Abstract
During animal development, regions of the embryo become committed to position-specific identities, which are determined by spatially restricted expression of Hox/homeotic genes. This expression pattern is initially established by the activity of the segmentation genes and is subsequently maintained during the proliferative stage through the action of transcription factors encoded by the trithorax (trx) and Polycomb (Pc) groups of genes. trithorax (trx)and ash1 (absent, small, or homeotic 1) are members of the Drosophila trx group. Their products are associated with chromosomes and are believed to activate transcription of target genes through chromatin remodeling. Recently, we reported molecular studies indicating that TRX and ASH1 proteins act in concert to bind simultaneously to response elements located at close proximity within the same set of target genes. Extension of these and other studies to mammalian systems required identification and cloning of the mammalian homologue of ash1 (the mammalian homologue of trx, ALL-1, was previously cloned). We have identified a human expressed sequence tag (EST) clone with similarity to the SET domain of Drosophila ASH1, and used it to clone the human gene. huASH1 resides at chromosomal band 1q21. The gene is expressed in multiple tissues as an ≈10.5-kb transcript and encodes a protein of 2962 residues. The protein contains a SET domain, a PHD finger, four AT hooks, and a region with homology to the bromodomain. The last region is not present in Drosophila ASH1, and as such might confer to the human protein a unique additional function. Using several anti-huASH1 Ab for immunostaining of cultured cells, we found that the protein is distributed in intranuclear speckles, and unexpectedly also in intercellular junctions. Double-immunofluorescence labeling of huASH1 and several junctional proteins localized the huASH1 protein into tight junctions. The significance of huASH1 dual location is discussed. In particular, we consider the possibility that translocation of the protein between the junctional membrane and the nucleus may be involved in adhesion-mediated signaling.
Specificity of body segment identities is defined during embryogenesis by the activities of homeotic genes (HOM-C/HOX). Transcription of these genes is highly regulated. It is initially determined at the blastoderm stage by the transiently expressed segmentation genes and is subsequently maintained by the combined activity of the trithorax group (trxG) and Polycomb group (PcG) of genes. Genes of these two groups, such as trithorax and polycomb, act as transcriptional activators and repressors, respectively (reviewed in refs. 1–3). trxG and PcG proteins function in part by assembly into multiprotein complexes that modulate chromatin structure. Drosophila complexes that have been already characterized include SWI/SNF, containing the trxG proteins BRAHMA, SNR1, and MOIRA (4); the NURF complex, containing the GAGA protein encoded by trxG Trl (5); the PCR1 complex, encompassing the products of PcG Polycomb, posterior sex combs, polyhomeotic, and sex combs on midleg (6); and a complex containing PcG E(Z) and ESC (7). SWI/SNF and PCR1 appear to compete with each other for binding to the nucleosomal template (6). The binding of the protein products of Drosophila trxG and PcG genes to many sites on salivary gland polytene chromosomes (8–11) suggests that their targets are numerous and not limited to homeotic genes.
Many vertebrate homologues of trxG and PcG genes have been cloned and characterized during the past several years (reviewed in refs. 12–14). Mice mutated in these genes show classical homeotic transformations affecting the skeleton, as well as a variety of hematopoietic defects altering proliferation/survival of blood cells (reviewed in refs. 12–14). Disruption of mammalian trxG/PcG genes directly affects transcription of their targets, such as HOX gene clusters. An additional target of the PcG bmi-1 gene (15) is the INK4α locus, which encodes the tumor suppressors and cell cycle inhibitors p16 and p19Arf, regulating the Rb and p53 genes. The human homologue of Drosophila trx, ALL-1, is directly involved in human leukemia through mechanisms involving gene fusion or partial duplication (16–18).
Drosophila absent, small, or homeotic 1 (ash1) is a trxG gene whose product shares several motifs with the TRX protein (11) and is present within a distinct multiprotein complex (4). Recently, we showed that the ASH1 protein physically interacts with TRX, colocalizes with it on polytene chromosomes, and targets a response element located at close proximity to trx response element within the homeotic gene ubx (19). These results suggested that Drosophila ASH1 and TRX are working in concert. As a first step to begin similar investigation in mammalian species we set out to clone the human homologue of ash1. Here we describe molecular characterization of the gene as well as the surprising finding that the protein product of human ASH1 (huASH1) is present not only in the nucleus but also in intercellular tight junctions.
Materials and Methods
Cell Lines.
Human cell lines studied included the epithelial lines Caco2 and MCF7, both purchased from the American Type Culture Collection, the ovarian carcinoma IGROV1 (20), kindly provided by A. Bershadsky at the Weizmann Institute, and primary foreskin fibroblasts (HFF), which were originally cultured by S. Yamada (National Institutes of Health).
Cloning huASH1.
The SET domain of Drosophila ASH1 (11) spanning residues 1366–1514, was used as a query in a computer search for expressed sequence tag (EST) clones. This search hit three human clones, one of which (I.M.A.G.E. Consortium number 745996) was used as a probe to screen a cDNA library derived from K562 cells (16). Nucleotide sequence of isolated cDNAs was determined on an Applied Biosystems automatic sequencer and analyzed by utilization of the GCG programs blast, fasta, tfasta, motifs, and bestfit.
Northern Blot Analysis.
A human multiple tissue Northern blot kit was purchased from CLONTECH and probed with a huASH1 cDNA spanning 7300–7750 nt. The results were confirmed by using additional cDNAs as probes.
Chromosomal Mapping.
Two specific oligonucleotide primers, 5′-AAC TTC AAA GGC AGG CCA-3′ and 5′-TCA GGA CTG AGG TGC AGT-3′, were designed on the basis of huASH1 sequence information. The primers were examined for PCR amplification of genomic DNAs included in the Genebridge 4 radiation hybrid panel (Research Genetics, Huntsville, AL). In parallel, fluorescence in situ hybridization (FISH) analysis on normal metaphases was performed by using as probes bacterial artificial chromosome (BAC) clones spanning genomic huASH1. The BAC clones were obtained by utilization of the primers mentioned above to screen a human BAC library (Research Genetics).
Immunohistochemical Analysis.
Segments of huASH1 cDNA spanning residues 8–146, 1612–1767, 2296–2407, and 2574–2780, as well as a cDNA of ALL-1 encompassing amino acids 904–995 were inserted into pET vectors (Novagen) and expressed in Escherichia coli. The encoded polypeptides (designated 337, 296, 4273, 4312, and 267, respectively), linked to a tail of 6 histidines, were purified by absorption to a Ni-NTA resin (Qiagen, Chatsworth, CA) and used to immunize rabbits (polypeptide 296 was injected into guinea pigs as well). Abs were purified by absorption and subsequent elution from polypeptide affinity columns or from protein G columns. mAb against β-catenin or γ-catenin were purchased from Transduction Laboratories (Lexington, KY). mAb against cingulin, ZO-1, and desmoplakin were kind gifts of S. Citi and W. Franke.
For fluorescence staining, cells on coverslips were simultaneously fixed and permeabilized in 3% paraformaldehyde and 0.5% Triton X-100 in PBS for 2 min, and postfixed in 3% paraformaldehyde for 20 min. FITC- and Cy3-labeled goat Abs to mouse and rabbit immunoglobulins (Jackson ImmunoResearch) were used as secondary Abs. Stained cultures were examined with an Axiophot microscope equipped with a ×100 1.3NA Plan Apochromat objective. In experiments in which the nuclear speckled patterns of huASH1 and ALL-1 were compared, we applied a system for computerized microscopy and fluorescence ratio imaging (21).
Results
Cloning and Structural Characterization of huASH1.
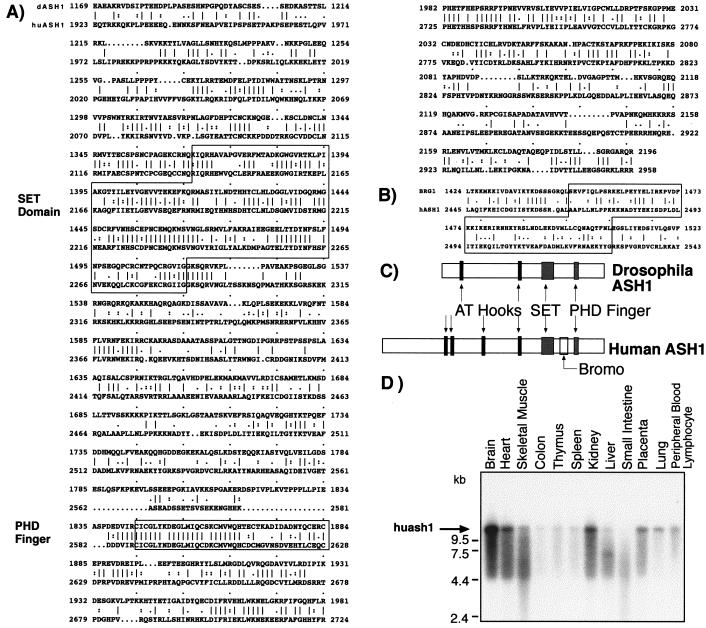
Using a human EST clone homologous to Drosophila ASH1 SET domain we screened a cDNA library and isolated a series of overlapping cDNAs. These and other cDNAs obtained by “walking,” spanned 10.5 kb corresponding to the size of the gene's transcripts (Fig. 1D). Within the cDNA, we identified a single ORF (PenBank accession no. AF257305) delineated between bp 320 and 9280 and encoding 2962 residues. (The residues are shown in Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org.) Applying the bestfit program to compare the predicted amino acid sequences of the human and Drosophila proteins, we found a significant homology between the C-terminal regions spanning 1036 and 1027 residues of the human and fly proteins, respectively (shown in supplemental data and Fig. 1A). The sequence located upstream to the major homology region is not conserved, yet it contains four and two AT hooks in the human and Drosophila proteins, respectively. The two proteins show 66% and 77% similarity in their SET and PHD finger domains, respectively (Fig. 1A). The sequence similarity between the two proteins is not limited to recognizable motifs but extends over the entire C-terminal regions. This sequence similarity and the colinearity of the major motifs (Fig. 1C) strongly suggest that the cDNA we cloned corresponds to the human homologue of Drosophila ash1. Further database search indicated similarity between huASH1 segments spanning residues 38–299 and 771-1987 and the human SET-binding protein (European Molecular Biology Laboratory (EMBL) accession number AB022660). In addition, the region encompassing residues 2445–2543 shows significant similarity to the consensus bromodomain (22, 23). Comparison to this domain within the BRG1 protein (23) is shown in Fig. 1B. Seven of the 10 highly conserved residues of bromodomains (24) are retained in huASH1 [P (2475), Y (2483), P (2490), D (2492), Y (2505), D (2514), and N (2541)]. No homology to the bromodomain was found in Drosophila ASH1.
Figure 1.
Motifs and homologies within huASH1 protein, and transcription analysis. (A) Comparison between the human and Drosophila sequences within the major homology region. (B) huASH1 bromodomain compared with the same motif in BRG1. The region corresponding to the classical bromodomain is boxed. (C) Predicated motifs in the human and Drosophila proteins. (D) Northern blot analysis of huASH1.
Expression and Chromosome Localization.
Northern analysis indicated that huASH1 is expressed as a 10.5-kb RNA in all tissues examined, with the highest levels in brain, kidney, and heart (Fig. 1D).
Radiation hybrid mapping placed the human ash1 gene on chromosome 1, 1.71 cR from D1S305 (logarithm of odds (lod) > 3). The Whitehead Institute database (www-genome.wi.mit.edu/) places D1S305 at 164.1 cM on chromosome 1, within an interval bound by D1S514 (157 cM) and D1S26359 (170 cM). This interval contains several ESTs showing >98% homology to the human ASH1 cDNA and located on 1q12–21 (ncibi.nlm.nih.gov/). FISH analysis on normal human metaphases was performed to verify the location. As probes, we used either of the BAC clones 331M14 and 341F3, spanning the huASH1 locus. This analysis placed the gene at 1q21 (not shown). This region is rearranged in a number of human malignancies including acute leukemias, non-Hodgkin's lymphoma and several solid tumors (25).
Intracellular Distribution of the huASH1 Protein.
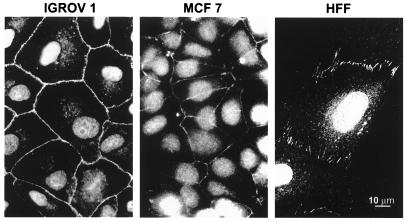
To study the product of huASH1, we raised Ab directed against four segments of the protein (see Materials and Methods). The Abs were used to immunostain several types of human cells, including the epithelial lines Caco2, MCF7, and IGROV1, as well as primary foreskin fibroblasts. Fig. 2 shows the staining of the cell lines with Ab raised in guinea pig against a polypeptide spanning residues 1612–1767. huASH1 protein was detected in two distinct cellular compartments: (i) it was present in a pattern of small speckles uniformly distributed throughout the nucleoplasm; and (ii) it colocated with cell–cell junctions. Similar patterns were obtained when using rabbit Ab directed against huASH1 polypeptides spanning amino acids 8–146, 1612–1767, or 2296–2407 (not shown). Rabbit Ab raised against a polypeptide containing residues 2574–2780 reacted consistently with nuclei but at lower frequency or less brightly with cell junctions. This might be due to masking of the relevant huASH1 epitope in cell junctions.
Figure 2.
Distribution of huASH1 protein in cultured human epithelial (IGROV1 and MCF7) and mesenchimal (HFF) cells. Anti-huASH1 Ab against a polypeptide encompassing residues 1612–1767 was raised in guinea pig.
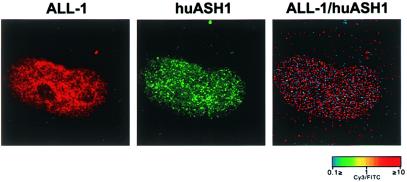
Previously, we showed that the ALL-1 protein distributes in a nuclear punctate pattern (26). That pattern resembles that of huASH1. Therefore, it was of interest to examine whether the two patterns overlap. The large number of dots observed for either protein prevented simple visual comparison of double-stained cells and required the application of the system for computerized microscopy and fluorescence ratio imaging (21). Double immunofluorescence staining was performed with rabbit anti-ALL-1 Ab and guinea pig anti-huASH1 Ab. Secondary Ab were FITC-conjugated goat anti-rabbit Ab and Cy3-conjugated goat anti-guinea pig Ab. FITC and Cy3 images and the ratio image (the ratio of the intensity values for the two proteins in each pixel of the picture) are shown in Fig. 3. In this figure, red and blue dots correspond to sites of ALL-1 and huASH1, respectively; colocalized sites appear yellow. The results indicated no appreciable colocalization of the ALL-1 and huASH1 speckles.
Figure 3.
Distribution of ALL-1 and huASH1 speckles in nuclei of human foreskin fibroblasts (HFF). Cells were double-stained with rabbit and guinea pig Ab against ALL-1 and huASH1, respectively. The relative distributions were examined by fluorescence ratio imaging using digital fluorescence microscopy. The localization of the two proteins is largely mutually exclusive as evidenced by the very small number of yellow spots representing colocalization.
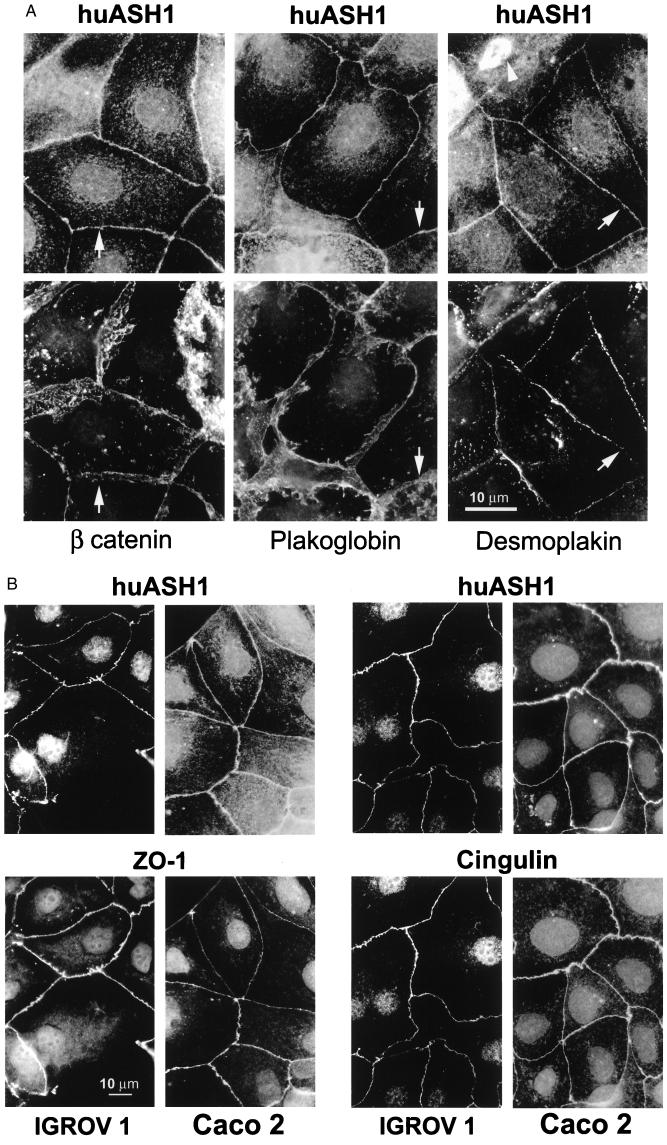
The identification of huASH1 in cell junctions raised the question as to the specific type of junction involved. Cell–cell adhesion junctions are classified into tight junctions that seal cells so as to prevent water and small molecules from leaking from one side of the epithelial sheet to the other and into two types of junctions that link adjacent cells by cadherens-type proteins associated in the cytoplasm with actin filaments (adherens) or with intermediate filaments (desmosomes). These three types of junctions are distinct prominent structures located at close proximity to form the well-established “junctional complex” (27).
To determine the particular cell junction with which huASH1 is associated, we compared the distribution of the latter to that of other proteins present in specific junctions. These proteins included β- and γ-catenin associated with adherens junctions, desmoplakin present in desmosomes, and ZO-1 and cingulin, which are tight junction components. Caco2 or MCF7 cells were double-stained with rabbit anti-huASH1 Ab and with mAb to the aforementioned proteins. The unique staining patterns of each of the junctions enabled direct visual comparison with the huASH1 pattern (Fig. 4). The conclusions drawn were verified by image merging. The distribution of huASH1 in cell junctions was sharp and uninterrupted. It varied from that of β-catenin and plakoglobin, which was wider, and from that of desmoplakin, which was discontinuous (see arrows indicating the regions of variance in Fig. 4A). In contrast, the pattern of huASH1 precisely matched that of ZO-1 and cingulin (Fig. 4B). We conclude that huASH1 localizes to tight junctions. Finally, we notice that, in dividing cells, huASH1 localizes to the mitotic spindle (Fig. 4A, Top Right, pointed by arrow).
Figure 4.
huASH1 protein localizes specifically to tight junctions. (A) Caco2 cells were double-stained with guinea pig Ab against huASH1 together with mAb against the adherens junction proteins β-catenin and plakoglobin, or together with mAb against the desmosomal protein desmoplakin. Note that all junction proteins (including those in B) are localized in the vicinity of each other, being components of the subapical junctional complex. Nevertheless, the distribution of huASH1 varies from that of the other proteins. Regions with the clearest variations are indicated by arrows (see text). It is noteworthy that, in mitotic cells, huASH1 is associated with the mitotic spindle (arrow at Right Top corner). (B) IGROV1 and Caco2 cells double-stained with guinea pig Ab against huASH1 and with mAb against the tight junction proteins Z0-1 or cingulin. Here, the membranal staining patterns appear identical.
Discussion
huASH1 is the fourth mammalian trxG gene to be cloned (14). The protein contains an AT hook, a SET domain and a PHD motif, and as such resembles most closely ALL-1. By analogy to Drosophila ash1 product, the human protein is likely to interact on chromatin with target genes including HOX loci. Unlike its Drosophila homologue, the human protein contains a bromodomain. That domain has been identified in dozens of proteins from yeast to man (23), including every nuclear histone acetylase identified to date. Recently, the three-dimensional structure of P/CAF bromodomain was solved, and the domain was found to interact specifically with acetylated lysin (24). In parallel, a biochemical study showed association between Gcn5 bromodomain and the amino-terminal tails of histones H3 and H4 (28). These results suggested that acetylated lysines in histones or other proteins would target bromodomain-containing proteins and associated complexes, acting in transcriptional control. Variant amino acids in bromodomains could determine the specificity of binding to particular chromatin or protein targets (29). The presence of the bromodomain in human, but not in Drosophila, ASH1 suggests additional protein–protein interaction of the former. That interaction would possibly result in more target genes or in finer tuning of huASH1 activity. This situation is reminiscent of the presence of transcriptional activating and repressing motifs [the latter interacting with histone deacetylase (30)] in ALL-1, but not in its Drosophila homologue TRX (31).
The huASH1 protein was detected in nuclei. This location was expected because its Drosophila homologue is associated with polytene chromosomes (11, 19). Also, the motifs identified within huASH1 are associated with DNA binding (AT hooks) and with chromatin-linked proteins (SET, PHD fingers, and bromodomain). Within the nucleus, huASH1 is distributed in a pattern of many (>100) small speckles. This pattern varies from most known punctate patterns (32) but resembles that of ALL-1 (26) and is comparable to that of some Drosophila PcG proteins (33), and occasionally to that of several human PcG proteins (34). Because the Drosophila homologs of huASH1 and ALL-1 appear to act in concert (19), it was not unreasonable to expect that the two human proteins would colocalize in the speckles. However, colocalization was not observed. In this context, we note that the function of speckles and whether they are associated with chromatin is not known (32). In addition, in contrast to PcG proteins colocalized in speckles, and also present within a single multiprotein complex (6, 33, 34), huASH1 and ALL-1 are components of different multiprotein complexes (T.N. and S.T., unpublished data).
Our most interesting finding is that huASH1 protein is present not only in nuclei but also in cell–cell tight junctions. Tight junctions are found in epithelial and endothelial cells (35) and serve two primary functions: sealing the space between adjacent cells, thereby restricting the movement of molecules across cell sheets, and acting as a boundary within the membrane to separate apical and basolateral domains, which are differentiated to allow active transport across the sheet. Ten proteins have been identified as components of tight junctions (reviewed in refs. 36 and 37). In addition, the junctions are attached to actin filaments. Three of the components, Z0-1, Z0-2, and Z0-3, are members of the MAGUK protein family (reviewed in refs. 37 and 38). Based on analysis of some of these family members, in particular DlgA in Drosophila and LIN2 in Caenorhabditis elegans, the MAGUK proteins are thought to act as molecular scaffolds for spatial organization of signal transduction pathways (37, 38). The nature of these pathways in vertebrates has not been elucidated yet. The identification of huASH1 in tight junctions makes it a candidate to be involved in that hypothetical pathway. It could be translocated after adhesion-mediated signaling from the membrane to the nuclei and there act to directly activate transcription of target genes. Conversely, it could be translocated away from the nucleus to attenuate its activity there and/or to participate in assembly or organization of tight junctions in the membrane. The function of huASH1 in tight junctions could be determined in the future by generating mutant mice or by overexpressing mutant forms of the protein in cultured cells. We note that two other tight junctions proteins, symplekin and ZO-1, were also found to reside within both the membrane and the nucleus (39, 40). Moreover, the intranuclear dot patterns of these two proteins are similar to that of huASH1. It will be of interest to determine whether the three proteins are physically associated in cell junctions and/or in nuclei, and to examine whether they colocalize in nuclear speckles.
A scenario in which huASH1 is translocated from cell junctions to the nuclei would be comparable to the situation involving the Notch protein. The latter is an evolutionary conserved transmembrane receptor that regulates cell fate decisions executed through intercellular communications (reviewed in ref. 41). On ligand activation, Notch intracellular domain is cleaved and migrates into the nucleus to bind and activate DNA-binding transcription factors (42, 43).
An alternative interpretation for huASH1 dual location is that it has two unrelated functions. Thus, the protein might act in most or all cells as a transcription factor, and also can be recruited for the assembly of tight junctions in epithelial and endothelial cells. Examples for proteins of that type appear to be β-catenin and the highly related γ-catenin (plakoglobin) (reviewed in refs. 44–46). Thus, β-catenin is detected mainly in adherens junctions where it links (via α-catenin) the membrane-anchored cadherin to actin filaments. Cytoplasmic β-catenin is present within a complex containing APC, axin, and the GSK kinase. On phosphorylation of β-actin by GSK, it is degraded by the ubiquitin-proteosome system. When Wnt signaling is activated by binding of an extracellular Wnt ligand to a membrane receptor, GSK is inhibited, and cytoplasmic β-catenin is stabilized. Increased levels of the latter lead to its nuclear translocation and subsequent binding to and transactivation of the LEF/TCF transcription factor.
Supplementary Material
Acknowledgments
We are indebted to Dr. Alexander Bershadsky for his continuous interest and help. These studies were supported by National Cancer Institute Grant CA50507 and by grants from the Israel Academy of Science, Israel Cancer Research Fund, The Louis and Fannie Tolz Fund for collaboration between The Weizmann Institute of Science and Jefferson Medical College, Deutsches Krebsforschungszentrum (DKFZ), the Minerva Foundation, and the Yad Abraham Center for Cancer Diagnosis and Therapy.
Abbreviation
- BAC
bacterial artificial chromosome
- EST
expressed sequence tag
References
- 1.Kennison A J. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 2.Simon J. Curr Opin Cell Biol. 1995;7:376–395. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 3.Pirrota V. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- 4.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 5.Tsukiyama T, Daniel C, Tamkun J, Wu C. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 6.Shao Z, Raible F, Mollaaghababa R, Guyon J R, Wu C-t, Bender W, Kingston R E. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 7.Jones C A, Ng J, Peterson A J, Morgan K, Simon J, Jones R S. Mol Cell Biol. 1998;18:2825–2834. doi: 10.1128/mcb.18.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franke A, Decamillis M, Zink D, Cheng S M, Brock H W, Paro R. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rastelli L, Chan C S, Pirotta V. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuzin B, Tillib S, Sedkov Y, Mizrokhi L, Mazo A. Genes Dev. 1994;8:2478–2490. doi: 10.1101/gad.8.20.2478. [DOI] [PubMed] [Google Scholar]
- 11.Tripoulas N, Lajeunesse D, Gildea J, Shearn A. Genetics. 1996;143:913–928. doi: 10.1093/genetics/143.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gould A. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 13.Schumacher A, Magnuson T. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 14.Van Lohuizen M. Curr Opin Genet Dev. 1999;9:355–361. doi: 10.1016/s0959-437x(99)80053-7. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs J J, Keiboom K, Marino S, De Pinho R A, van Lohuizen M. Nature (London) 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 16.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 17.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 18.Schichmann S A, Canaani E, Croce C M. J Am Med Assoc. 1995;273:571–576. [PubMed] [Google Scholar]
- 19.Rozovskaia T, Tillib S, Smith S, Sedkov Y, Rozenblatt-Rosen O, Petruk S, Yano T, Nakamura T, Ben-simchon L, et al. Mol Cell Biol. 1999;19:6441–6447. doi: 10.1128/mcb.19.9.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma J, Maliepaard M, Kolker H J, Verweij J, Schellens J H. Cancer Chemother Pharmacol. 1998;41:186–192. doi: 10.1007/s002800050727. [DOI] [PubMed] [Google Scholar]
- 21.Zamir E, Katz B-Z, Aota S I, Yamada K M, Geiger B, Kam Z. J Cell Sci. 1999;112:1655–1669. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
- 22.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. Nucleic Acids Res. 1992;20:2063. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeanmougin F, Wurtz J-M, Le Douarin B, Chambon P, Losson R. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 24.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M-M. Nature (London) 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 25.Mitelman F. Catalog of Chromosome Aberrations in Cancer. 5th Ed. New York: Wiley–Liss; 1994. [Google Scholar]
- 26.Yano T, Nakamura T, Blechman J, Sorio C, Dang C V, Geiger B, Canaani E. Proc Natl Acad Sci USA. 1997;94:7286–7291. doi: 10.1073/pnas.94.14.7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular Biology of the Cell. 3rd Ed. New York: Garland; 1995. [Google Scholar]
- 28.Ornagh P, Ballario P, Lena A M, Gonzalez A, Filetici P. J Mol Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 29.Winston F, Allis C D. Nat Struct Biol. 2000;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 30.Fuks F, Burgers W A, Brehn A, Hughes-Davies L, Kouzarides T. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 31.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce C M, Canaani E. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamond A I, Earnshaw W C. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 33.Buchenau P, Hodgson J, Strutt H, Arnalt-Jovin D J. J Cell Biol. 1998;141:489–481. doi: 10.1083/jcb.141.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saurin A J, Shiels C, Williamson J, Satijn D P E, Otte A P, Sheer D, Freemont P S. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farquhar M G, Palade G E. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson B R, Keon B H. Annu Rev Cell Dev Biol. 1998;14:89–109. doi: 10.1146/annurev.cellbio.14.1.89. [DOI] [PubMed] [Google Scholar]
- 37.Mitic L L, Anderson J M. Annu Rev Physiol. 1998;60:121–142. doi: 10.1146/annurev.physiol.60.1.121. [DOI] [PubMed] [Google Scholar]
- 38.Dimitratos S D, Woods D F, Stathakis D G, Bryant P J. Bioassays. 1999;21:912–921. doi: 10.1002/(SICI)1521-1878(199911)21:11<912::AID-BIES3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 39.Keon B H, Schäfer S, Kuhn C, Grund C, Franke W W. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottard C J, Arpin M, Fanning A, Louvard D. Proc Natl Acad Sci USA. 1996;93:10779–10784. doi: 10.1073/pnas.93.20.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kopan R, Turner D L. Curr Opin Neurobiol. 1996;6:594–601. doi: 10.1016/s0959-4388(96)80090-0. [DOI] [PubMed] [Google Scholar]
- 42.Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–388. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 43.Kidd S, Lieber T, Young M W. Genes Dev. 1998;12:3728–3790. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Ze'ev A, Geiger B. Curr Opin Cell Biol. 1998;10:629–639. doi: 10.1016/s0955-0674(98)80039-2. [DOI] [PubMed] [Google Scholar]
- 45.Willert K, Nusse R. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 46.Cox R T, Peifer M. Curr Biol. 1998;8:R140–R144. doi: 10.1016/s0960-9822(98)70081-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.