Abstract
Numerical zymotaxonomy and variability of the internal transcribed spacers (ITS) between the small and large subunits of the rRNA genes were used to examine strain variation and relationships in natural populations of Leishmania (Viannia) braziliensis. A total of 101 strains from distinct hosts and Brazilian geographic regions were assigned to 15 zymodemes clustered in two major genetic groups. The great number of isolates (48.5%) placed in zymodeme IOC/Z-27 were collected on the Atlantic coast. The high molecular diversity found in populations in the Amazon Basin was related to the great number of sandfly vector(s) in that region. The results of the restriction fragment length polymorphism analysis of the ITS depicted considerable intraspecific variation. Genotypic groups A, B, and C contained 39, 40, and 22 isolates, which were divided into 16, 10, and 15 genotypes, respectively. The genetic polymorphism observed demonstrates the degree of diversity of L. (V.) braziliensis strains from different regions where they are endemic. The results reinforce the clonal theory for Leishmania parasites showing the genetic diversity of this pathogen and an association of L. (V.) braziliensis genotypes with specific transmission cycles, probably reflecting an adaptation of different clones to the vector species involved.
Infections with the parasitic protozoan Leishmania (Viannia) braziliensis Vianna 1911 (Kinetoplastida: Trypanosomatidae) or strain variants are recognized as causing human illness in many areas of (sub)tropical America (at least 15 countries), where it constitutes a significant public health problem. Many of these parasites seem to have a unique life cycle, with different phlebotomine sandfly (Diptera: Psychodidae) vectors and/or animal reservoirs and a different geographic distribution (13). This pathogen is capable of producing a variety of clinical manifestations, such as (i) cutaneous leishmaniasis (CL), which may heal spontaneously; (ii) mucosal leishmaniasis (ML), a hyperergic invasive ulcerative form that progresses in the absence of any apparent cellular defect (15); and (iii) disseminated CL (4).
Most of the environmental factors affecting the epidemiology of the various leishmaniases are still poorly understood. Wild mammals serve as reservoirs for most of the New World Leishmania spp. (21), but there is increasing evidence that some of the human pathogenic Leishmania strains can be maintained in both sylvan and urban cycles. In the case of L. (V.) braziliensis, the principal vertebrate hosts in the sylvan cycle have not been identified, but there is evidence that dogs, horses, and donkeys may serve as reservoir hosts of this parasite (15). The existence of an urban cycle involving peridomestic sandfly species for L. (V.) braziliensis reflects the ability of these parasites and their vectors to adapt to changes in their original forested habitats with important public health implications. Studies using molecular techniques to characterize L. (V.) braziliensis populations from different regions have shown a relationship between level of similarity among the parasite populations (12, 24) and their geographic range, but recent data have also indicated that the considerable variability detected among these parasites is more probably related to the sandfly vector(s) and/or animal reservoir(s) involved in the transmission cycles (18).
Pathogens that produce many different genetic variants are more prone to infect multiple hosts (37). Although several studies have discussed the polymorphism observed in natural populations of different Leishmania species (8, 25, 34), until now there has been little information available about the genetic variability of the parasites and the correlation with ecoepidemiological features of the disease (16). The risk factors for clinical leishmaniasis are still poorly understood and probably are influenced by host and parasite features.
Considering the public health importance of leishmaniasis caused by L. (V.) braziliensis in Brazil and the role of the genetic polymorphism of the parasites in the epidemiology of the disease, we applied multilocus enzyme electrophoresis (MLEE) and the analysis of the restriction fragment length polymorphism (RFLP) of the internal transcribed spacers (ITS) of the rRNA genes as typing methods to determine the level of genetic variation in natural population of L. (V.) braziliensis derived from different endemic areas. MLEE are rapidly evolving markers and have been the most universally accepted means of characterizing and differentiating the many diverse types of leishmanial parasite (5, 30). The electrophoretic mobilities and banding patterns of several different enzymes are usually checked and compared to produce enzyme profiles that designate zymodemes. The relationships among the zymodemes are often verified by numerical taxonomic methods (5, 22, 30, 36). Recently, the ITS of the rRNA genes were reported to be useful markers to type Leishmania parasites (6, 33, 34). These regions are flanking the ribosomal genes, which are found in all eukaryotic organisms, and are clustered in tandem arrays on different chromosomes. The ITS regions of the rRNA genes evolve more rapidly than the functional domains that they flank, and phylogenetic studies of several organisms have demonstrated that they are useful to infer about the relationship among closely related populations of organisms (17).
The observed genetic variability and the relationships among the isolates were the basis for investigating whether the genetic differences in L. (V.) braziliensis reflect distinct ecoepidemiological features of the infection since these strains are endemic in the distinct areas studied, thus bringing to light fundamental information for future control programs of the disease.
MATERIALS AND METHODS
Leishmanial strains.
The parasites used in the present study and the sources of original stocks are listed in Table 1. We analyzed Leishmania (V.) braziliensis isolates from four geographic localities in Brazil that are regions where cutaneous leishmanisis is endemic: Rio de Janeiro, Espírito Santo, and Pernambuco, as well as in the Amazonian region. Leishmania (V.) braziliensis reference strains and isolates used in the present study are maintained at DIFIOCRUZ Leishmania Type Culture Collection (registration no. 731 [WFCC World Data Center on Microorganisms Directory]).
TABLE 1.
Results of molecular typing and provenance details of 101 L. (V.) braziliensis strains collected over a 23-year period that were studied for their genetic relatedness
| Zymodemea | Genotypic typeb (no. of stocks) | Geographic originc | Clinical formd |
|---|---|---|---|
| Z-27 (MHOM/BR/66/M2903) | B6(1) | RJ, Angra dos Reis | CL |
| B6 (1) | RJ, Itacoara | ML | |
| B6 (1) | RJ, Maricá | ML | |
| B5 (1), B6 (8), B9 (1) | RJ, Mesquita | CL | |
| B6 (3) | RJ, Paraty | CL | |
| B9 (1) | RJ, Quiçamã | CL | |
| B5 (1), B6 (5), B7 (1), B8 (1) | RJ, Rio de Janeiro | CL | |
| B6 (1) | RJ, São Gonçalo | CL | |
| B6 (1) | RJ, Teresópolis | CL | |
| A12 (1), A16 (1) | ES, Afonso Cláudio | CL | |
| A1 (1) | ES, Áqua Doce do Norte | CL | |
| B4 (1) | ES, Aracruz | CL | |
| A16 (1) | ES, Castelo | CL | |
| B3 (1) | ES, Fundão | CL | |
| A9 (1) | ES, Itaguaçu | CL | |
| A7 (2) | ES, Itarana | CL | |
| A13 (1), A16 (1) | ES, Muniz Freire | CL | |
| B2 (4) | ES, Santa Leopoldina | CL | |
| B10 (2) | ES, Serra | CL | |
| B1 (5) | ES, Viana | CL, ML | |
| Z-31 (IPAR/BR/80/IM231) | C13 (1) | RO, BR-319Rd, Km866 | |
| Z-32 (ICAR/BR/86/IM2978) | C1 (1) | RO, Cachoeira Samuel | |
| Z-34 (MHOM/BR/88/IM3476) | C2 (1) | AM, Tapauã River | CL |
| Z-35 (MHOM/BR/88/IM3483) | C3 (1) | AM, Tapauã River | CL |
| C5 (1), C6 (2), C7 (4), C8 (3), C9 (2) | AM, Coari-Urucu River | CL | |
| C4 (1) | AM, Manaus | ML | |
| Z-45 (MHOM/BR/91/JRS) | A4 (4), A6 (2), A8 (1), A10 (1), A11 (1), A14 (6), A15 (1) | PE, Amaraji | CL |
| Z-53 (MAGO/BR/92/IN154) | C15 (1) | RO, Porto Velho | |
| Z-63 (MHOM/BR/91/IM3708) | C10 (1) | AM, Coari-Urucu River | CL |
| Z-64 (MHOM/BR/91/IM3711) | C11 (1) | AM, Coari-Urucu River | CL |
| Z-65 (MHOM/BR/91/IM3723) | C12 (1) | AM, Coari-Urucu River | CL |
| Z-69 (MDID/BR/95/IM4159) | C14 (1) | AM, Barcelos | |
| Z-72 (MHOM/BR/98/AFS) | A4 (1), A6 (3), A11 (1), A14 (1) | PE, Amaraji | CL |
| Z-73 (MHOM/BR/98/ASB) | A4 (3), A13 (2) | PE, Amaraji | CL, ML |
| Z-74 (MHOM/BR/98/EJS) | A3 (1), A5 (1) | PE, Amaraji | CL |
| Z-75 (MHOM/BR/98/FJS) | A2 (1) | PE, Amaraji | CL |
The zymodeme number of the Instituto Oswaldo Cruz (classification was established by numerical zymotaxonomic analysis) is given. The reference strain designation code is given in parentheses as follow host (I for Insecta [CAR, Psychodopygus carrerai; PAR, Psychodopygus paraensis;] M for Mammalia [AGO, Agouti paca; DID, Didelphis marsupialis; HOM, Homo sapiens])/country of origin/year of isolation/original code.
Denotes the identity between isolates by using all polymorphic rDNA results.
Codes for states in Brazil (AM, Amazonas; ES, Espírito Santo; PE, Pernambuco; RJ, Rio de Janeiro; RO, Rondônia) are given.
Leishmaniasis lesions in humans (CL, cutaneous; ML, mucosal).
MLEE.
The methods used to prepare samples and study the electrophoretic mobility of some enzymes in agarose gels were performed as described elsewhere (5, 19). Allelic variation for the following enzymes was assessed: aconitate hydratase (EC 4.2.1.3), glucose-6-phosphate dehydrogenase (EC 1.1.1.49), glucose phosphate isomerase (EC 5.3.1.9), isocitrate dehydrogenase NAD and NADP (EC 1.1.1.42), malate dehydrogenase (EC 1.1.1.37), malic enzyme (EC 1.1.1.40), mannose phosphate isomerase (EC 5.3.1.8), nucleosidase (EC 3.2.2.1), 6-phosphogluconate dehydrogenase (EC 1.1.1.43), phosphoglucomutase (EC 1.4.1.9), Leu-Pro dipeptidase (EC 3.4.13.9), and Leu-Gly dipeptidase (EC 3.4.11.1). The bands produced on the gels were numbered according to enzymatic mobility, and the enzyme profiles were subjected to numerical analysis and used to group the samples in zymodemes. Leishmanial strains with the same electromorphs were classified into the same zymodeme (5). One strain from each zymodeme was chosen as the reference for that zymodeme (Table 1).
Intergenic region typing (IRT).
Genomic RFLP analysis of the ITS between the small and large subunits of the ribosomal DNA (rDNA) locus (PCR-RFLP ITSrDNA) of leishmanial parasites used in the present study (Table 1) was done, as described elsewhere (6). The ITS were PCR amplified and digested with restriction enzymes (AluI, BstUI, EcoRI, FspI, HaeIII, HhaI, RsaI, and TaqI). Digestion products were separated by high-resolution electrophoresis in a 12% polyacrylamide gel by using the Genephor apparatus (Amersham Pharmacia Biotech). The fragment sizes were estimated relative to molecular weight markers and the banding patterns were used to group the isolates in genotypes (each genotype grouped isolates with the same banding pattern for all of the restriction enzyme used) and were also subjected to numerical analysis.
Numerical analysis of MLEE or PCR-RFLP ITSrDNA data.
For this numerical analysis, a matrix with the presence or absence of the bands was built and analyzed by using the NTSYS software program (version 2.1; Exeter Software). The similarity matrices constructed by using the simple matching coefficient (Ssm = m/n, where m is the number of matches and n is the total number of characters) were transformed into phenograms by the UPGMA (unweighted pair-group method with arithmetic averages) algorithm (35). The confidence of the groups was tested applying a bootstrap analysis with 1,000 replicates using FreeTree software (version 0.9.1.50). The average and standard deviation of the similarities observed among individuals from each area (considering the clusters observed by the phenetic analysis) where the infection is endemic were calculated as a parameter of genetic diversity.
RESULTS
MLEE.
Fourteen enzyme loci of 101 stocks of L. (V.) braziliensis from different hosts and Brazilian geographic areas were analyzed, and the electromorphic profiles were compared to reference strains revealing 15 zymodemes (Table 1). The most common zymodeme (Z-27) grouped 49 isolates from Espírito Santo and Rio de Janeiro States. In contrast, 31 strains collected in a rural area in Pernambuco State were classified into five zymodemes. Furthermore, most of the 22 stocks from the Amazon and Rondônia States were individually assigned to separate zymodemes. However, the analysis of allelic variation occurring in stocks compared to the reference strain MHOM/BR/1966/M2903 of L. (V.) braziliensis revealed that some zymodemes differ from each other in only one enzymatic locus position (data not shown).
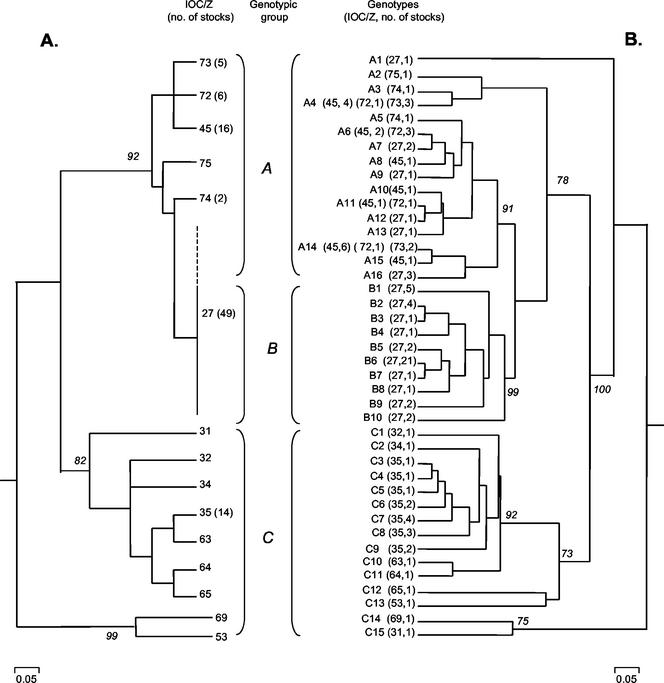
Affinities between zymodemes were calculated by using the simple matching similarity coefficient and were transformed into a phenogram by using mean distances between groups (Fig. 1A). The 15 L. (V.) braziliensis zymodemes could be clustered into two major groups, with high level of statistic support as demonstrated by the bootstrap value obtained for each group (Fig. 1A). Each group clustered strains collected on either the Atlantic coast or the Amazon region. One group (comprising isolates from CL or ML patients) was less diverse compared to the other group (comprising isolates from humans and various wild animals and sylvan sandflies). Furthermore, the later group contained two parasites (Z-53 and Z-69) that clustered independently of other zymodemes. These two zymodemes were assigned to the second group based on their geographic origin and because they presented a higher level of similarity with the zymodemes of this group than to the ones of the other group.
FIG. 1.
(A) MLEE phenogram showing the similarity between groups of L. (V.) braziliensis zymodemes. Unless indicated otherwise by the number in parentheses, IOC/Z contains a unique isolate. (B) Molecular tree showing the clustering of L. (V.) braziliensis genotypes classified in this work (PCR-RFLP ITSrDNA). The bootstrap values (italic numbers) are shown for the principal clusters. Details about strains are given in Table 1. The three main clusters represent parasites collected from different hosts and geographic areas in Brazil (i.e., the Atlantic coast [A and B] or the Amazon region [C]). Isolates characterized as IOC/Z-27 (outlined with continuous and dashed lines) presented genotypes in either group A and B. A, Strains collected in rural localities (Espírito Santo and Pernambuco) where different sandfly and animal species are involved in the transmission cycle; B, strains from urban areas (Espírito Santo and Rio de Janeiro) where transmission is related with a cycle involving domestic animals and preidomestic sandfly species; and C, strains from the Amazon region (in this case, the parasites are maintained in an enzootic cycle involving various wild animals and sylvan sand flies).
Genotyping.
The IRT allowed the examination of polymorphisms among L. (V.) braziliensis zymodemes (Fig. 2). All leishmanial isolates indicated in Table 1 were analyzed. It was possible to characterize 41 genotypes, each representing parasites with unique ITS fragment profiles. By using numerical analysis, the strains were studied for their genetic relatedness (Fig. 1B). Three statistically supported groups were observed. Genotypic groups A, B, and C comprised 39, 40, and 22 isolates, which were divided into 16, 10, and 15 related genotypes, respectively. It was observed that zymodemes into which more than one isolate was assigned are represented by more than one genotype: Z-27 included 49 samples and 16 genotypes, Z-35 included 14 samples and 7 genotypes, Z-45 included 16 samples and 7 genotypes, and Z-72 included 6 samples and 4 genotypes (Table 1). Except for Z-27, all of the other zymodemes representing large clusters included isolates from the same geographic locality.
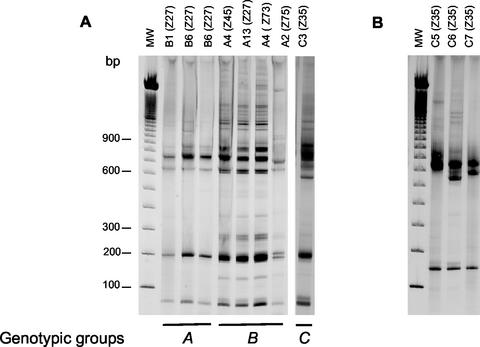
FIG. 2.
Acrylamide (12%) gel electrophoresis comparison of ITS fragment patterns generated with the restriction enzymes BstUI (A) and HhaI (B), among selected strains of L. (V.) braziliensis from Brazil. The codes in each lane represent the genotypes and the respective zymodemes (IOC/Z). The genotypic groups correspond to those described in Fig. 1. (A) Differences among groups A, B, and C; (B) differences among isolates from the same zymodeme.
Isolates characterized as the same zymodeme (Z-27) were clustered into two independent clusters (A and B). Cluster A comprised isolates from rural localities in Pernambuco and Espírito Santo. Cluster B accounted for strains related with domestic animals and peridomestic sandflies that were assigned to the same zymodeme (Z-27). Group C included only isolates from the Amazonian region. This group was more diverse, presenting some genotypes (C12, C13, C14, and C15) that clustered independently from the others, but with more affinity for the genotypes of group C than for the ones of group A or B. A clonal population (genotype B6) classified in this group contained 21 isolates from different geographic locations in the Espírito Santo and Rio de Janeiro states. In contrast, eight isolates characterized as genotype A4 were assigned to separate zymodemes. Furthermore, parasites showing a high degree of genetic similarity were found to cause either CL or ML in humans (Table 1).
Geographic distribution and genetic diversity of the genotypes.
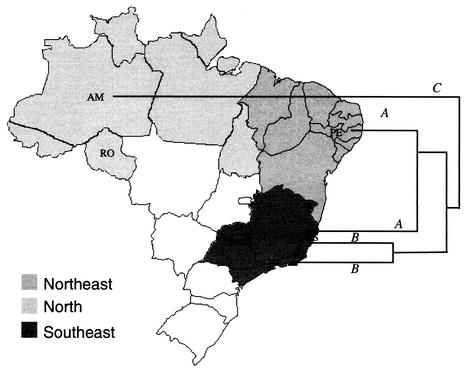
A geographic structuring was observed for some genotypes but not for others that was related to genotypes from more distant areas where the organisms were endemic (Fig. 1 and 3). The genetic similarity average based on the MLEE and IRT data was calculated for each locality, and it was assumed as a parameter to evaluate the level of genetic diversity. We observed that the isolates from the Amazonian region presented the highest level of genetic diversity and that the isolates from Rio de Janeiro were more homogeneous (Fig. 3). The isolates from Espírito Santo were considered as being involved in two distinct transmission cycles. The Espírito Santo isolates from urban areas grouped together with the isolates from Rio de Janeiro (group B) and those from rural areas clustered with isolates from Pernambuco (group A) (Fig. 3).
FIG. 3.
Map of Brazil showing the areas from which the L. (V.) braziliensis isolates originated. A schematic and summarized dendrogram showing the relationship among the genotypic groups (A, B, and C as defined in Fig. 1) was superimposed. The level of genetic diversity found among the isolates from each area where the parasites were endemic was estimated by using the average of similarities as follows (averages ± the standard deviations of MLEE and IRT, respectively): Amazonia (0.75 ± 0.082 and 0.76 ± 0.095), Pernambuco (0.92 ± 0.01 and 0.83 ±0.082), Espírito Santo A (1.0 and 0.89 ± 0.05), Espírito Santo B (1.0 and 0.83 ± 0.11), and Rio de Janeiro (1.0 and 0.93 ± 0.035). The Brazilian states where the isolates were collected included Amazonia, Rondônia, Pernambuco, Espírito Santo, and Rio de Janeiro (see Table 1).
DISCUSSION
Advances in molecular technology are facilitating the study of the ecology of Leishmania clonal populations by providing information on (i) sources of infection, (ii) transmission patterns, (iii) response to treatment, and (iv) the importance of immunity in preventing reinfection (27). In the New World, the Leishmania (Viannia) parasites (7, 8), the main causal agents of American CL, represent a biologically diverse group of microorganisms showing considerable intraspecies variability (5, 6, 12) that complicates taxonomic classification and epidemiological studies.
The present systematic molecular study revealed L. (V.) braziliensis strains to be an extremely diverse population. The multilocus data on a diverse collection of strains from different hosts and ecologic regions in Brazil indicate a clonal population structure, with some zymodemes (e.g., IOC/Z-27) widely distributed and others seemingly unique and localized to a particular endemic focus (e.g., IOC/Z-35 and IOC/Z-45). The IRT analysis resulted in a very high degree of discrimination among isolates from diverse regions, confirming the ability of this technique to discriminate between closely related parasites (6). The three distinct genotypic groups described herein may represent important genetic variation within L. (V.) braziliensis, which could explain the plasticity of these parasites and of their ability to adapt to changing ecological conditions. Changes in the ITSrDNA locus, which is a noncoding region, should not reflect the ability to adapt to ecological changes. However, because of the high level of polymorphism in the composition of the ITSrDNA locus, several molecular epidemiological studies have been conducted by using this tool. Analysis of the ITS regions of the nuclear rRNA operon for different parasites has demonstrated the utility of this marker in detect genetic diversity and the association of this with several epidemiological aspects (9, 26)
The present study also indicates that most of the genotypes are specific to some geographic areas (e.g., genotype B1 contains isolates from one geographic location, in Espírito Santo, that were collected over a distinct period of time [between 1982 and 1995]), showing the importance of clonal dissemination versus sexual reproduction for this organism in nature, at least occurring in some regions. The stability of this genotype adapting to some environmental modifications through years in the same area is a clear proof of the aforementioned statement.
L. (V.) braziliensis can infect vertebrate hosts of different species and orders (21) and has been associated with a number of different sandfly species (15). Our analyses indicated that the molecular diversity found in parasites from the Amazon Basin is apparently related to the great number of sandfly vector(s) and/or animal reservoir(s) involved in the transmission cycles (13, 14). In contrast, the L. (V.) braziliensis populations circulating in the Brazilian Atlantic coast (in the present study, representing 81.2% of the isolates collected from human beings and dogs in old established communities in nonforested areas) showed a lower level of heterogeneity than did the Amazonian strains and have peridomestic sandfly species such as Lutzomyia intermedia and Lutzomyia whitmani as the principal suspected vectors (1, 3, 10). The presence of the same genotypes in humans and dogs in the same area suggests that specific transmission cycles could be defined by using the proposed methodologies.
A total of 49% of the parasites collected in areas (Rio de Janeiro or Espírito Santo) where sylvan animals are probably not involved in the transmission cycle were clustered together into the same zymodeme (IOC/Z-27) of the L. (V.) braziliensis reference strain (MHOM/BR/1966/M2903), which was isolated from a patient with American CL in the Serra dos Carajás, Pará State. In this case, a sylvan sandfly, Psychodopygus wellcomei, has been determined to be the vector (20, 31). In contrast, isolates from Pernambuco, where grass mice (Bolomys lasiurus) and black rats (Rattus rattus) have been found to be parasite reservoirs (S. P. Brandão-Filho, M. E. F. Brito, F. G. Carvalho, E. A. Ishikawa, E. Cupolillo, and J. J. Shaw, Abstr. Wordleish 2000, abstr. 100, 2000), were grouped into five distinct zymodemes. Factors that predispose pathogens to infect multiple hosts include high levels of genetic diversity and opportunities for cross-species transmission (37). The extent of host adaptation is therefore linked to and limited by the genetic variability. Pathogens that contain more accumulating mutations should produce more genetic variants and are more likely to be generalists. An alternative explanation to the extent of host adaptation of some organisms is to consider that the infection of a given host is only related to opportunity; therefore, parasites that infect several different hosts are more likely to be isolated in different transmission cycles.
The RFLP-ITSrDNA data show that parasites from urban areas (Espírito Santo and Rio de Janeiro), where transmission is associated with Lutzomyia intermedia, cluster together. Another cluster comprises parasites from rural localities in Espírito Santo and Pernambuco, where Lutzomyia whitmani is the principal vector species. Other studies using different molecular markers have demonstrated a geographic structuring of Leishmania parasites (2, 16, 23, 33, 34), and populations of L. (V.) braziliensis isolates collected from geographically closed areas seem to be more genetically similar (12, 24). However, the present study indicates that the geographic structuring of L. (V.) braziliensis reflects the plasticity of these parasites to adapt to different vector species (or populations of this vectors) involved in the transmission cycle. Furthermore, our results indicate an association between the diversity of vectors involved in the transmission of L. (V.) braziliensis and the genetic diversity of the parasites. Evaluation of the phosphoglycan composition of these leishmanial populations is fundamental to an understanding of the vector-parasite interaction (32). Another important approach is to study the population structure of the sandfly vectors in areas where the genetic diversity of L. (V.) braziliensis has been determined.
It was recently suggested that the dispersion of Leishmania clones is associated with the vector and reservoir movements (18). However, what is important to note is the plasticity of leishmanial parasites, their ability to adapt to changes in ecologic conditions, and a possible evolutionary relationship between them and their vectors as observed in the Old World (28).
The clinical expression of leishmanial infection is dependent on a number of different factors (15). The identification of virulence factors that contribute to the pathogenesis of L. (V.) braziliensis infections are poorly understood, mainly because of the lack of good in vivo models. A better understanding of the genetic basis of pathogenesis in ML is needed, as are studies on the role of host factors in immune susceptibility to this form of the disease. Our genotyping studies did not identify a pathogenic clone specifically associated with the development of ML. However, in other analyses we were able to detect distinct genotypic types of a single L. (V.) braziliensis isolate from a patient with ML (unpublished data). Competitive interactions among coinfecting clones could affect host immune responses and play a role in determining the immunopathology of the progressive and destructive form of ML. Some underlying causes of its hyperergic response might include the following factors: (i) the resistance of some parasite clones to elimination (27) and the persistence of “allergic” antigen that evokes hypergic hypersensitivity inflammatory responses and (ii) autoimmune phenomena related to antigens cross-reactive between leishmanial parasites and host tissues (11, 29).
In conclusion, the combination of MLEE and IRT-ITSrDNA is useful for studies of the interface of population genetic and epidemiology. The application of these molecular techniques could allow us to better determine the genetic diversity of various Leishmania parasites circulating in nature. This information is expected to yield new approaches to mitigate parasite transmission and virulence in humans and eventually to improve parasite control.
Acknowledgments
We thank Luiz Eduardo de Carvalho Paes for technical assistance and Hooman Momen and Octavio Fernandes for the critical revision of the manuscript.
This work was supported by Fundação Oswaldo Cruz, Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro FAPERJ, Conselho Nacional de Desenvolvimento Científico e Tecnológico CNPq, and PRONEX III/CNPq/MCT (Brazil).
REFERENCES
- 1.Aguilar, C. M., E. F. Rangel, G. Grimaldi, Jr., and H. Momem. 1987. Human, canine and equine leishmaniasis caused by Leishmania braziliensis braziliensis in an endemic area in the State of Rio de Janeiro. Mem. Inst. Oswaldo Cruz 82:143. [DOI] [PubMed] [Google Scholar]
- 2.Akman, L., H. S. Z. Aksu, R.-Q. Wang, S. Ozensoy, Y. Ozbel, Z. Alkan, M. A. Ozcel, G. Culha, K. Ozcan, S. Uzun, H. R. Memisoglu, and K.-P. Chang. 2000. Multi-site polymorphism analyses of Leishmania isolates define their genotypes predicting clinical epidemiology of leishmaniasis in a specific region. J. Eukaryot. Microbiol. 47:545-554. [DOI] [PubMed] [Google Scholar]
- 3.Brandão-Filho, S. P., D. Campbell-Lendrum, M. E. F. Brito, J. J. Shaw, and C. R. Davies. 1999. Epidemiological surveys confirm an increasing burden of cutaneous leishmaniasis in north-east Brazil. Trans. R. Soc. Trop. Med. Hyg. 93:488-494. [DOI] [PubMed] [Google Scholar]
- 4.Carvalho, E. M., A. Barral, J. M. Costa, A. Bittencourt, and P. Marsden. 1994. Clinical and immunopathological aspects of disseminated cutaneous leishmaniasis. Acta Trop. 56:315-325. [DOI] [PubMed] [Google Scholar]
- 5.Cupolillo, E., G. Grimaldi, Jr., and H. Momen. 1994. A general classification of New World Leishmania using numerical zymotaxonomy. Am. J. Trop. Med. Hyg. 50:296-311. [DOI] [PubMed] [Google Scholar]
- 6.Cupolillo, E., G. Grimaldi, Jr., H. Momen, and S. M. Beverley. 1995. Intergenic region typing (IRT): a rapid molecular approach to the characterization and evolution of Leishmania. Mol. Biochem. Parasitol. 73:145-155. [DOI] [PubMed] [Google Scholar]
- 7.Cupolillo, E., G. Grimaldi, Jr., and H. Momen. 1997. Genetic diversity among Leishmania (Viannia) parasites. Ann. Trop. Med. Parasitol. 91:617-626. [DOI] [PubMed] [Google Scholar]
- 8.Cupolillo, E., H. Momen, and G. Grimaldi, Jr. 1998. Genetic diversity in natural populations of New World Leishmania. Mem. Inst. Oswaldo Cruz 93:663-668. [DOI] [PubMed] [Google Scholar]
- 9.Dengjel, B., M. Zahler, W. Hermanns, K. Heinritzi, T. Spillmann, A. Thomschke, T. Löscher, R. Gothe, and H. Rinder. 2001. Zoonotic potential of Enterocytozoon bieneusi. J. Clin. Microbiol. 39:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falqueto, A., J. R. Coura, G. C. Barros, G. Grimaldi Filho, P. A. Sessa, V. R. Carias, A. C. de Jesus, and J. T. de Alencar. 1986. Participation of the dog inthe cycle of transmission of cutaneous leishmaniasis in the municipality of Viana, State of Espirito Santo, Brazil. Mem. Inst. Oswaldo Cruz 81:155-156. [DOI] [PubMed] [Google Scholar]
- 11.Galvão-Castro, B., J. A. As Ferreira, K. F. Marzochi, M. C. A. Marzochi, S. G. Coutinho, and P. H. Lambert. 1984. Polyclonal B-cell activation, circulating immune complexes and autoimmunity in human American visceral leishmaniasis. Clin. Exp. Immunol. 56:58-66. [PMC free article] [PubMed] [Google Scholar]
- 12.Gomes, R. F., A. M. Macedo, S. D. J. Pena, and M. N. Melo. 1995. Leishmania (Viannia) braziliensis: genetic relationships between strains isolated from different areas of Brazil as revealed by DNA fingerprinting and RAPD. Exp. Parasitol. 80:681-687. [DOI] [PubMed] [Google Scholar]
- 13.Grimaldi, G., Jr., R. B. Tesh, and D. McMahon-Pratt. 1989. A review of the geographic distribution and epidemiology of leishmaniasis in the New World. Am. J. Trop. Med. Hyg. 41:687-725. [DOI] [PubMed] [Google Scholar]
- 14.Grimaldi, G., Jr., H. Momen, R. D. Naiffi, D. McMahon-Pratt, and T. V. Barret. 1991. Characterization and classification of leishmanial parasites from humans, wild mammals, and sandflies in the Amazon region of Brazil. Am. J. Trop. Med. Hyg. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 15.Grimaldi, G., Jr., and R. B. Tesh. 1993. Leishmaniases of the New World: current concepts and implications for future research. Clin. Microbiol. Rev. 6:230-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerbouj, S., I. Guisani, N. Speybroeck, D. Le Ray, and J. C. Dujardin. 2001. Genomic polymorphism of Leishmania infantum: a relationship with clinical pleomorphism? Infect. Genet. Evol. 1:49-59. [DOI] [PubMed] [Google Scholar]
- 17.Hillis, D. M., and M. T. Dixon. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 66:411-453. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa, E. A. Y., F. T. Silveira, A. L. P. Magalhães, R. B. Guerra, Jr., M. N. Melo, R. Gomes, T. G. Silveira, and J. J. Shaw. 2002. Genetic variation in populations of Leishmania species in Brazil. Trans. R. Soc. Trop. Med. Hyg. 96:111-121. [DOI] [PubMed] [Google Scholar]
- 19.Jaffe, C. L., G. Grimaldi, Jr., and D. Mcmahon-Pratt. 1984. The cultivation and cloning of Leishmania, p. 47-91. In C. M. Morel (ed.), Genes and antigens of parasites. UNDP/World Bank/World Health Organization-FINEP-CNPQ-FIOCRUZ, Rio de Janeiro, Brazil.
- 20.Lainson, R., J. J. Shaw, R. D. Ward, and H. Fraiha. 1973. Leishmaniasis in Brazil. IX. Considerations of the Leishmania braziliensis complex: importance of sandflies of the genus Psychodopygus (Mangabeira) in the transmission of L. braziliensis braziliensis in North Brazil. Trans. R. Soc. Trop. Med. Hyg. 67:184-196. [DOI] [PubMed] [Google Scholar]
- 21.Lainson, R., and J. J. Shaw. 1987. Evolution, classification, and geographical distribution, p. 1-120. In W. Peters and R. Killick-Kendrick (ed.), The leishmaniases in biology and epidemiology, vol. 1. Academic Press, Ltd., London, England.
- 22.Le Blancq, S. M., R. E. Cibulskis, and W. Peters. 1986. Leishmania in the Old World. 5. Numerical analysis of isoenzyme data. Trans. R. Soc. Trop. Med. Hyg. 80:517-524. [DOI] [PubMed] [Google Scholar]
- 23.Lewin, S., G. Schonian, N. El Tai, L. Oskam, P. Bastien, and W. Presber. 2002. Strains typing in Leishmania donovani by using sequence-confirmed amplified region analysis. Int. J. Parasitol. 32:1267-1276. [DOI] [PubMed] [Google Scholar]
- 24.Macedo, A. M., M. N. Melo, R. F. Gomes, and S. D. Pena. 1992. DNA fingerprints, a tool for identification and determination of the relationships between species and strains of Leishmania. Mol. Biochem. Parasitol. 53:63-70. [DOI] [PubMed] [Google Scholar]
- 25.Mauricio, I. L., M. W. Gaunt, J. R. Stothard, and M. A. Miles. 2001. Genetic typing and phylogeny of the Leishmania donovani complex by restriction analysis of PCR amplified gp63 intergenic regions. Parasitology 122:393-403. [DOI] [PubMed] [Google Scholar]
- 26.Nimri, L. F., I. N. S. Moura, L. Huang, C. Del Rio, D. Rimland, J. S. Duchin, E. M. Dotson, and C. B. Beard. 2002. Genetic diversity of Pneumocystis carinii f. sp. hominis based on variation in nucleotide sequences of internal transcribed spacers of rRNA genes. J. Clin. Microbiol. 40:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Read, A. F., and L. H. Taylor. 2001. The ecology of genetically diverse infections. Science 292:1099-1102. [DOI] [PubMed] [Google Scholar]
- 28.Ready, P. D. 2000. Sand fly evolution and its relationship to Leishmania transmission. Mem Inst. Oswaldo Cruz 95:589-590. [DOI] [PubMed] [Google Scholar]
- 29.Ridley, D. S. 1987. Pathology, p. 666-701. In W. Peters and R. Killick-Kendrick (ed.), The leishmaniases in biology and medicine, vol. 2. Academic Press, Ltd., London, England.
- 30.Rioux, J.-A., G. Lanotte, E. Serres, F. Pratlong, P. Bastien, and J. Perieres. 1990. Taxonomy of Leishmania. using isoenzymes: suggestions for a new classification. Ann. Parasitol. Hum. Comp. 65:111-125. [DOI] [PubMed] [Google Scholar]
- 31.Ryan, L., R. Lainson, and J. J. Shaw. 1987. Leishmaniasis in Brazil. XXIV. Natural flagellate infections of sandflies (Diptera: Psychodidae) in Para State, with particular reference to the role of Psychodopygus wellcomei as a vector of L. braziliensis braziliensis in the Serra dos Carajas. Trans. R. Soc. Trop. Med. Hyg. 81:353-359. [DOI] [PubMed] [Google Scholar]
- 32.Sacks, D. L., G. Modi, E. Rowton, G. Spath, L. Epstein, S. J. Turco, and S. M. Beverley. 2000. The role of phosphoglycans in Leishmania-sand fly interactions. Proc. Natl. Acad. Sci. USA 97:406-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schonian, G., H. Akuffo, S. Lewin, K. Maasho, S. Nylén, F. Pratlong, C. L. Eisenberg, L. F. Schnur, and W. Presber. 2000. Genetic variability within the species Leishmania aethiopica does not correlate with clinical variations of cutaneous leishmaniasis. Mol. Biochem. Parasitol. 106:239-248. [DOI] [PubMed] [Google Scholar]
- 34.Schonian, G., L. Schnur, M. El Fari, L. Oskam, A. A. Kolesnikov, W. Sokolowska-Kohler, and Presber, W. 2001. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Trans. R. Soc. Trop. Med. Hyg. 95:217-224. [DOI] [PubMed] [Google Scholar]
- 35.Sneath, P. H., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman & Co., San Francisco, Calif.
- 36.Thomaz-Soccol, V., G. Lanotte, J. A. Rioux, F. Pratlong, A. Martini-Dumas, and E. Serres. 1993. Phylogenetic taxonomy of New World Leishmania. Ann. Parasitol. Hum. Comp. 68:104-106. [PubMed] [Google Scholar]
- 37.Woolhouse, M. E., L. H. Taylor, and D. T. Haydon. 2001. Population biology of multihost pathogens. Science 292:1109-1112. [DOI] [PubMed] [Google Scholar]