: 10.1128/mcb.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 26.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 27.Weaver B K, Kumar K P, Reich N C. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baeuerle P A, Baltimore D. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 29.Eisenbeis C F, Singh H, Storb U. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 30.Mamane Y, Sharma S, Petropoulos L, Lin R, Hiscott J. Immunity. 2000;12:129–140. doi: 10.1016/s1074-7613(00)80166-1. [DOI] [PubMed] [Google Scholar]