Abstract
The actinomycete Rhodococcus equi is an important pathogen of horses and an emerging opportunistic pathogen of humans. Identification of R. equi by classical bacteriological techniques is sometimes difficult, and misclassification of an isolate is not uncommon. We report here on a specific PCR assay for the rapid and reliable identification of R. equi. It is based on the amplification of a fragment of the choE gene encoding cholesterol oxidase. The choE-based PCR was assessed by using a panel of strains comprising 132 isolates from different sources and of different geographical origins, all initially identified biochemically as R. equi, and 30 isolates of representative non-R. equi actinomycete species, including cholesterol oxidase producers. The expected 959-bp amplicon was observed only with R. equi isolates, as confirmed by sequencing of a variable region of the 16S RNA gene from a random sample of 20 PCR-positive isolates. All R. equi isolates gave a positive choE-based PCR result, which correlated with a high degree of conservation of the choE gene. Three of the 132 strains originally identified as R. equi were negative for the choE gene, and subsequent analysis of their 16S RNA gene sequences confirmed that they belonged to other bacterial species (Dietzia maris, Mycobacterium peregrinum, and Staphylococcus epidermidis). All non-R. equi isolates were negative by the choE-based PCR. ATCC 21387, the only known isolate of Brevibacterium sterolicum, gave a 959-bp amplicon whose DNA sequence was virtually identical to that of R. equi choE. Comparison of the 16S RNA genes indicated that ATCC 21387 should be considered an R. equi isolate.
The nocardioform actinomycete Rhodococcus equi is a primary pathogen of horses. In foals, R. equi causes severe pyogranulomatous pneumonia, often accompanied by ulcerative enteritis and mesenteric lymphadenitis (29). In recent years, R. equi has emerged as an important opportunistic pathogen in humans, causing potentially life-threatening infections in severely immunocompromised people, in particular, human immunodeficiency virus-infected patients (43). In humans, R. equi causes a lung disease reminiscent of pulmonary tuberculosis. R. equi can also infect cattle, in which it has been associated with ulcerative lymphangitis (34), and it has also been recovered from inflamed tonsils and the cervical lymph nodes of pigs (21, 34). The natural habitat of R. equi is the soil, especially that enriched with fecal material from domestic and wild animals (37).
R. equi infections are diagnosed by culturing and subsequent phenotypic analysis of the isolates by means of classical morphological and biochemical tests (9). However, the colony characteristics, cellular morphology, and reaction to acid-fast staining differ between R. equi isolates (32). Although the API Coryne multisubstrate identification system (bio-Merieux), a commercial kit widely used in clinical microbiology laboratories, includes R. equi in its database, its reliability for the biochemical identification of rhodococcal isolates is limited (8, 35). These inconsistent test results for R. equi frequently result in misidentification, in which R. equi is mistaken as other rhodococcal species or even corynebacteria or other actinomycetes (10, 16, 36). Accurate identification of Rhodococcus isolates to the species level is possible on the basis of chemotaxonomic properties (11). However, these techniques are excessively laborious, time-consuming, and expensive for routine use in clinical microbiology laboratories.
We recently identified the R. equi choE gene, a chromosomal locus encoding cholesterol oxidase (22), an enzyme believed to be a major virulence factor of R. equi (13). Mutational analysis indicated that ChoE is the membrane-damaging factor responsible for the typically shovel-shaped synergistic hemolysis (CAMP-like) reaction elicited by R. equi in the presence of sphingomyelinase C-producing bacteria, such as Listeria ivanovii, Bacillus cereus, and Staphylococcus aureus (22). This CAMP-like reaction can be used as a phenotypic marker for the rapid presumptive identification of R. equi (30; unpublished data). We describe here a new PCR method for the rapid and specific identification of R. equi based on the detection of choE sequences. This assay accurately differentiated R. equi from other closely related actinomycetes and correctly reassigned strains initially incorrectly identified as R. equi to other species on the basis of morphological and biochemical characteristics. It also identified as R. equi a well-known cholesterol oxidase-producing strain, Brevibacterium sterolicum ATCC 21387 (7).
MATERIALS AND METHODS
Bacterial strains and microbiological procedures.
A total of 132 isolates from different geographical areas (Spain, France, Germany, the Dominican Republic, and Australia), identified at the source as R. equi, were included in the study. The 132 strains comprised 34 strains from pneumonic foals, 49 strains from human AIDS patients, and 49 strains from nonclinical samples (46 strains from soil and 3 strains from foal feces). Three R. equi reference strains, strains 103 (5), ATCC 6939 (the type strain of R. equi, also known as the Magnusson strain [18]), and ATCC 33701 (28), as well as 30 isolates of representative actinomycetes, were included as controls (Table 1). The bacteria were routinely cultured at 37°C for 48 h by using brain heart infusion as the base medium. Viable counts were determined by plating 10-fold serial dilutions of the initial bacterial suspension. CAMP-like synergistic hemolysis tests were performed on sheep blood agar plates with Columbia base medium (bioMérieux) and L. ivanovii ATCC 19119 as the indicator strain, as described previously (22, 30).
TABLE 1.
Non-R. equi strains used in the study
| Species | Strain |
|---|---|
| Corynebacterium sp. | ATCCa 14665 |
| Corynebacterium kutscheri | ATCC 15677 |
| Corynebacterium variabilis | ATCC 33010 |
| Corynebacterium xerosis | NCIMBb 9956 |
| Corynebacterium pseudotuberculosis | ATCC 19410 |
| Corynebacterium bovis | CAc 23 |
| Corynebacterium aquaticum | CA 29 |
| Corynebacterium urealyticum | ATCC 43042 |
| Mycobacterium sp. | NCIMB 11678 |
| Mycobacterium phlei | ATCC 11758 |
| Mycobacterium smegmatis | ATCC 14468 |
| Mycobacterium bovis BCG | ATCC 35744 |
| Mycobacterium xenopi | CA 78931 |
| Mycobacterium kansasii | CA 70243 |
| Mycobacterium tuberculosis | CA 1908 |
| Mycobacterium terrae | CA 52828 |
| Mycobacterium chelonae | CA 71765 |
| Mycobacterium lentiflavum | CA 33805 |
| Brevibacterium sp. | CA 157 |
| Brevibacterium sterolicum | ATCC 21387 |
| Nocardia sp. | CA 158 |
| Nocardia asteroides | ATCC 14759 |
| Rhodococcus sp. | NCIMB 9457 |
| Rhodococcus erythropolis | ATCC 11048 |
| Gordonia sp. | CA 93049 |
| Gordonia bronchialis | ATCC 25592 |
| Tsukamurella pulmonis | CIPd 104791T |
| Dietzia maris | ATCC 35013T |
| Turicella otitidis | CA 62716 |
| Streptomyces sp. | CA 163 |
ATCC, American Type Culture Collection.
NCIMB, National Collections of Industrial and Marine Bacteria.
CA, Collection of Actinomycetes of the Department of Biochemistry and Molecular Biology of the University of Cantabria. All strains were originally isolated from animal samples in the Animal Health Regional Laboratory, or from human patients in the Marqués de Valdecilla Hospital, Cantabria, Spain. They were identified by phenotypic analysis and 16S rDNA sequencing.
CIP, Collection of Institut Pasteur, Paris, France.
General DNA procedures.
Bacterial genomic DNA was routinely prepared by suspending one colony in 50 μl of distilled water; the same volume of Instagene matrix (Bio-Rad) was added, and the mixture was heated at 80°C for 10 min. The samples were centrifuged at 10,000 × g for 5 min, and the supernatants were used for PCR amplification. Bacterial dilutions used in PCR sensitivity tests were processed in the same way. Highly purified, concentrated genomic DNA for use in sensitivity tests was obtained as described previously (22). DNA concentrations were determined both by agarose gel electrophoresis and spectrophotometrically. Both strands of the PCR products were sequenced with an Applied Biosystems 377 apparatus. The sequences were analyzed by using the BLAST (1) network service from the National Center for Biotechnology Information (Bethesda, Md.) and were aligned by use of the AlignX program (InforMax, Bethesda, Md.). For restriction fragment length polymorphism (RFLP) analysis, the choE amplicons were purified by use of the Gel Extraction Purification kit (Qiagen) and were subsequently digested with the following restriction enzymes: AvaI, BamHI, BglI, HinPI, PvuI, XhoI, and XmnI (New England Biolabs, Beverly, Mass.). The digestion products were resolved by electrophoresis on 1.2% agarose gels in Tris borate buffer and were visualized by ethidium bromide staining.
Primers and PCR conditions.
The oligonucleotide primers were synthesized by TIB MOLBIOL (Berlin, Germany). The reaction mixture for PCR amplification consisted of 10 μl of DNA template, 4 U of Taq DNA polymerase (Bioline, London, United Kingdom), 1.5 mM MgCl2, 10 μl of 10× PCR amplification buffer [160 mM (NH4)2SO4, 670 mM Tris-HCl (pH 8.8), 0.1% Tween 20], 40 pmol of each primer, 0.2 mM each deoxynucleoside triphosphate (Bioline), and double-distilled water to a final volume of 100 μl. The DNA was first denatured at 95°C for 5 min and was then subjected to 30 amplification cycles under the following conditions: denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. After the final cycle, the reactions were terminated by an extra run at 72°C for 10 min, and then the mixtures were kept at 4°C until they were analyzed.
All strains tested in this study were amplified at least three times.
Nucleotide sequence accession numbers.
The 16S rRNA gene (rDNA) sequences from strains 20 and 21 are registered in the GenBank database under accession numbers AF537361 and AF537362, respectively.
RESULTS
R. equi-specific PCR based on choE.
Oligonucleotide primers were designed after alignment of the sequence of choE from R. equi strain 103, recently determined in our laboratory (22), with other known or putative cholesterol oxidase gene sequences available in databases (Mycobacterium tuberculosis, GenBank accession no. X99343; Mycobacterium leprae, GenBank accession no. NC002677; Streptomyces coelicolor, GenBank accession no. AL161755; Streptomyces sp. choA, GenBank accession no. M31939; Streptomyces sp. choM, GenBank accession no. U13981; Pimelobacter simplex, GenBank accession no. AF247810; and Burkholderia cepacia, GenBank accession no. AB051407) to identify areas of similarity and difference. On the basis of that analysis we defined the 21-mer R. equi-specific primers COX-F (5′-GTCAACAACATCGACCAGGCG), corresponding to positions 1221 to 1241 (forward primer), and COX-R (5′-CGAGCCGTCCACGACGTACAG), complementary to the sequence spanning positions 2160 to 2180 (reverse primer) (coordinates are according to the sequence of the choE region deposited in the EMBL data bank under accession no. AJ242746). These primers are predicted to give an amplicon of 959 bp, according to the sequence of R. equi strain 103.
Genomic DNA from R. equi reference strains 103, ATCC 6939, and ATCC 33701 and from 132 isolates from different sources and of different geographical origins presumptively identified as R. equi were screened by PCR with the COX primers described above. For the 3 reference strains and for 129 of the presumptive R. equi isolates, the PCR resulted in the production of the expected 0.95-kbp product (Fig. 1). A region of the 16S rDNA comprising species-specific sequences was amplified by PCR with primers DG74 and PL06 (12) from a random sample of 20 of the choE-based PCR (choE PCR)-positive isolates and sequenced. These 20 strains included both human and animal clinical isolates and environmental isolates. Their sequences were compared to the 16S rRNA sequences in databases, which confirmed that all the isolates belonged to R. equi. Only 3 of the 132 isolates (isolates 11, 20, and 21) gave a negative choE PCR result. The complete sequence of the 16S rDNA of each of these negative strains was determined after PCR amplification with primers 16S-F (5′-AGAGTTTGATCCTGGCTCAG) and 16S-R (5′-AGGAGGTGATCCAGCCGC). Strain 11 was identified as S. epidermidis (99% identity with S. epidermidis ATCC 14990T 16S rDNA), strain 20 was identified as D. maris (99% identity with D. maris ATCC 35013T), and strain 21 was identified as M. peregrinum (98% identity with M. peregrinum ATCC 14467).
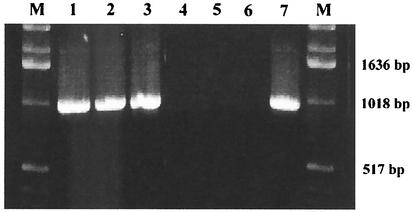
FIG. 1.
Representative choE PCR amplification products obtained from genomic DNA samples of R. equi. Lanes: 1, strain 103; 2, ATCC 6939; 3, isolate 70; 4, isolate 11; 5, isolate 20; 6, isolate 21; 7, B. sterolicum ATCC 21387; M, DNA size marker.
The specificity of the choE PCR was verified by testing 30 strains from representative actinomycete species (Table 1). These strains included bacteria known to express cholesterol oxidase activity or to encode putative cholesterol oxidase enzymes, such as Brevibacterium spp., Mycobacterium spp., Nocardia asteroides, Rhodococcus erythropolis, and Streptomyces spp. (17). No amplification product was detected for any of these strains except one, Brevibacterium sterolicum ATCC 21387, for which a 0.95-kbp product, i.e., a product of the same size as that obtained with the R. equi isolates, was observed (Fig. 1). A further detailed analysis of the B. sterolicum strain indicated that it was missclassified and belonged to the R. equi taxon (see below).
In vitro sensitivity of choE PCR for R. equi.
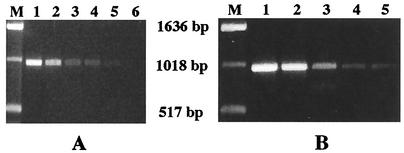
The in vitro sensitivity of the PCR test was assessed by determining both the minimum amount of DNA and the minimum number of bacterial cells required for detection of a 959-bp choE amplicon. For these tests, 10-fold dilutions of highly purified DNA or an exponential-phase culture (optical density at 600 nm = 0.5) of R. equi reference strains ATCC 6939 and ATCC 33701 were used. A positive result was obtained with a minimum of 0.02 ng of DNA or 3 R. equi CFU (Fig. 2).
FIG. 2.
Sensitivity of the choE PCR. (A) choE amplicon obtained with various amounts of purified R. equi ATCC 6939 genomic DNA. Lane M, DNA size marker; lanes 1 to 6, serial 10-fold dilutions of the DNA preparation (lane 1, 200 ng; lane 6, 2 pg). (B) choE amplicons obtained with various quantities of R. equi ATCC 6939 bacterial cells. Lane M, DNA size marker; lanes 1 to 5, 3 × 104, 3 × 103, 3 × 102, 30, and 3 CFU, respectively.
Conservation of choE sequences in R. equi.
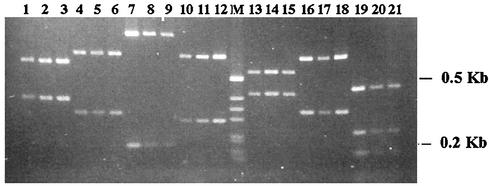
The choE gene of strain 103 maps to a 2.3-kbp PstI chromosomal fragment (22). Southern blot analyses revealed this same hybridization pattern in type strain ATCC 6939 and in the 20 choE PCR-positive isolates used for the 16S rDNA cataloguing described above (data not shown), suggesting a high degree of conservation of the choE sequence in R. equi. We confirmed this by DNA sequencing and RFLP analysis. The complete sequences of the choE genes from 3 of the 20 R. equi strains, selected on the basis of their different degrees of cholesterol oxidase and cognate CAMP-like activities (strain 2, isolated from a diseased foal, had a high level of activity; strain 3, from a human patient, had an intermediate level of activity; and strain 70, also from a human patient, was exceptional, in that it presented no detectable activity), were determined. These three choE sequences diverged only minimally (sequence identities, >99.5%). RFLP analyses of the 959-bp choE amplicons from the 20 R. equi isolates previously analyzed by Southern blotting also revealed identical patterns for each of the seven restriction enzymes used (Fig. 3).
FIG. 3.
RFLP patterns of choE amplicons from three R. equi isolates (strains 15, 65, and 115, of equine, human, and environmental origin, respectively, in that order for the digestions with each of the enzymes) digested with PvuI (lanes 1 to 3), XhoI (lanes 4 to 6), XmnI (lanes 7 to 9), AvaI (lanes 10 to 12), BamHI (lanes 13 to 15), BglI (lanes 16 to 18), and HinPI (lanes 19 to 21). Lane M, DNA size marker.
Reclassification of B. sterolicum ATCC 21387 as R. equi.
Comparison of the cholesterol oxidase gene sequences of R. equi 103 (choE) and B. sterolicum ATCC 21387 (choB) (22) revealed they were almost identical, consistent with the positive result obtained with B. sterolicum ATCC 21387 by PCR with the COX primers (see above). This high degree of sequence conservation between choE and choB derived from two presumptively different bacterial species was surprising in view of the substantial degree of divergence (81%) that exists between cholesterol oxidase genes from different actinomycetes, even for those belonging to the same genus (as, for example, the case of Streptomyces sp. choA and choM alleles, which are only 86% identical). Given the strong sequence conservation of choE among R. equi isolates, the identity between choE and the reported choB sequence could be explained either (i) by a recent horizontal gene transfer event between R. equi and B. sterolicum or (ii) by the fact that ATCC 21387 is an R. equi isolate. To discriminate between these two possibilities, we determined the complete sequence of the 16S rDNA of ATCC 21387 after PCR amplification with primers 16S-F and 16S-R (see above). The sequence was 100% identical to that of R. equi type strain ATCC 6939 (GenBank accession no. X80603), indicating that B. sterolicum ATCC 21387 is in fact an R. equi strain. ATCC 21387 gives a CAMP-like reaction with L. ivanovii. Furthermore, the biochemical profiles of ATCC 21387 determined with the API Coryne system are fully compatible with those of R. equi and less similar to those of Brevibacterium.
DISCUSSION
In this work, we studied the usefulness of the R. equi cholesterol oxidase gene, choE, recently identified and characterized in our laboratory (22), as a target for the development of a species-specific PCR method for the rapid identification of this pathogenic nocardioform actinomycete. Routine application of a PCR-based method requires that the target sequence be highly specific for the microorganism and that it be highly conserved in all strains of that organism. Our results show that the choE target sequence used fulfills these requirements. Sequences complementary to the COX primers were present in all of the isolates included in a large collection of R. equi strains from different sources and of different geographical origins, suggesting that they are universally conserved in this bacterial species. This is consistent with the production of extracellular choE-derived cholesterol oxidase activity and of its associated phenotypic marker, the CAMP-like reaction with L. ivanovii, by virtually all isolates of R. equi (unpublished data). On the other hand, no choE amplicon was detected in any of the 29 actinomycetes used as negative controls, which included relevant pathogenic species, such as N. asteroides and M. tuberculosis, that also produce cholesterol oxidase activity or that carry choE-related genes (17). Most importantly, these negative control strains included cholesterol oxidase-producing rhodococci, demonstrating the species specificity of our PCR method. The choE gene homologs carried by these bacteria were sufficiently divergent to prevent positive amplification with our COX primers. The specificities of these primers for R. equi were corroborated by sequence analysis of the 16S rDNA from a representative sample of the choE PCR-positive strains.
Interestingly, one R. equi isolate from the panel of isolates tested did not produce detectable CAMP-like activity but gave a positive choE PCR result. This illustrates the validity of the molecular method that we developed as a tool to identify R. equi, as it gives positive results even for rhodococcal strains that do not express the choE-associated phenotypic marker. The cholesterol oxidase-nonproducing R. equi isolate, strain 70, acquired the capacity to produce an active enzyme (and CAMP-like reactivity) upon complementation with a plasmid carrying a wild-type choE gene, indicating that that strain bears a nonfunctional choE allele. Analysis of the strain 70 choE sequence revealed the presence of a 1-bp insertion in the 5′ region of the gene that produced a frame shift and, subsequently, the loss of cholesterol oxidase activity (unpublished results).
The choE PCR with the COX primers also identified three isolates that had incorrectly been identified as R. equi according to the negative PCR results, thus further illustrating the value of the assay. These three isolates were confirmed to be non-R. equi isolates by 16S rDNA sequencing. One, an isolate from the bone marrow of a human patient with human immunodeficiency virus infection, was identified as D. maris, a halophilic actinomycete previously reported as being involved in human infections only on two occasions (4, 27). The second isolate was identified as M. peregrinum, a species also rarely found as a cause of opportunistic infections in humans (25, 31). The third was a strain of S. epidermidis, a coagulase-negative Staphylococcus sp., which was possibly identified as R. equi on the basis of positivity by Gram staining, the coccoid aspect often presented by R. equi, and the resemblance of old R. equi colonies to those of staphylococci.
B. sterolicum ATCC 21387 was the only presumptive non-R. equi strain included in our study which yielded a 0.95-kb PCR product with the COX primers. ATCC 21387 is a well-known strain used for the industrial production of cholesterol oxidase and the extensive genetic and biochemical characterization of this enzyme (20, 23, 24). Indeed, the three-dimensional crystal structure of the cholesterol oxidase protein was determined by using the purified enzyme from B. sterolicum ATCC 21387 (15). Our previous data showed that the sequence of the cholesterol oxidase gene from B. sterolicum ATCC 21387, designated choB, was almost identical to that of choE from R. equi (22). Here we found that the 16S rDNA sequence of ATCC 21387 is also identical to that of R. equi. This finding, together with the strong conservation of the choE gene sequence among R. equi isolates, indicates that ATCC 21387, the only known strain of the species “B. sterolicum,” was misclassified and is R. equi.
Only a few rapid molecular methods have been developed for R. equi identification. Clinical isolates from foals can be identified by detection of the VapA antigen, with monoclonal antibodies, or alternatively, by PCR detection of its gene, vapA, which is present on an 85-kb virulence plasmid (38, 39, 40). However, the virulence plasmid is not present in all strains of human and environmental origin (in general, in nonequine isolates) (41), thus limiting the general usefulness of the vapA-based identification of R. equi by PCR. Other PCR-based molecular methods have been used to amplify a chromosomal segment of unknown function (2) or the 16S rDNA (3, 33), but their validities have been assessed with only a small number of strains. Although 16S rDNA sequencing is accepted as a general means for species differentiation, some heterogeneity can exist between different isolates of the same species (14). A recent study of the 16S rDNA sequences of several representative strains showed that R. equi is a very heterogeneous taxon, with variations in 16S rDNAs of up to 4% (19). On the other hand, closely related species may have identical or almost identical 16S rDNA sequences (6, 26). Therefore, it is useful to have other species-specific targets, such as choE, to undertake an assay for the reliable identification of a bacterial species. Finally, a PCR-RFLP method targeting a 65-kDa heat shock protein gene and primarily devised for the identification of mycobacteria was shown to discriminate R. equi strains (42). However, this assay is too laborious and lengthy because, due to the conserved nature of the 65-kDa heat shock protein gene, the amplification product is of the same size for all actinomycete species and R. equi can be discriminated only by restriction analysis of the amplicon.
In summary, we describe a new PCR assay which can be usefully applied for the rapid, sensitive, and reliable identification of R. equi isolates and their differentiation from isolates of other pathogenic and nonpathogenic actinomycetes.
Acknowledgments
We gratefully acknowledge all the colleagues who kindly provided us with bacterial strains for this study (T. Chakraborty, M. de Pablos, A. Enríquez, V. García, J. M. García Arenzana, J. L. Hernández, A. Kodjo, C. Lammler, M. Lantero, P. Martín Rabadán, A. M. Martín Sánchez, A. Morton, and J. F. Prescott). We also thank C. Polidura and Z. Madrazo for their contribution in identifying some of the bacterial strains and I. Andrés for helpful discussions.
This work was supported by grants from the Spanish Ministry for Science and Technology (grant PB97-0327-C03) and Fundación Marqués de Valdecilla (grant A/30/01).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arriaga, J. M., N. D. Cohen, J. N. Derr, M. K. Chaffin, and R. J. Martens. 2002. Detection of Rhodococcus equi by polymerase chain reaction using species-specific nonproprietary primers. J. Vet. Diagn. Investig. 14:347-353. [DOI] [PubMed] [Google Scholar]
- 3.Bell, K. S., J. C. Philp, N. Christofi, and D. W. Aw. 1996. Identification of Rhodococcus equi using the polymerase chain reaction. Lett. Appl. Microbiol. 23:72-74. [DOI] [PubMed] [Google Scholar]
- 4.Bemer-Melchior, P., A. Haloun, P. Riegel, and H. B. Drugeon. 1999. Bacteremia due to Dietzia maris in an immunocompromised patient. Clin. Infect. Dis. 29:1338-1340. [DOI] [PubMed] [Google Scholar]
- 5.De La Peña-Moctezuma, A., J. F. Prescott, and M. Goodfellow. 1996. Attemps to find phenotypic markers of the virulence plasmid of Rhodococcus equi. Can. J. Vet. Res. 60:29-33. [PMC free article] [PubMed] [Google Scholar]
- 6.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 7.Fujishiro, K., T. Ohta, M. Hasegawa, K. Yamaguchi, T. Mizukami, T. Uwajima, and T. Ohta. 1990. Isolation and identification of the gene of cholesterol oxidase from Brevibacterium sterolicum ATCC 21387, a widely used enzyme in clinical analysis. Biochem. Biophys. Res. Commun. 172:721-727. [DOI] [PubMed] [Google Scholar]
- 8.Funke, G., F. N. Renaud, J. Freney, and P. Riegel. 1997. Multicenter evaluation of the updated and extended API (RAPID) Coryne database 2.0. J. Clin. Microbiol. 35:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giguere, S., and J. F. Prescott. 1997. Clinical manifestations, diagnosis, treatment, and prevention of Rhodococcus equi infections in foals. Vet. Microbiol. 56:313-334. [DOI] [PubMed] [Google Scholar]
- 10.Gómez, C., A. Martínez, A. Cano, A. Altuna, A. Lafuente, J. M. Artero, J. M. Prieto, and F. Martin Luengo. 1994. Neumonía cavitada de evolución torpida en paciente con infección por VIH. Enf. Infecc. Microbiol. Clin. 12:39-40. [PubMed] [Google Scholar]
- 11.Goodfellow, M. 1986. Genus Rhodococcus, p. 2362-2371. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 4. The William & Wilkins Co., Baltimore, Md.
- 12.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hondalus, M. 1997. Pathogenesis and virulence of Rhodococcus equi. Vet. Microbiol. 56:257-268. [DOI] [PubMed] [Google Scholar]
- 14.Kirschner, P., and E. Bottger. 1992. Microheterogeneity within rRNA of Mycobacterium gordonae. J. Clin. Microbiol. 30:1049-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, J., A. Vrielink, P. Brick, and D. M. Blow. 1993. Crystal structure of cholesterol oxidase complexed with a steroid substrate: implications for flavin adenine dinucleotide dependent alcohol oxidases. Biochemistry 32:11507-11515. [PubMed] [Google Scholar]
- 16.Macias, J., J. A. Pineda., F. Borderas, J. A. Gallardo, J. Delgado, M. Leal, A. Sánchez-Quijano, and E. Lissen. 1996. Rhodococcus or Mycobacterium? An example of misdiagnosis in HIV infection. AIDS 11:253-254. [PubMed] [Google Scholar]
- 17.MacLachlan, J., A. T. Wotherspoon, R. O. Ansell, and C. J. Brooks. 2000. Cholesterol oxidase: sources, physical properties and analytical applications. J. Steroid Biochem. Mol. Biol. 72:169-195. [DOI] [PubMed] [Google Scholar]
- 18.Magnusson, H. 1923. Spezische Infektiese Pneumonie beim Fohlen. Ein neuer Eitererreger beim Pferde. Arch. Wiss. Prakt. Tierheilkd. 50:22-38. [Google Scholar]
- 19.McMinn, E. J., G. Alderson, H. I. Dodson, M. Goodfellow, and A. C. Ward. 2000. Genomic and phenomic differentiation of Rhodococcus equi and related strains. Antonie Leeuwenhoek 78:331-340. [DOI] [PubMed] [Google Scholar]
- 20.Motteran, L., M. S. Pilone, G. Molla, S. Ghisla, and L. Pollegioni. 2001. Cholesterol oxidase from Brevibacterium sterolicum. The relationship berween covalent flavinylation and redox properties. J. Biol. Chem. 276:18024-18030. [DOI] [PubMed] [Google Scholar]
- 21.Mutimer, M. D., and J. B. Woolcock. 1980. Corynebacterium equi in cattle and pigs. Tijdschr. Diergeneeskd. 105:25-27. [PubMed] [Google Scholar]
- 22.Navas, J., B. Gonzalez-Zorn, N. Ladron, P. Garrido, and J. A. Vazquez-Boland. 2001. Identification and mutagenesis by allelic exchange of choE, encoding a cholesterol oxidase from the intracellular pathogen Rhodococcus equi. J. Bacteriol. 183:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta, T., K. Fujishiro, K. Yamaguchi, Y. Tamura, K. Aisaka, T. Uwajima, and M. Hasegawa. 1991. Sequence of gene choB encoding cholesterol oxidase of Brevibacterium sterolicum: comparison with choA of Streptomyces sp. SA-COO. Gene 15:93-96. [DOI] [PubMed] [Google Scholar]
- 24.Ohta, T., K. Fujishiro, K. Yamaguchi, T. Uwajima, K. Aisaka, and M. Hasegawa. 1992. Hyperexpression and analysis of choB encoding cholesterol oxidase of Brevibacterium sterolicum in Escherichia coli and Streptomyces lividans. Biosci. Biotechnol. Biochem. 56:1786-1791. [DOI] [PubMed] [Google Scholar]
- 25.Pagnoux, C., X. Nassif, C. Boitard, and J. Timsit. 1998. Infection of continuous subcutaneous insulin infusion site with Mycobacterium peregrinum. Diabetes Care 21:191-192. [DOI] [PubMed] [Google Scholar]
- 26.Patel, R., K. E. Piper, M. S. Rouse, J. M. Steckelberg, J. R. Uhl, P. Kohner, M. K. Hopkins, F. R. Cockerill, and B. C. Kline. 1998. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. J. Clin. Microbiol. 36:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pidoux, O., J. N. Argenson, V. Jacomo, and M. Drancourt. 2001. Molecular identification of a Dietzia maris hip prosthesis infection isolate. J. Clin. Microbiol. 39:2634-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prescott, J. F. 1981. Capsular serotypes of Corynebacterium equi. Can. J. Comp. Med. 45:130-134. [PMC free article] [PubMed] [Google Scholar]
- 29.Prescott, J. F. 1991. Rhodococcus equi: an animal and human pathogen. Clin. Microbiol. Rev. 4:20-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ripio, M. T., C. Geoffroy, G. Domínguez, J. E. Alouf, and J. A. Vazquez-Boland. 1995. The sulphydril-activated cytolysin and a sphingomyelinase C are the major membrane-damaging factors involved in cooperative (CAMP-like) haemolysis of Listeria spp. Res. Microbiol. 146:303-313. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Gancedo, M. B., T. Rodríguez-González, G. Yague, P. I. Valero Guillen, and M. Segovia-Hernández. 2001. Mycobacterium peregrinum bacteremia in an immunocompromised patient with a Hickman catheter. Eur. J. Clin. Microbiol. Infect. Dis. 20:589-590. [DOI] [PubMed] [Google Scholar]
- 32.Scott, M. A., B. S. Graham, R. Verrall, R. Dixon, W. Schaffner, and K. T. Tham. 1995. Rhodococcus equi, an increasingly recognized opportunistic pathogen. Report of 12 cases and review of 65 cases in the literature. Am. J. Clin. Pathol. 103:649-655. [DOI] [PubMed] [Google Scholar]
- 33.Sellon, D. C., K. Walker, M. Suyemoto, and C. Altier. 1997. Nucleic acid amplification for rapid detection of Rhodococcus equi in equine blood and tracheal wash fluids. Am. J. Vet. Res. 58:1232-1237. [PubMed] [Google Scholar]
- 34.Soedarmanto, I., R. Oliveira, C. Lammler, and H. Durrling. 1997. Identification and epidemiological relationship of Rhodococcus equi isolated from cases of lymphadenitis in cattle. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 286:457-467. [DOI] [PubMed] [Google Scholar]
- 35.Soto, A., J. Zapardiel, and F. Soriano. 1994. Evaluation of API Coryne system for identifying coryneform bacteria. J. Clin. Pathol. 47:756-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutor, G. C., C. Fibich, P. Kirscher, M. Kuske, R. E. Schmidt, I. Schedel, and H. Deicher. 1996. Poststenotic cavitating pneumonia due to Rhodococcus equi in HIV infection. AIDS 10:339-351. [DOI] [PubMed] [Google Scholar]
- 37.Takai, S., K. Narita, K. Ando, and S. Tsubaki. 1986. Ecology of Rhodococcus (Corynebacterium) equi in soil on a horse-breeding farm. Vet. Microbiol. 12:169-177. [DOI] [PubMed] [Google Scholar]
- 38.Takai, S., T. Morishita, Y. Nishio, Y. Sasaki, S. Tsubaki, T. Higuchi, S. Hagiwara, H. Senba, M. Kato, N. Seno, T. Anzai, and M. Kamada. 1994. Evaluation of a monoclonal antibody-based colony blot test for rapid identification of virulent Rhodococcus equi. J. Vet. Med. Sci. 56:681-684. [DOI] [PubMed] [Google Scholar]
- 39.Takai, S., T. Ikeda, Y. Sasaki, Y. Watanabe, T. Ozawa, S. Tsubaki, and T. Sekizaki. 1995. Identification of virulent Rhodococcus equi by amplification of gene coding for 15- to 17-kilodalton antigens. J. Clin. Microbiol. 33:1624-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takai, S., S. A. Hines, T. Sekizaki, V. M. Nicholson, D. A. Alperin, M. Osaki, D. Takamatsu, M. Nakamura, K. Suzuki, N. Ogino, T. Kakuda, H. Dan, and J. F. Prescott. 2000. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect. Immun. 68:6840-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takai, S. Y., Y. Imai, N. Fukunaga, Y. Uchida, K. Kamisawa, Y. Sasaki, S. Tsubaki, and T. Sekizaki. 1995. Identification of virulence-associated antigens and plasmids in Rhodococcus equi from patients with AIDS. J. Infect. Dis. 172:1306-1311. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, T. B., C. Patterson, Y. Hale, and W. W. Safranek. 1997. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J. Clin. Microbiol. 35:79-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinstock, D. M., and A. E. Brown. 2002. Rhodococcus equi: an emerging pathogen. Clin. Infect. Dis. 34:1379-1385. [DOI] [PubMed] [Google Scholar]