Abstract
We recently developed a new PCR-restriction fragment length polymorphism (RFLP)-based assay using the miniexon sequence from the genus Leishmania. Here we report the application of this new genotyping method to naturally infected clinical samples for the differentiation of New and Old World Leishmania species. Of the newly developed assay and four currently applied diagnostic tests (i.e., in vitro cultivation, serology, and two other molecular assays using either the small subunit-internal transcribed spacer sequence or a repetitive genomic sequence), the miniexon assay showed the highest sensitivity, 89.7%, compared to 70.6, 57.1, 51.7, and 79.3%, respectively. Species differentiation was robust and reliable compared with that by two other Leishmania genotyping techniques. The assay provides a valuable tool for the identification of Leishmania directly from clinical samples and enables determination of the infecting species by a facile technique with high discrimination power. Since Leishmania causes a broad spectrum of diseases distinguished by different parasite and host factors, detection and characterization of the infecting species is crucial for the confirmation of a diagnosis as well as the establishment of the clinical prognosis and the initiation of an adequate therapeutic approach. The miniexon PCR-RFLP assay will facilitate such determination and might improve diagnosis and treatment of leishmaniasis.
Diagnosis of leishmaniasis can be made on the basis of clinical and epidemiological data but has to be confirmed by the demonstration of the parasite to avoid potential misdiagnosis (29). Because of differences among the Leishmania species in levels of virulence and in responses to the various chemotherapeutic regimens, correct identification is essential in order to determine the clinical prognosis and prescribe an appropriate species-specific therapeutic regimen (3).
At present, the classical methods used for the direct detection of the parasite include the visualization of amastigotes by microscopic examination of Giemsa-stained smears and in vitro culture of the parasite. Though microscopic examination is rapid, cheap, and easy to perform, it lacks sensitivity due to the generally low number of parasites in tissue samples (42). Culture techniques are more sensitive but require a sophisticated laboratory setup, are time-consuming, and harbor the risk of contamination (3). Indirect methods based on the analysis of specific immune responses cannot distinguish between past and current infections, and depending on the antigens used in serological assays, diagnosis can be complicated by cross-reaction of antibodies with other pathogens. Another drawback is the limited sensitivity because antibody titers may vary with the infecting species, tissue tropism, and the immunocompetence of the host (1).
Species discrimination is important not only for epidemiological reasons but also for clinical reasons. Leishmania species were originally classified by their geographical distributions and the clinical presentations of the diseases they cause, as well as by epidemiology, sand fly vectors, or animal reservoirs (20). Since morphological discrimination of Leishmania species is not possible, a variety of biochemical, immunological, and molecular tools have been developed for the differentiation of the pathogenic species, such as isoenzyme, serodeme, and schizodeme analysis and hybridization techniques with species-specific DNA probes (16, 43).
With the advances in molecular techniques, a number of molecular markers and PCR protocols for the detection or identification of Leishmania on different taxonomical levels (genus, complex, and species) have been reported (11, 43). Target sequences for characterization include either nuclear DNA, such as the small subunit rRNA (SSU rRNA) gene (40), a repetitive genomic sequence (30), the miniexon (spliced leader) gene repeat (18), the beta-tubulin gene region (22), the gp63 gene locus (41), and internal transcribed spacer (ITS) regions (9); microsatellite DNA (36, 37); or kinetoplast DNA, such as minicircle sequences (34).
While all the above-mentioned approaches provide a multitude of valid taxonomic characters for differentiation, they are accompanied by a number of factors which limit their use in a routine diagnostic laboratory. Some of the assays are restricted to the identification of Leishmania at the genus level (5, 25) and do not allow species differentiation, whereas others are adapted for individual complex characterization (10, 13), which limits their applicability to restricted geographical areas. Other molecular approaches involve the combination of PCR amplification with DNA probe hybridization to enhance the sensitivity of the assay (33, 35), an elaborate and time-consuming procedure. The use of random amplified polymorphic DNA analysis of genomic DNA (12, 38) is limited since it requires purified parasite DNA and highly standardized PCR conditions to guarantee specificity. In addition, the results may be difficult to interpret. Assays based on the multiplex PCR technique (17) represent yet another approach but harbor the risk of reduced sensitivity caused by primer dimer formation.
The miniexon gene of kinetoplastid protozoa, which is involved in the transsplicing process of nuclear mRNA, is present as 100 to 200 tandemly repeated copies per nuclear genome. Each repeat consists of three major parts: a transcribed region comprising a highly conserved 39-nucleotide exon; a moderately conserved intron, approximately 55 to 101 bp; and a nontranscribed, intergenic spacer of variable length (51 to 1,350 bp) depending on the genus and species (39). These features allow the miniexon to be utilized as a genotyping marker. Based on these characteristics, we recently established a PCR assay similar to that described by Fernandes et al. (14). This PCR assay amplifies all the miniexon sequences in a single reaction using universal primers, allowing preliminary discrimination between the major complexes (i.e., Old World Leishmania, New World Leishmania, and New World Viannia complexes) as a result of the variability in sizes of the amplification products. After restriction enzyme digestion of the PCR product, a characteristic restriction fragment length polymorphism (RFLP) pattern is produced that depends on size variations in the polymorphic spacer regions as well as mutations in the recognition sites of a chosen restriction enzyme. The miniexon PCR-RFLP genotyping scheme was validated with cultured World Health Organization reference strains of Leishmania and cultured isolates from patients attending the polyclinic of the Swiss Tropical Institute (STI). It has proven to be a valuable tool showing high resolution, sensitivity, and an ability to discriminate between all clinically relevant Leishmania species (24).
The aim of this study was to optimize the miniexon PCR assay for routine diagnosis of infections in naturally infected clinical samples without the need for cultivation and to compare our results with those obtained by using current methods for diagnosis of leishmaniasis. Moreover, genotyping results of the miniexon RFLP system were validated by parallel analysis of the samples with two other PCR-RFLP typing schemes which are based on an uncharacterized repetitive genomic sequence of Old World Leishmania (30) or the ITS region of the rRNA gene (37a).
MATERIALS AND METHODS
Clinical samples.
A total of 50 samples from patients presumed to have leishmaniasis were used in this study. The samples were either sent to our diagnostic unit or obtained from patients returning from areas where the disease is endemic who attended the STI polyclinic with either cutaneous signs or a tentative diagnosis of visceral leishmaniasis. Three samples were from patients with a concurrent HIV infection. The samples consisted of 25 skin biopsy specimens, 16 bone marrow biopsy specimens, 2 lesion aspirates, 4 blood samples (anticoagulated with EDTA), 2 duodenal biopsy specimens, and 1 gastric biopsy specimen. Four of the 25 skin biopsy specimens arrived as paraffin-embedded tissue samples. Samples were split on arrival; one part was used for in vitro cultivation, except in the case of paraffin-embedded tissue samples, and the other part was processed for PCR analysis. Corresponding serum samples were available from 16 patients, and they were tested for Leishmania-specific antibodies by an indirect fluorescent-antibody test (IFAT). Of these, four samples were derived from patients who had traveled to the Americas, six samples were derived from patients for whom there was no information on travel history, and 19 were derived from patients with a history of travel in the Old World. In this study, a sample was considered positive when the results of at least one of the assays performed were positive. Assays were repeated independently to exclude contamination.
Specimen processing and DNA preparation.
Skin, duodenal, and gastric biopsy specimens arrived in 0.9% NaCl or another physiological transport medium and were first cut into smaller pieces with a sterile scalpel in a petri dish. They were then transferred with the medium into a 2-ml tube, pelleted by centrifugation, and finally resuspended in 500 μl of lysis buffer (10 mM Tris-HCl [pH 8.0], 5 mM EDTA, 0.5% sodium dodecyl sulfate).
Lesion aspirates were centrifuged at 8,000 × g for 10 min and resuspended in 500 μl of lysis buffer.
Anticoagulated bone marrow samples were centrifuged for 15 min at 800 × g. The buffy coat was transferred to a new tube, washed with phosphate-buffered saline (PBS), pH 7.4, and resuspended in 500 μl of lysis buffer. Coagulated clots of bone marrow samples were treated like skin biopsy samples as described above.
EDTA-anticoagulated blood was mixed with an equal volume of PBS, pH 7.4, layered over Ficoll-Paque PLUS (density, 1.077 g/liter; Amersham Biosciences Europe GmbH, Dübendorf, Switzerland), and spun for 30 min at 400 × g The interphase containing peripheral blood mononuclear cells was carefully transferred to a fresh tube, washed twice with PBS, pH 7.4, and resuspended in 500 μl of lysis buffer.
Small sections (not more than 25 mg) of paraffin-embedded tissue samples were scratched with a sterile scalpel into a 1.5-ml tube. Paraffin was removed by being washed once with 1.2 ml of xylene and two times with 1.2 ml of ethanol. Residual ethanol was evaporated by incubating the tube for 15 min in a heating block at 37°C. The pellet was resuspended in 180 μl of buffer ATL (QIAamp DNA Mini Kit; Qiagen AG, Basel, Switzerland), and DNA was extracted and purified by using the QIAamp DNA Mini Kit according to the QIAamp DNA Mini Tissue protocol.
Generation of the positive control template.
A 498-bp fragment of the cysteine protease cpb2 gene from Leishmania mexicana (accession no. AJ319727; nucleotides 1302 to 1800), designated as the positive internal control in the miniexon PCR assay (6) was amplified by PCR by using primers cpb2fw (5′-TGA TCA CCA CGA AGG AGT GC-3′) and cpb2rev (5′-ACG CGA GGG CAC GCG AGG-3′). Miniexon primer sites were introduced by subsequent nested PCR with the composite primers cpb2Mifw (5′-TAT TGG TAT GCG AAA CTT CCG TGA TCA CCA CGA AGG AGT GC-3′) and cpbMirev (5′-ACA GAA ACT GAT ACT TAT ATA GCG ACG CGA GGG CAC GCG AGG-3′). PCR mixtures (50 μl) and cycling conditions for all the reactions were the same as those for the miniexon PCR described below. PCR products were purified by size-selective polyethylene glycol precipitation and cloned by using the pGEM-T vector system (Promega, Madison, Wis.) according to the manufacturer's instructions. Plasmid DNA containing the positive control template was purified by using the CONCERT Rapid Plasmid Miniprep system (Invitrogen Life Technologies, Basel, Switzerland) according to the manufacturer's manual. Purified plasmid DNA was stored in Tris-EDTA, pH 8.0, containing 0.05% W-1 (Invitrogen Life Technologies).
PCR assays.
For repetitive sequence PCR (Leishmania genotyping [LEG] PCR) (28), primers T2 (5′-CGG CTT CGC ACC ATG CGG TG-3′) and B4 (5′-ACA TCC CTG CCC ACA TAC GC-3′) were used to amplify a repetitive DNA sequence from Old World Leishmania. Two microliters of DNA solution was amplified in a 100-μl reaction mixture containing 50 mM PCR buffer, 0.2 mM deoxynucleoside triphosphates, 1.5 mM MgCl2, 0.5 μM (each) primers, and 0.5 U of Taq DNA polymerase (Invitrogen Life Technologies). PCR amplification was carried out in a DNA Thermal Cycler 480 (Perkin Elmer, Boston, Mass.). After an initial denaturation step (5 min at 96°C), 40 cycles of 96°C for 30 s, 56°C for 45 s (annealing), and 72°C for 60 s (extension) were performed. For miniexon PCR, primers Fme (5′-TAT TGG TAT GCG AAA CTT CCG-3′) and Rme (5′-ACA GAA ACT GAT ACT TAT ATA GCG-3′) were used to amplify the nontranscribed spacer region of the miniexon sequence of Leishmania (24). Two microliters of DNA solution was amplified in a 50-μl reaction mixture containing 50 mM PCR buffer, 0.2 mM deoxynucleoside triphosphates, 1.5 mM MgCl2, 40 mM tetramethylammonium chloride, 12% dimethyl sulfoxide, 0.5 μM (each) primers, and 0.5 U of Taq DNA polymerase. The positive control template was added to a final concentration of 250 fg (corresponding to approximately 70,000 plasmid copies) per reaction mix. The PCR amplification was carried out in a Mastercycler gradient 5331 (Eppendorf, Hamburg, Federal Republic of Germany). After an initial denaturation (5 min at 94°C), 35 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 45 s were performed.
Genotyping.
Typing of the Leishmania species was achieved by RFLP analysis of the amplified miniexon gene. Ten microliters of the miniexon PCR products was digested with the restriction enzymes according to the genotyping scheme described elsewhere (24).
In order to confirm our genotyping results, DNA aliquots from all the samples found to be positive by PCR were analyzed by two other PCR-RFLP genotyping schemes based on the sequence differences in the genomic repeat of Old World Leishmania (30) and the ITS region of the SSU rRNA gene (37a).
Cultivation of parasites.
The culture medium, a 1:1 mixture of SDM-79 (7) and SM (8) complemented with 15% heat-inactivated fetal bovine serum, was supplemented with an anticontamination cocktail consisting of penicillin G (60 μg/ml), kanamycin (100 μg/ml), chloramphenicol (10 μg/ml), and flucytosine (50 μg/ml). The medium was directly inoculated with the patient specimens and incubated at 27°C without gas phase in T-25 tissue culture flasks (Falcon, BD Biosciences, Allschwil, Switzerland). Cultures were checked for the presence of promastigote forms of Leishmania by examination with an inverted microscope. Microscopic examination was done every 2 days for up to 15 days.
Serology.
Serum samples were tested for anti-Leishmania antigen antibodies by using an IFAT. Twelve-well slides were coated with imprint preparations of an L. donovani-infected hamster spleen and stored at −80°C. Slides were defrosted, fixed with acetone for 10 min at room temperature, and dried under ventilation. The wells on the slides were covered with 25 μl of serial twofold dilutions (1:40 to 1:1,280 in PBS, pH 7.2) of the sera and incubated in a humid chamber for 30 min at 37°C. After three washes with PBS, pH 7.2, 25 μl of fluorescein isothiocyanate-labeled goat anti-human immunoglobulin G (Biomérieux, Marcy l'Etoile, France) diluted 1:80 in Evansblue (0.1 mg/ml of PBS, pH 7.2) was applied to each well. After incubation in a humid chamber for 30 min at 37°C and three washes with PBS, pH 7.2, the slides were dried under ventilation, mounted with 50% (vol/vol) glycerol-PBS, and covered with a coverslip. Staining of the samples was assessed by examination with a fluorescence microscope and compared with that of positive and negative controls. Samples showing fluorescence with serum dilutions higher than 1:80 were regarded as positive.
RESULTS
Application of the miniexon PCR assay to clinical samples.
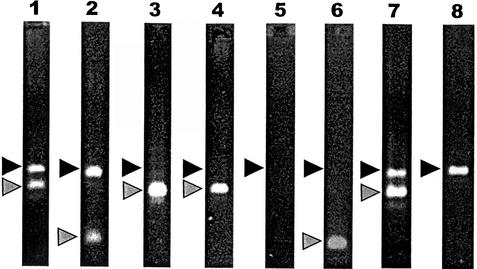
In order to identify inhibitory effects in the PCR due to impurities in the sample and to assess tube-to-tube variations in PCR efficiencies, the test was set up with an internal positive control. The sensitivity of the assay was not compromised by the coamplification of the positive control template (data not shown), and a concentration of 250 fg of plasmid DNA (corresponding to about 70,000 plasmid copies) per reaction mix produced a clear band in the PCR assay. From a total number of 50 clinical samples, DNA was purified and tested with the miniexon PCR assay. DNA from each sample was coamplified along with the positive control template. Of the 26 DNA samples which gave a positive result, the majority outcompeted the positive control template (Fig. 1, lanes 3 and 4), whereas in the remaining samples the signals of both the Leishmania and the positive control were visible on the agarose gel (Fig. 1, lanes 1 and 2). The other 24 samples were negative in the assay (Fig. 1, lane 8). PCR inhibition was observed in 6 of the 50 samples (Fig. 1, lane 5). Inhibition was resolved in two samples after a 10-fold dilution, after which one sample became positive (Fig. 1, lane 6) and one was determined to be negative. The remaining four samples had to be purified again by using the QIAamp DNA Mini Kit, after which two became positive (Fig. 1, lane 7) and two were determined to be negative.
FIG. 1.
Coamplification of Leishmania DNA from clinical samples and a positive control template. Black triangles denote the 540-bp band of the positive control template, and grey triangles correspond to the Leishmania-specific PCR signal. Lanes 1 and 2, positive samples. The PCR products of the target DNA and the positive control template are visible on the agarose gel. Lanes 3 and 4, positive samples. Target DNA outcompeted the positive control template. Lane 5, PCR inhibition is indicated because the positive control template is not visible. Lane 6, previously inhibited sample after DNA dilution. Lane 7, previously inhibited sample after subsequent purification. Lane 8, negative sample with only control template visible.
Comparison of the diagnostic assays.
The miniexon PCR assay was compared to the methods currently applied at the STI for the diagnosis of leishmaniasis (i.e., in vitro cultivation and serology) and a PCR assay which is based on the amplification of a repetitive genomic sequence of Old World Leishmania (LEG PCR assay) (28). All tissue samples, which were obtained from patients with a tentative diagnosis of leishmaniasis on clinical grounds, were tested with the LEG and the miniexon PCR assays, and 38 of them were inoculated into culture. Corresponding serum samples, available from 16 patients, were analyzed for anti-Leishmania antigen antibodies by IFAT. Results are summarized in Table 1. For a limited comparison, only positive samples were tested by the PCR assay using the ITS region of the SSU rRNA gene (37a).
TABLE 1.
Diagnostic results for clinical samples
| Specimen type | Total no. of positive samplesa | No. of samples positive by:
|
||||
|---|---|---|---|---|---|---|
| Culture | LEG PCR | Miniexon PCR | Serol- ogyb | SSU-ITS PCR | ||
| Skin biopsy | 12 | 6 | 8 | 12 | 4 | 6 |
| Bone marrow | 7 | 2 | 7 | 4 | 3 | 3 |
| Lesion aspirate | 1 | 0 | 1 | 1 | 0 | 1 |
| Bloodc | 2 | 2 | 2 | 2 | 0 | 1 |
| Duodenal biopsyd | 2 | 1 | 2 | 2 | 0 | 1 |
| Gastric biopsyd | 1 | 1 | 1 | 1 | 0 | 1 |
| Paraffin-embedded skin biopsy | 4 | 0 | 2 | 4 | 1 | 1 |
| Total | 29 | 12 | 23 | 26 | 8 | 15 |
Leishmaniasis confirmed by at least one of the diagnostic tests.
For patients from whom a corresponding serum sample was available.
Supplemented with EDTA.
From patients positive for HIV.
No single test could serve as a “gold standard” against which the different assays could be evaluated. Therefore, assessment of test performance (Table 2) was based on the assumptions that (i) a patient was positive for leishmaniasis when the infection was detected by at least one of the diagnostic assays and (ii) a patient was negative for leishmaniasis when results of all four assays (including serology) were negative. A Leishmania infection was diagnosed in 29 patients. Two infections were detected by all four methods, 12 infections were detected by three methods, and 10 infections were detected by two methods. In five patients, the presence of a Leishmania infection was confirmed by one method only (four cases were confirmed by the miniexon PCR assay, and one was confirmed by the LEG PCR assay).
TABLE 2.
Comparison of the diagnostic assays
| Assay (total)a | No. of positiveb samples/no. of tested samples
|
Sensitivity (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Skin biopsy specimens | Bone marrow samples | Blood samples | Duodenal biopsy specimensc | Gastric biopsy specimens | Lesion aspirates | Total | ||
| In vitro cultivation (38) | 6/9 | 3/5 | 1/1 | 1/1 | 1/1 | NA | 12/17 | 70.59 |
| Serology (16) | 5/9 | 3/4 | 0/1 | NA | NA | NA | 8/14 | 57.14 |
| LEG PCR (50) | 10/16d | 7/7 | 2/2 | 2/2 | 1/1 | 1/1 | 23/29 | 79.31 (100)d |
| SSU-ITS PCR (29)e | 7/16 | 3/7 | 1/2 | 2/2 | 1/1 | 1/1 | 15/29 | 51.71 |
| Miniexon PCR (50) | 16/16 | 4/7 | 2/2 | 2/2 | 1/1 | 1/1 | 26/29 | 89.66 |
Total number of samples subjected to each assay.
Leishmaniasis confirmed by at least one of the diagnostic tests.
NA, not applicable.
All negative samples were subsequently shown to be infected with New World species; therefore, sensitivity for Old World species was adjusted to 100%.
Only samples that were previously identified as positive by other assays were tested.
In vitro cultivation was performed for a total of 38 samples. No cultures were set up from the four paraffin-embedded specimens, and results for eight specimens are missing because either they were lost due to contamination or there was no request for this test. Twelve of 17 Leishmania-positive samples were detected by cultivation, of which one infection was confirmed by one other test, 13 were confirmed by two other tests, and three were confirmed by three other tests, resulting in a sensitivity of 70.6%. The remaining 21 samples with a negative culture result were also negative by both PCR assays, giving a specificity of 100%.
Sixteen serum samples were available to test by IFAT for anti-Leishmania antigen antibodies. This test identified eight positive samples from 14 patients whose infections were confirmed by other assays (two were confirmed by three other tests, three were confirmed by two other tests, and three were confirmed by one other test) but failed to diagnose the infection in six samples determined to be positive by other assays, giving a sensitivity of 57.1%. Two serum samples available from patients without an infection, as confirmed by all other tests, were also negative (specificity, 100%).
The LEG PCR detected Leishmania in 23 of the 29 positive samples, giving a sensitivity of 79.1%, but failed to diagnose the infection in six specimens that were confirmed to be positive by other tests. Parasites in all six samples for which no PCR product was obtained were subsequently identified by miniexon RFLP analysis as Leishmania species from the New World, resulting in a sensitivity of 100% for Old World species only. Specificity of the LEG PCR assay was 100% with all samples which were also negative by all other tests, but one sample was positive only with this test and was not confirmed to be positive by any of the other tests.
The PCR-RFLP genotyping assay based on the ITS region of the SSU rRNA gene detected a Leishmania infection in 15 of the 29 samples and gave a sensitivity of 51.7%. Because we tested only samples previously identified as positive, we were not able to determine the specificity of the assay.
The miniexon PCR detected Leishmania in 26 of the 29 positive samples and gave a sensitivity of 89.7% but failed to diagnose the infection in three specimens, of which two infections were confirmed by the LEG PCR assay and positive serology and one was confirmed by the LEG PCR assay only. It is worth mentioning that these three DNA samples were all derived from bone marrow specimens and were freeze-thawed several times. All the 21 Leishmania-negative patients were negative by the miniexon PCR assay (specificity, 100%), but it should be noted that four of the 29 positive results were not confirmed by any other test.
Genotyping of Leishmania spp. by RFLP analysis.
All samples positive by the miniexon PCR assay were subjected to RFLP analysis of the amplification product (24). No unusual or ambiguous RFLP patterns occurred, and Leishmania from all positive samples could be assigned to a given species according to the typing scheme. The species in 15 of the 26 positive samples was identified as L. infantum, that in four samples was identified as L. major, those in one sample each were identified as L. tropica and L. mexicana, that in three samples was identified as L. braziliensis, and that in two samples was identified as L. guyanensis.
Species in the 23 samples positive by the LEG PCR assay were typed by RFLP analysis as belonging to the Old World species L. infantum (n = 14), L. donovani (n = 4), L. major (n = 4), and L. tropica (n = 1).
DNA aliquots of the samples from the 29 Leishmania-positive patients were also analyzed by the PCR-RFLP genotyping assay, which is based on sequence variations in the ITS region of the SSU rRNA gene (37a). With this assay, species from positive samples were typed at the complex level. They were assigned to the L. donovani (n = 7), L. major (n = 6), L. tropica (n = 1), and L. braziliensis (n = 1) complexes.
A summary of the genotyping results is given in Table 3. There was a good correlation between the results obtained with the LEG and miniexon genotyping schemes. However, the LEG PCR system cannot detect any of the New World species, and a species from four samples typed as L. donovani with the LEG PCR system was typed as L. infantum with the miniexon system. There was also a good correlation between the results of the miniexon and ITS genotyping schemes, except that a species from two samples typed as L. infantum by the miniexon system was assigned to the L. major complex by the ITS system. These two samples were two of the four samples with a species typed as L. donovani by the LEG PCR system.
TABLE 3.
Results from three PCR-RFLP genotyping schemes for Leishmania spp.
| No. of patients | Species or complex identified bya:
|
||
|---|---|---|---|
| LEG-RFLP typing | Miniexon RFLP typing | ITS-RFLP typing | |
| 1 | ND | L. braziliensis | L. braziliensis complex |
| 2 | ND | L. braziliensis | ND |
| 2 | ND | L. guyanensis | ND |
| 1 | ND | L. mexicana | ND |
| 1 | L. tropica | L. tropica | L. tropica complex |
| 4 | L. major | L. major | L. major complex |
| 7 | L. infantum | L. infantum | L. donovani complex |
| 4 | L. infantum | L. infantum | ND |
| 3 | L. infantum | ND | ND |
| 2b | L. donovani | L. infantum | ND |
| 2b | L. donovani | L. infantum | L. major complex |
ND, not determined (PCR test failed to detect the infection).
Discordant results.
DISCUSSION
Detection of Leishmania parasites in a clinical sample is necessary to confirm a suspected case of leishmaniasis. Most commonly used methods for the direct detection of the parasite (e.g., microscopic examination of Giemsa-stained smears and in vitro cultivation) lack sensitivity because of the paucity of Leishmania parasites in some specimens (42) or are hampered by the problem of contamination (3). Indirect methods (e.g., detection of anti-Leishmania antigen antibodies by serological methods) are also limited in sensitivity and are not able to discriminate between past and current infections. The advent of the PCR technology has opened new avenues in the diagnosis of leishmaniasis, and several approaches have been developed during the last two decades (11, 43). Most of these assays are based on highly repetitive genomic gene loci or extrachromosomal kinetoplast DNA sequences, but their use is confined to the detection of the parasite at genus (5, 25) or complex (10, 13) level, and therefore, their application is limited to restricted geographical areas and diagnostic settings.
Our assay was developed for use in a diagnostic setting where patients with a history of global travel are seen. It was the aim of this study to optimize and evaluate a recently developed PCR-RFLP-based genotyping assay for Leishmania species. The assay targets the miniexon gene of the parasite and has proven to be simple and cheap; i.e., the detection of all Leishmania spp. is accomplished by one PCR with all-encompassing primers. It has a sufficient sensitivity, high specificity, and the ability to differentiate between all clinically relevant Leishmania species by RFLP analysis of the PCR product (24).
The inclusion of a positive control template makes it possible to detect inhibitory effects in the PCR mix and also controls for tube-to-tube variation both within and between different PCR experiments. Several methods are available for the design and construction of an internal positive control, some of which were successfully used in PCR assays for Leishmania. These include the coamplification of the human β-globin gene in a multiplex PCR approach (27), the use of a competitive PCR assay with heterologous plasmid sequences (25), and the use of synthetic probes in the reaction (4). We used a competitive PCR assay targeting a 498-bp fragment of the cpb2 gene of L. mexicana. This fragment is slightly larger than our target sequence, has a G+C content similar to that of the miniexon sequence, and is free of any restriction sites required for subsequent RFLP analysis of the miniexon sequence. In all assays, this positive control template yielded a clearly distinguishable PCR product, and coamplification did not affect assay sensitivity or compromise amplification of the miniexon sequence and never interfered with subsequent RFLP analysis (data not shown).
Fifty patient samples were available for the validation of the assay, and samples were subjected in parallel to a second PCR assay (LEG PCR) (28). When possible, samples were cultured, and serum samples were tested for Leishmania-specific antibodies. As expected, PCR assays were by far more sensitive than the currently applied diagnostic methods. Successful culture is dependent on the presence of viable organisms, and growth requirements are known to differ between species (3). IFAT failed to detect six infections (those of four patients with cutaneous manifestations and those of two patients suspected to have visceral leishmaniasis) which were all confirmed by at least two other tests. This failure might reflect the generally lower sensitivity of serological assays for detection of cutaneous forms of the disease (21) or might be due to a lack of cross-reactivity with the antigens on which the assay is based (26). Furthermore, humoral responses in patients with a concurrent HIV infection or other causes of immunosuppression may be moderate or even absent (1). Indeed, all three samples obtained from HIV-positive patients were negative by the serological assay.
The LEG PCR assay was tailored for the characterization of Old World Leishmania and consequently failed to detect six infections by parasites subsequently typed as New World species by the miniexon assay. On the other hand, the miniexon assay failed to detect three infections by parasites typed as Old World species by the LEG assay, which might be due to lower sensitivity of the miniexon PCR. However, storage of DNA from these three patients might also have compromised the results of the miniexon PCR.
Proper sampling is crucial for the establishment of a reliable diagnostic assay for leishmaniasis, and basic questions, such as what the optimal site of sample collection is (32) and whether blood samples are sufficient for a reliable diagnosis, remain open. Hematogenous dissemination has been described for L. donovani, L. infantum, L. tropica, and L. braziliensis (J. Kubar, J. F. Quaranta, P. Marty, A. Lelievre, Y. Le Fichoux, and J. P. Aufeuvre, Letter, Nat. Med. 3:368, 1997), and PCR-based assays on blood samples have proven useful for the diagnosis and monitoring of visceral leishmaniasis in adults and children in the Mediterranean region, as well as in HIV-coinfected patients (15, 19). The four blood samples in our study were processed only after the request for an additional tissue sample, and their analysis confirmed the results obtained with the other analyzed specimens.
The miniexon PCR assay was more sensitive than the conventional diagnostic methods and capable of detecting infection in a wide range of clinical samples, including paraffin-embedded tissue samples. This is of advantage since tissue biopsy specimens are often fixed and embedded in paraffin for histological examinations. Application of the PCR assay to paraffin-embedded specimens prevents repeated invasive sampling if a differential diagnosis of leishmaniasis is required and might be useful for the investigation of archived samples. It is noteworthy that other PCR-based assays have reported a higher sensitivity. Yet, these assays detect the presence of Leishmania parasites rather than individual Leishmania species. Although we found the miniexon assay to show a high sensitivity, other assays might be preferable for samples in which very low densities might be expected.
Prior to the development of biochemical or molecular tools for the characterization of Leishmania, diagnosis of leishmaniasis relied on clinical and epidemiological criteria. Over the past few years, the reports of atypical clinical manifestations have accumulated, challenging this traditional classification. This includes reports of visceral leishmaniasis caused by the dermotropic species L. tropica and L. mexicana (23) and the occurrence of cutaneous manifestations caused by L. chagasi and L. infantum, which usually cause visceral leishmaniasis (31; J. A. Rioux and G. Lanotte, Letter, Trans. R. Soc. Trop. Med. Hyg. 84:898, 1990).
Therefore, correct classification of infecting Leishmania species can provide important complementary information for the clinician that may result in modified clinical prognosis, as well as initiation of species-specific therapeutic approaches (3). Furthermore, identification of the infecting species is important in monitoring therapy. In areas where different species are sympatrically transmitted, discrimination between relapse of a latent infection and reinfection by another species is important in cases of reemerging leishmaniasis.
In order to appraise the validity of the miniexon genotyping scheme, we typed the samples with two other PCR-RFLP-based methods, the LEG and the ITS system. Although no unusual patterns were encountered with any of the genotyping schemes and the correlation between the results of all the systems was satisfying (Table 3), some discrepancies were observed. Because the biological species concept is difficult to apply to Leishmania and there is as yet no agreement on the definition of the species boundaries within the genus (2), none of the genotyping systems could actually serve as a gold standard for the evaluation of the others. We observed four discrepancies, two in which data obtained with the miniexon PCR suggested the presence of L. infantum but LEG PCR identified L. donovani. Although Leishmania has been thought to be clonal (2), hybrid forms within the L. donovani complex might occur. Using independent single marker genes cannot exclude this possibility. Discrepant identification of L. infantum and L. major in two cases cannot be explained but might reflect a laboratory artifact. Previously, the miniexon genotyping system has been tested with both World Health Organization reference strains and culture isolates from the STI, all independently characterized by isoenzyme analysis, and the results were in good yet not perfect concordance with the species definitions (24).
In conclusion, the miniexon PCR-RFLP-based genotyping assay has proven to be a reliable test for the detection of Leishmania in a wide range of clinical samples obtained from a travel clinic. The standardization of the test as a competitive assay including the coamplification of an internal positive control template has turned out to be a valuable tool for the assessment of PCR performance. We have demonstrated that the assay is quick and easy to perform and shows in this study a higher sensitivity than conventional diagnostic methods. Moreover, it enables the characterization of the parasite at species level by a simple method within a reasonable time frame and provides a useful tool for the characterization of clinically relevant Leishmania species, which can help the clinician choose an adequate therapeutic approach.
Acknowledgments
The project was supported by the European Community grant no. FAIR-CT 98-4104. This work also obtained support from the Schweizerische Nationalversicherung.
We thank all the members of the Diagnostic Centre at the Swiss Tropical Institute for their collaboration and support.
REFERENCES
- 1.Alvar, J., C. Canavate, B. Gutierrez-Solar, M. Jimenez, F. Laguna, R. Lopez-Velez, R. Molina, and J. Moreno. 1997. Leishmania and human immunodeficiency virus coinfection: the first 10 years. Clin. Microbiol. Rev. 10:298-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banuls, A. L., M. Hide, and M. Tibayrenc. 1999. Molecular epidemiology and evolutionary genetics of Leishmania parasites. Int. J. Parasitol. 29:1137-1147. [DOI] [PubMed] [Google Scholar]
- 3.Berman, J. D. 1997. Human leishmaniasis: clinical, diagnostic, and chemotherapeutic developments in the last 10 years. Clin. Infect. Dis. 24:684-703. [DOI] [PubMed] [Google Scholar]
- 4.Bezold, G., M. Lange, K. Gethoffer, H. Pillekamp, H. Reindl, C. Richter, G. Schonian, L. Weber, and R. U. Peter. 2001. Competitive polymerase chain reaction used to diagnose cutaneous leishmaniasis in German soldiers infected during military exercises in French Guiana. Eur. J. Clin. Microbiol. Infect. Dis. 20:421-424. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya, R., K. Das, S. Sen, S. Roy, and H. K. Majumder. 1996. Development of a genus specific primer set for detection of Leishmania parasites by polymerase chain reaction. FEMS Microbiol. Lett. 135:195-200. [DOI] [PubMed] [Google Scholar]
- 6.Brooks, D. R., H. Denise, G. D. Westrop, G. H. Coombs, and J. C. Mottram. 2001. The stage-regulated expression of Leishmania mexicana CPB cysteine proteases is mediated by an intercistronic sequence element. J. Biol. Chem. 276:47061-47069. [DOI] [PubMed] [Google Scholar]
- 7.Brun, R., and M. Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 8.Cunningham, I. 1977. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J. Protozool. 24:325-329. [DOI] [PubMed] [Google Scholar]
- 9.Davila, A. M., and H. Momen. 2000. Internal-transcribed-spacer (ITS) sequences used to explore phylogenetic relationships within Leishmania. Ann. Trop. Med. Parasitol. 94:651-654. [DOI] [PubMed] [Google Scholar]
- 10.De Bruijn, M. H., and D. C. Barker. 1992. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop. 52:45-58. [DOI] [PubMed] [Google Scholar]
- 11.Degrave, W., O. Fernandes, D. Campbell, M. Bozza, and U. Lopes. 1994. Use of molecular probes and PCR for detection and typing of Leishmania—a mini-review. Mem. Inst. Oswaldo Cruz 89:463-469. [DOI] [PubMed] [Google Scholar]
- 12.Eisenberger, C. L., and C. L. Jaffe. 1999. Leishmania: identification of Old World species using a permissively primed intergenic polymorphic-polymerase chain reaction. Exp. Parasitol. 91:70-77. [DOI] [PubMed] [Google Scholar]
- 13.Eresh, S., S. M. McCallum, and D. C. Barker. 1994. Identification and diagnosis of Leishmania mexicana complex isolates by polymerase chain reaction. Parasitology 109(Pt. 4):423-433. [DOI] [PubMed]
- 14.Fernandes, O., V. K. Murthy, U. Kurath, W. M. Degrave, and D. A. Campbell. 1994. Mini-exon gene variation in human pathogenic Leishmania species. Mol. Biochem. Parasitol. 66:261-271. [DOI] [PubMed] [Google Scholar]
- 15.Fisa, R., C. Riera, E. Ribera, M. Gallego, and M. Portus. 2002. A nested polymerase chain reaction for diagnosis and follow-up of human visceral leishmaniasis patients using blood samples. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl. 1):S191-S194. [DOI] [PubMed] [Google Scholar]
- 16.Grimaldi, G., Jr., J. R. David, and D. McMahon-Pratt. 1987. Identification and distribution of New World Leishmania species characterized by serodeme analysis using monoclonal antibodies. Am. J. Trop. Med. Hyg. 36:270-287. [DOI] [PubMed] [Google Scholar]
- 17.Harris, E., G. Kropp, A. Belli, B. Rodriguez, and N. Agabian. 1998. Single-step multiplex PCR assay for characterization of New World Leishmania complexes. J. Clin. Microbiol. 36:1989-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan, M. Q., A. Ghosh, S. S. Ghosh, M. Gupta, D. Basu, K. K. Mallik, and S. Adhya. 1993. Enzymatic amplification of mini-exon-derived RNA gene spacers of Leishmania donovani: primers and probes for DNA diagnosis. Parasitology 107(Pt. 5):509-517. [DOI] [PubMed] [Google Scholar]
- 19.Lachaud, L., J. Dereure, E. Chabbert, J. Reynes, J. M. Mauboussin, E. Oziol, J. P. Dedet, and P. Bastien. 2000. Optimized PCR using patient blood samples for diagnosis and follow-up of visceral leishmaniasis, with special reference to AIDS patients. J. Clin. Microbiol. 38:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lainson, R., and J. J. Shaw. 1987. Evolution, classification and geographical distribution, p. 1-120. In W. Peters and R. Killick-Kendrick (ed.), The leishmaniases in biology and medicine. Academic Press, Orlando, Fla.
- 21.Liew, F. Y., and C. A. O'Donnell. 1993. Immunology of leishmaniasis. Adv. Parasitol. 32:161-181. [DOI] [PubMed]
- 22.Luis, L., A. Ramirez, C. M. Aguilar, S. Eresh, D. C. Barker, and A. Mendoza-Leon. 1998. The genomic fingerprinting of the coding region of the beta-tubulin gene in Leishmania identification. Acta Trop. 69:193-204. [DOI] [PubMed] [Google Scholar]
- 23.Magill, A. J., M. Grogl, R. A. Gasser, Jr., W. Sun, and C. N. Oster. 1993. Visceral infection caused by Leishmania tropica in veterans of Operation Desert Storm. N. Engl. J. Med. 328:1383-1387. [DOI] [PubMed] [Google Scholar]
- 24.Marfurt, J., I. Niederwieser, D. Makia, H. P. Beck, and I. Felger. 2003. Diagnostic genotyping of Old and New World Leishmania species by PCR-RFLP. Diagn. Microbiol. Infect. Dis. 46:115-124. [DOI] [PubMed]
- 25.Mathis, A., and P. Deplazes. 1995. PCR and in vitro cultivation for detection of Leishmania spp. in diagnostic samples from humans and dogs. J. Clin. Microbiol. 33:1145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medrano, F. J., C. Canavate, M. Leal, C. Rey, E. Lissen, and J. Alvar. 1998. The role of serology in the diagnosis and prognosis of visceral leishmaniasis in patients coinfected with human immunodeficiency virus type-1. Am. J. Trop. Med. Hyg. 59:155-162. [DOI] [PubMed] [Google Scholar]
- 27.Meredith, S. E., E. E. Zijlstra, G. J. Schoone, C. C. Kroon, G. J. Van Eys, K. U. Schaeffer, A. M. el Hassan, and P. G. Lawyer. 1993. Development and application of the polymerase chain reaction for the detection and identification of Leishmania parasites in clinical material. Arch. Inst. Pasteur Tunis 70:419-431. [PubMed] [Google Scholar]
- 28.Minodier, P., R. Piarroux, F. Gambarelli, C. Joblet, and H. Dumon. 1997. Rapid identification of causative species in patients with Old World leishmaniasis. J. Clin. Microbiol. 35:2551-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson, R. D., A. De Queiroz Sousa, and S. M. B. Jeronimo. 2001. Leishmania species: visceral (kala-azar), cutaneous and mucosal leishmaniasis, p. 2831-2845. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Churchill Livingstone, New York, N.Y.
- 30.Piarroux, R., M. Fontes, R. Perasso, F. Gambarelli, C. Joblet, H. Dumon, and M. Quilici. 1995. Phylogenetic relationships between Old World Leishmania strains revealed by analysis of a repetitive DNA sequence. Mol. Biochem. Parasitol. 73:249-252. [DOI] [PubMed] [Google Scholar]
- 31.Ponce, C., E. Ponce, A. Morrison, A. Cruz, R. Kreutzer, D. McMahon-Pratt, and F. Neva. 1991. Leishmania donovani chagasi: new clinical variant of cutaneous leishmaniasis in Honduras. Lancet 337:67-70. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez, J. R., S. Agudelo, C. Muskus, J. F. Alzate, C. Berberich, D. Barker, and I. D. Velez. 2000. Diagnosis of cutaneous leishmaniasis in Colombia: the sampling site within lesions influences the sensitivity of parasitologic diagnosis. J. Clin. Microbiol. 38:3768-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramos, A., D. A. Maslov, O. Fernandes, D. A. Campbell, and L. Simpson. 1996. Detection and identification of human pathogenic Leishmania and Trypanosoma species by hybridization of PCR-amplified mini-exon repeats. Exp. Parasitol. 82:242-250. [DOI] [PubMed] [Google Scholar]
- 34.Rodgers, M. R., S. J. Popper, and D. F. Wirth. 1990. Amplification of kinetoplast DNA as a tool in the detection and diagnosis of Leishmania. Exp. Parasitol. 71:267-275. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez, N., B. Guzman, A. Rodas, H. Takiff, B. R. Bloom, and J. Convit. 1994. Diagnosis of cutaneous leishmaniasis and species discrimination of parasites by PCR and hybridization. J. Clin. Microbiol. 32:2246-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi, V., P. Wincker, C. Ravel, C. Blaineau, M. Pages, and P. Bastien. 1994. Structural organisation of microsatellite families in the Leishmania genome and polymorphisms at two (CA)n loci. Mol. Biochem. Parasitol. 65:271-282. [DOI] [PubMed] [Google Scholar]
- 37.Russell, R., M. P. Iribar, B. Lambson, S. Brewster, J. M. Blackwell, C. Dye, and J. W. Ajioka. 1999. Intra and inter-specific microsatellite variation in the Leishmania subgenus Viannia. Mol. Biochem. Parasitol. 103:71-77. [DOI] [PubMed] [Google Scholar]
- 37a.Schonian, G., A. Nasereddin, N. Dinse, C. Schweynoch, D. F. H. Schallig, W. Presber, and C. L. Jaffe. A PCR method for the diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn. Microbiol. Infect. Dis., in press. [DOI] [PubMed]
- 38.Schonian, G., C. Schweynoch, K. Zlateva, L. Oskam, N. Kroon, Y. Graser, and W. Presber. 1996. Identification and determination of the relationships of species and strains within the genus Leishmania using single primers in the polymerase chain reaction. Mol. Biochem. Parasitol. 77:19-29. [DOI] [PubMed] [Google Scholar]
- 39.Ullu, E., C. Tschudi, and A. Günzl. 1996. Trans-splicing in trypanosomatid protozoa, p. 115-133. In D. F. Smith and M. Parsons (ed.), Molecular biology of parasitic protozoa. IRL Press at Oxford University Press, Oxford, England.
- 40.Van Eys, G. J., G. J. Schoone, N. C. Kroon, and S. B. Ebeling. 1992. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol. Biochem. Parasitol. 51:133-142. [DOI] [PubMed] [Google Scholar]
- 41.Victoir, K., A. L. Banuls, J. Arevalo, A. Llanos-Cuentas, R. Hamers, S. Noel, S. De Doncker, D. Le Ray, M. Tibayrenc, and J. C. Dujardin. 1998. The gp63 gene locus, a target for genetic characterization of Leishmania belonging to subgenus Viannia. Parasitology 117(Pt. 1):1-13. [PubMed] [Google Scholar]
- 42.Weigle, K. A., M. de Davalos, P. Heredia, R. Molineros, N. G. Saravia, and A. D'Alessandro. 1987. Diagnosis of cutaneous and mucocutaneous leishmaniasis in Colombia: a comparison of seven methods. Am. J. Trop. Med. Hyg. 36:489-496. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, S. M. 1995. DNA-based methods in the detection of Leishmania parasites: field applications and practicalities. Ann. Trop. Med. Parasitol. 89(Suppl. 1):95-100. [PubMed] [Google Scholar]