Abstract
Swine influenza is an acute respiratory disease caused by type A influenza viruses. Before 1998, swine influenza virus isolates in the United States were mainly of the classical H1N1 lineage. Since then, phylogenetically distinct reassortant H3N2 viruses have been identified as respiratory pathogens in pigs on U.S. farms. The H3N2 viruses presently circulating in the U.S. swine population are triple reassortants containing avian-like (PA and PB2), swine-like (M, NP, and NS), and human-like (HA, NA, and PB1) gene segments. Recent sequence data show that the triple reassortants have acquired at least three distinct H3 molecules from human influenza viruses and thus form three distinct phylogenetic clusters (I to III). In this study we analyzed the antigenic and pathogenic properties of viruses belonging to each of these clusters. Hemagglutination inhibition and neutralization assays that used hyperimmune sera obtained from caesarian-derived, colostrum-deprived pigs revealed that H3N2 cluster I and cluster III viruses share common epitopes, whereas a cluster II virus showed only limited cross-reactivity. H3N2 viruses from each of the three clusters were able to induce clinical signs of disease and associated lesions upon intratracheal inoculation into seronegative pigs. There were, however, differences in the severity of lesions between individual strains even within one antigenic cluster. A correlation between the severity of disease and pig age was observed. These data highlight the increased diversity of swine influenza viruses in the United States and would indicate that surveillance should be intensified to determine the most suitable vaccine components.
Influenza in swine is caused by influenza A viruses. It is an acute respiratory disease whose severity depends on many factors, including host age, virus strain, and secondary infections (3). Influenza A viruses are isolated from a number of other animal host species, including birds, humans, horses, whales, and mink. They are generally host specific. However, although whole viruses rarely cross the species barrier, gene segments can cross this barrier through the process of genetic reassortment or genetic shift. Pigs have been postulated to play an important role in interspecies transmission by acting as the “mixing vessel” for reassortment between viruses specific to different host species (16). Swine support the replication of both human and avian influenza A viruses (10). The viral receptor sialyloligosaccharides on pig tracheal cells possess the chemical linkages preferential for human (N-acetylneuraminic acid-2,3-galactose) and for avian (N-acetylneuraminic acid-2,6-galactose) influenza viruses (6).
Three main subtypes of influenza virus are presently circulating in swine populations in the United States: H1N1, H3N2, and H1N2 (7, 9, 12, 13, 21-23). Most of the H1N1 viruses in North America and Asia belong to the classic swine lineage, which is genetically related to the human H1N1 viruses responsible for the 1918 Spanish influenza pandemic (18, 19). In 1998, influenza viruses of the subtype H3N2 were isolated from swine in multiple states in the United States. The initial North Carolina isolate contained gene segments similar to those of the human (HA, NA, and PB1) and classic swine (NS, NP, M, PB2, and PA) lineages (double reassortant H3N2 viruses). Subsequent isolates from other states contained genes from the human (HA, NA, and PB1), swine (NS, NP, and M), and avian (PB2 and PA) lineages (triple reassortant H3N2 viruses) (23). The double-reassortant H3N2 virus was isolated only from the initial North Carolina outbreak, whereas the triple-reassortant genotype spread throughout much of the country (21).
Sequence analysis of the triple-reassortant swine H3N2 viruses isolated during 1999 showed that their hemagglutinin (HA) molecules belonged to one of three recent phylogenetically distinct human-like HA lineages. On that basis, these H3N2 viruses could be separated into three distinct clusters (I to III) (21). The ongoing reassortment of swine influenza viruses with circulating human strains has implications for both the future surveillance of U.S. swine herds and for the efficacy of present vaccines. Additionally, if present swine vaccines are unable to control the spread of all H3N2 virus variants, these viruses may continue to circulate widely and form a lineage of virus that in the future may cross back into the human population.
Although they contained phylogenetically distinct HA molecules, it was not clear whether the H3N2 swine virus clusters were also antigenically distinct. In addition, there have been no comprehensive reports describing the disease caused by the triple-reassortant H3N2 viruses, and the impact of the different HA molecules on their pathogenic potential in pigs is unknown. Therefore, we sought to determine (i) whether the hyperimmune sera of pigs experimentally infected with representative viruses from each H3N2 cluster cross-react with viruses of the other clusters according to hemagglutination inhibition (HI) and neutralization tests and (ii) whether viruses from all three clusters replicate and induce clinical signs and influenza-specific lesions in intratracheally infected pigs of different age groups.
MATERIALS AND METHODS
Virus strains.
All viruses used in this study were obtained from the repository at St. Jude Children's Research Hospital, Memphis, Tenn. These viruses were A/Swine/Texas/4199-2/98 (Sw/TX/98 [H3N2]), a cluster I virus; A/Swine/Colorado/23619/99 (Sw/CO/99 [H3N2]), a cluster II virus; and A/Swine/Oklahoma/18089/99 (Sw/OK/99 [H3N2]) and A/Swine/Illinois/21587/99 (Sw/IL/99 [H3N2]), both cluster III viruses. Viruses for use as antigens in HI and virus neutralization assays were grown in the allantoic cavities of 11-day-old embryonated chicken eggs or in MDCK cells. Viruses used to inoculate pigs were obtained from intranasally infected (104 PFU/pig) 11-day-old, caesarian-derived, colostrum-deprived (CDCD) piglets. Bronchoalveolar lavage fluid (BALF) was harvested from the infected piglets 24 to 120 h postinfection (p.i.) for use as inoculum.
Hyperimmune pig sera.
Pig hyperimmune serum specific for the Sw/TX/98 H3N2 virus was obtained from the National Veterinary Services Laboratories in Ames, Iowa. Hyperimmune sera against the Sw/CO/99 and Sw/OK/99 viruses were produced in CDCD piglets intranasally inoculated with 104 PFU/pig when 11 days old. The animals were given a boost dose of 104 PFU/pig after 2 weeks and were sacrificed 9 days later. Blood was collected at necropsy, and aliquots of serum were stored at −20°C.
Serologic testing.
The collected sera were tested by HI assay (14) for the presence of antibodies to the surface glycoproteins of Sw/TX/98, Sw/OK/99, and Sw/CO/99. All sera were pretreated with the receptor-destroying enzyme from Vibrio cholerae (Denka Seiken, Tokyo, Japan) to prevent interference by nonspecific serum inhibitors. Virus was diluted to four agglutinating doses. For consistency with previously reported studies, HI titers of ≥40 were recorded as positive. For the virus neutralization tests, the allantoic fluid or MDCK cell supernatants containing the Sw/TX/98, Sw/CO/99, or Sw/OK/99 viruses were standardized to 100 50% tissue culture infective doses/reaction and were incubated for 60 min at 37°C with twofold serial dilutions (starting at 1:8) of hyperimmune serum. After being washed with medium, MDCK cells were exposed to the virus-antibody mixtures for 1 h at room temperature. Cell monolayers were then washed with medium, and cultivation medium containing 0.3% bovine serum albumin was added. Infected cells were incubated for 72 h at 37°C and were evaluated by light microscopy for cytopathic effects. The virus neutralization titer was established by the Reed and Muench method (15).
Infection of pigs.
Outbred specific-pathogen-free pigs were obtained from a commercial hog farm in Iowa. Groups of four to six pigs were housed in separate isolation rooms in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center, an American Association for Accreditation of Laboratory Animal Care-accredited facility. At the age of 4 or 12 weeks, pigs were anesthetized by intramuscular injection with a mixture of ketamine, xylazine, zolazepam, and tiletamine (11) before being inoculated intratracheally (via laryngoscope and tracheal tube) with 1 ml of virus solution containing 1 × 103, 1 × 104, or 7 × 104 PFU. Pigs were observed for 5 days p.i. and were then sacrificed.
Clinical observation and sampling.
For 5 days, pigs were monitored for clinical signs, including lethargy, anorexia, coughing, hyperpnea or dyspnea, and nasal and ocular discharge, and their body temperature was recorded daily. Blood, BALF, and nasal swabs were collected during necropsy. BALF was obtained after the lungs were removed from the thoracic cavity by rinsing the lungs (via trachea) with ca. 30 ml of McCoy's medium without serum. Nasal swabs and BALF were tested to determine virus burden.
Virus titration.
Tenfold serial dilutions of BALF, starting at 1:10, were prepared in cell culture infection medium (McCoy's medium without serum, supplemented with 5 μg of trypsin/ml). MDCK cells (with medium plus trypsin) were inoculated with the dilutions and were incubated in microtiter plates at 37°C for 72 h. The plates were examined for cytopathic effects after 72 h. Virus titers were calculated by the Reed and Muench method (15).
Necropsy and macroscopic examination of lungs.
Animals were euthanized 5 days p.i. by intravenous administration of pentobarbital (Fort Dodge Animal Health, Fort Dodge, Iowa), and their lungs were removed in toto. The percentage of the surface of each pulmonary lobe that was affected by macroscopic lesions was estimated visually. The percentages were averaged to obtain a mean value for the seven pulmonary lobes of each animal. The nasal turbinates, trachea, and right cardiac lobe were removed and were formalin fixed for further histopathologic evaluation.
Histopathologic evaluation.
The nasal turbinates and trachea were examined for epithelial changes and subepithelial inflammation. The lungs were examined for bronchiolar epithelial changes and peribronchiolar inflammation in large, medium, and small or terminal bronchioles. The alveoli were also evaluated for inflammatory changes. Because lesions were found most consistently in medium-sized airways, data obtained from the medium bronchioles were used for comparisons.
The medium bronchioles were graded on a scale of 0 to 3+ as follows: 0 (normal: lined by medium-to-tall columnar epithelial cells with ciliated apical borders and basal pseudostratified nuclei; minimal inflammation); 1+ (epithelial layer columnar even in outline with only slightly increased proliferation; cilia still visible on many cells); 2+ (prominent changes in the epithelial layer ranging from attenuation to marked proliferation; cells disorganized and layer outline irregular at the luminal border); and 3+ (epithelial layer markedly disrupted and disorganized with necrotic cells visible in the lumen; some bronchioles attenuated and others in marked reactive proliferation). One very experienced examiner was utilized for the evaluation of tissue sections. The examiner was unaware of the group of animal from which the tissues were derived.
Statistical analysis.
Mean body temperatures, extent of gross and histopathologic changes, and virus replication in infected and control groups were compared by using the two-sided Student's t test. P values of ≤0.05 were considered to indicate a statistically significant difference between groups.
RESULTS
Serologic cross-reactivity among swine H3N2 viruses of different clusters.
Webby et al. (21) reported that the newly emergent triple-reassortant swine influenza viruses are associated with three phylogenetically distinct human-like HA molecules that allow the viruses to be grouped into three clusters. To determine whether viruses of the three clusters are cross-reactive, we used HI and neutralization assays to test hyperimmune sera produced in CDCD piglets against viruses representative of the three clusters. Sw/TX/98, Sw/CO/99, and Sw/OK/99 hyperimmune sera reacted with their homologous virus antigens at high titers (Table 1). In contrast, the Sw/CO/99 hyperimmune sera failed to react with the Sw/TX/98 virus antigen (HI titer, <10) and reacted at a low HI titer with the Sw/OK/99 antigen (HI titer, 80). Similarly, hyperimmune sera from Sw/TX/98- and Sw/OK/99-immunzed pigs failed to react with Sw/CO/99 antigen (HI titers, <10/<10 and 20/40, respectively). On the other hand, sera from Sw/TX/98-immunized pigs reacted to a high titer with Sw/OK/99 virus antigen and vice versa. To verify these results we conducted virus neutralization tests, which produced results similar to those of the HI tests. Sw/TX/98, Sw/CO/99, and Sw/OK/99 hyperimmune sera reacted with the homologous viruses to high titers (≥256) in neutralization tests. When Sw/CO/99 serum was incubated with Sw/TX/98 or Sw/OK/99 viruses, neutralization activity was absent or low (≤8). Similarly, hyperimmune sera from Sw/TX/98- or Sw/OK/99-immunized pigs did not considerably neutralize the cluster II Sw/CO/99 virus. In contrast, a high titer of neutralizing activity was observed when sera from Sw/TX/98-immunized pigs were incubated with Sw/OK/99 virus (≥256) and vice versa (≥512). Therefore, cluster I and cluster III viruses appear to be antigenically similar, whereas cluster II virus is antigenically dissimilar to both cluster I and III viruses.
TABLE 1.
HI titers of different H3N2 swine influenza virus cluster-specific hyperimmune seraa
| Cluster | Virus antigen | Titer for:
|
||
|---|---|---|---|---|
| Sw/TX/98 | Sw/CO/99 | Sw/OK/99 | ||
| I | Sw/TX/98 | 320/320 | <10/<10 | 320/320 |
| II | Sw/CO/99 | 10/10 | 2,560/1,280 | 20/40 |
| III | Sw/OK/99 | 320/320 | 80/80 | 1,280/1,280 |
Numbers represent duplicate experiments with the same sera.
Infection of 4-week-old pigs with different clusters of H3N2 swine influenza virus.
Forty-five 4-week-old outbred pigs were purchased from a commercial hog farm in Iowa. HI tests with various H3N2 antigens and a classical H1N1 antigen showed all animals to be negative for influenza virus-specific antibodies. A group of 12 pigs was infected with each of the pig-adapted Sw/TX/98, Sw/OK/99, and Sw/IL/99 viruses; six pigs in each group received a “low” dose of 103 PFU, and six received a “high” dose of 104 PFU. Because limited amounts of Sw/CO/99 virus were available, only six pigs were infected with this virus (103 PFU per pig). Three animals were mock infected with medium only. The mean rectal temperature of each infected group increased to ≥40°C 1 to 3 days p.i. (Table 2). Clinical signs, which comprised respiratory distress, nasal secretion, conjunctivitis, and coughing, began on days 2 to 4 p.i. The prevalence of clinical signs differed with the viruses. Most of these signs were consistently observed in the groups infected with Sw/OK/99 and Sw/TX/98, and some of the signs were observed in the Sw/CO/99 and Sw/IL/99 cohorts.
TABLE 2.
Average rectal temperature of groups of six 4-week-old pigs infected with a low dose (103 PFU) of various H3N2 swine influenza viruses
| Cluster | Virus | Average rectal temp (°C) on day p.i.
|
|||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | ||
| I | Sw/TX/98 | 39.2 | 39.8 | 40.0 | 39.8 | 39.6 | 39.7 |
| II | Sw/CO/99 | 39.2 | 39.5 | 39.8 | 40.0 | 39.8 | 40.0 |
| III | Sw/OK/99 | 39.0 | 40.3 | 40.2 | 40.4 | 40.1 | 39.7 |
| III | Sw/IL/99 | 39.2 | 39.7 | 40.0 | 40.1 | 39.8 | 40.0 |
| None | Controla | 39.2 | 39.2 | 39.0 | 39.1 | 39.1 | 39.2 |
Control pigs were inoculated with medium only.
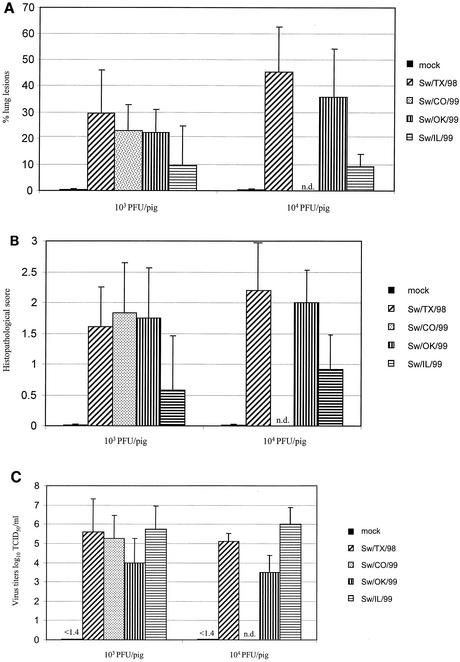
During necropsy, the percentage of each lung surface with macroscopic lesions was calculated. As shown in Fig. 1A, mock-infected control animals had no macroscopic lung lesions. Pigs infected with H3N2 cluster I, II, or III virus demonstrated macroscopic lung lesions in both the low-dose (103 PFU per pig) and high-dose (104 PFU per pig) groups. Sw/TX/99 virus-infected animals had the most extensive lesions in both dose groups; pigs given the high-dose inoculum had a higher percentage of lung surface affected by lesions, but we observed no significant difference between the dose groups (P = 0.18). Similarly, Sw/CO/99, Sw/OK/99, and Sw/IL/99 viruses induced a moderate-to-severe extent of lung disease, and again no significant difference was seen between the low-dose and high-dose animals. Interestingly, there were marked differences between the lung lesion profiles of the two cluster III prototype viruses Sw/OK/99 and Sw/IL/99. Sw/OK/99 virus infection caused noticeably more extensive lung lesions at both doses, and the difference was statistically significant at the high dose (P = 0.01). No statistically significant difference was seen between any of the groups infected with different cluster viruses, with the exception of Sw/TX/98 and Sw/IL/99. In the high-dose group, Sw/TX/98 virus-infected animals showed significantly more extensive lesions than did Sw/IL/99 virus-infected animals (P < 0.01). In general, the gross lesions that we observed were marked, plum colored, consolidated areas on individual lobes. The diaphragmatic lobes were less involved than the other lobes. The mediastinal lymph nodes were usually hyperemic and enlarged.
FIG. 1.
Infection of 4-week-old pigs with H3N2 cluster I, II, and III swine influenza viruses. (A) Mean percentage of lung surface with macroscopic lesions on day 5 p.i. (B) Microscopic lesions in the medium bronchioles on day 5 p.i. (C) Virus titers in BALF on day 5 p.i. TCID50, 50% tissue culture infective dose. n.d., not done.
The medium bronchioles were microscopically examined in tissue sections of the right cardiac pulmonary lobe and were histopathologically scored (Fig. 1B). Mock-infected animals showed no lesions, whereas lesions were detected in all animals infected with cluster I, II, or III virus (Fig. 2 through 5). Animals infected with low-dose or high-dose Sw/TX/99 and Sw/OK/99 virus and with low-dose Sw/CO/99 virus had high scores, reflecting disruption of the bronchial epithelial cell layer (characterized by acute epithelial necrosis or subsequent attenuation or reactive proliferation). However, no significant difference was detected between the histopathologic scores of the low- and high-dose Sw/TX/99 or Sw/OK/99 virus-infected groups (P ≥ 0.26). Animals infected with either low- or high-dose Sw/IL/99 virus had a moderate score, reflecting slightly increased proliferation of the bronchial epithelial layer; again, no significant difference was seen between low- and high-dose infections. As in the macroscopic lung lesions, Sw/IL/99 virus-infected pigs showed significantly less damage than did Sw/OK/99 virus-infected pigs in both the low-dose (P = 0.04) and high-dose (P < 0.01) cohorts, even though both viruses belong to the same phylogenetic cluster. In comparison to Sw/IL/99 virus-infected animals, Sw/TX/99 virus-infected animals had significantly higher histopathology scores at both doses (P ≤ 0.02), and Sw/CO/99 virus-infected animals had a significantly higher score at the low dose (P = 0.01).
FIG. 2.
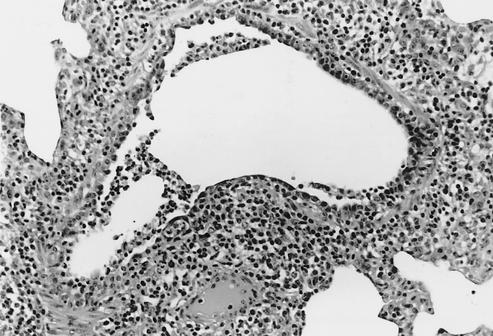
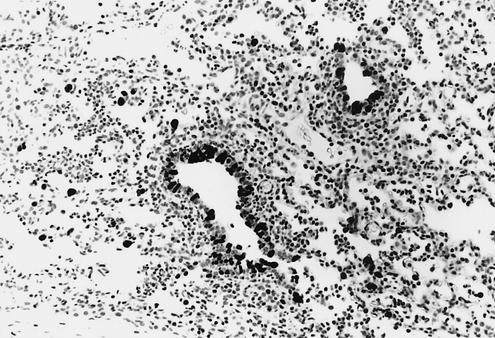
Medium-sized bronchiole from a 4-week-old pig 5 days p.i. with Sw/TX/98 virus (cluster I). The airway is lined irregularly with immature cuboidal epithelial cells. The lumen contains sloughed necrotic epithelial cells and a few neutrophils. A loose cuff of infiltrating lymphocytes surrounds the bronchiole (magnification, ×149).
FIG. 5.
Medium-sized bronchiole from 4-week-old pigs 5 days p.i. with Sw/OK/99 virus (cluster III). (A) Necrosis and sloughing of epithelial cells have resulted in prominent attenuation of the remaining epithelial cell layer. The lumen contains proteinaceous debris, sloughed necrotic epithelial cells, and a few leukocytes. Lymphocytes surround the airway (magnification, ×149). (B) Higher magnification of a bronchiole. Epithelial layer is severely disrupted by necrosis and sloughing of cells into airway lumen, which contains cellular debris, proteinaceous fluid, and a few leukocytes. At this stage of infection, much of the acute necrosis has already occurred, and attenuation and proliferation of the remaining epithelial cells and underlying connective tissue predominate (magnification, ×204).
Virus titers in BALF were also analyzed. As shown in Fig. 1C, all viruses replicated in the respiratory tract of pigs and no statistically significant difference was observed between the low- and high-dose groups (P ≥ 0.5). Interestingly, BALF virus titers were significantly higher in animals infected with Sw/IL/99 virus than in those infected with Sw/OK/99 virus, regardless of dose (P < 0.04). This finding is of particular interest because the Sw/IL/99 virus induced significantly less extensive gross and microscopic disease. In addition, in the high-dose group, the virus titers of the Sw/TX/99 virus-infected animals were significantly higher than those of the Sw/OK/99 virus-infected animals (P < 0.01); no significant differences were found between the low- and high-dose groups infected with the other viruses.
Infection of 12-week-old pigs with H3N2 swine influenza viruses of different clusters.
To determine the age-dependent pathogenesis of cluster I, II, and III H3N2 viruses, we purchased 39 12-week-old pigs from a commercial hog farm in Iowa. All animals tested negative by HI for antibodies to H3N2 viruses and to a classical H1N1 virus. Eleven pigs were infected with each CDCD piglet-adapted strain at a dose of 104 PFU/pig (Sw/TX/98) or 103 PFU/pig (Sw/CO/99). Postmortem evaluation revealed significantly less lung disease than was observed in 4-week-old pigs who received the same dose of inoculum (data not shown). Subsequently, 28 12-week-old pigs were infected with the Sw/TX/98, Sw/OK/99, and Sw/CO/99 viruses (seven pigs each) at a dose of 7 × 104 PFU/pig or were mock infected and were sacrificed 5 days p.i. To ensure that the same virus isolates were used in these experiments, the Sw/TX/98, Sw/CO/99, and Sw/OK/99 inocula were derived from high-titer BALF obtained from 4-week-old piglets during the previous experiment.
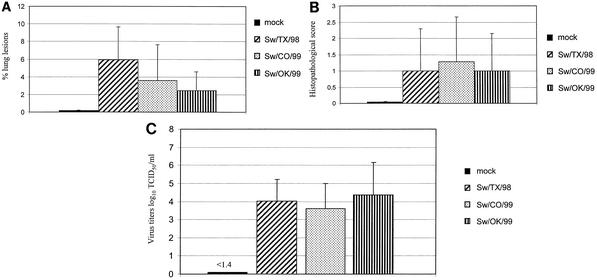
After intratracheal infection, the rectal temperature of the infected pigs rose moderately (≤40°C) 1 to 3 days p.i. Clinical signs of respiratory distress, coughing, and conjunctivitis were observed in some pigs. During necropsy, the extent of macroscopic surface lesions was averaged for the seven lobes of each lung. As shown in Fig. 6A, mock-infected control animals displayed only a negligible extent of macroscopic lung lesions. A moderate extent of macroscopic lung lesions was detected in pigs infected with each of the prototype H3N2 viruses. These lung lesions were significantly less extensive than those observed in the 4-week-old animals infected with the same virus isolate (P < 0.01). Similar results were obtained when the medium bronchioles of the right cardiac pulmonary lobe were examined histopathologically. Mock-infected control animals showed no microscopic lung lesions, whereas animals infected with H3N2 cluster I, cluster II, and cluster III viruses showed mild histologic changes (Fig. 6B). Again, these changes were less severe than those observed in infected 4-week-old animals, although no statistically significant difference was detected. Figure 6C shows the virus titers in the BALF of 12-week-old pigs infected with three H3N2 swine influenza viruses. All viruses replicated in the respiratory tract of the 12-week-old pigs, though to lower titers than in the 4-week-old animals. A significant difference in virus titers between the two age groups was seen only in Sw/CO/99 virus-infected animals (P = 0.05).
FIG. 6.
Infection of 12-week-old pigs with H3N2 cluster I, II, and III swine influenza viruses. (A) Mean percentage of lung surface with macroscopic lesions on day 5 p.i. (B) Microscopic lesions in the medium bronchioles on day 5 p.i. (C) Virus titers in BALF on day 5 p.i. TCID50, 50% tissue culture infective dose.
DISCUSSION
Since the emergence of H3N2 viruses in 1998, the epidemiology of swine influenza virus infections in U.S. pigs has changed rapidly. These newly emergent H3N2 viruses have continued to reassort with human viruses, allowing the creation of three phylogenetically distinct clusters. We found that cluster I and cluster III viruses cause cross-reactive antibody responses, whereas cluster II virus does not cross-react with either cluster I or cluster III prototype virus. It had already been noted by Webby and colleagues (21) that p.i. antiserum specific for Sw/TX/98 virus showed only limited HI cross-reactivity with Sw/CO/99 virus. This study confirmed and extended the initial finding by testing the cross-reactivity of all three clusters. Cluster III hyperimmune serum did not react well with the cluster II prototype virus Sw/CO/99, and the newly produced cluster II hyperimmune serum did not react to high titers with the cluster I and cluster III viruses or antigens.
Antigenic drift of H3N2 swine influenza viruses away from the H3N2 strain used in present commercial swine influenza vaccines has been described in The Netherlands and Belgium (5), and replacement of the vaccine H3N2 strain with a more recent swine H3N2 isolate has been considered. When animals vaccinated with the commercial swine influenza vaccine were challenged with a recent Dutch field strain, the vaccine prevented fever and the transmission of virus to the unchallenged group mates. However, vaccinated pigs excreted influenza virus for a short period after challenge; therefore, the vaccine's protection was suboptimal (5). In a study investigating whether antigenic evolution within H1N1 swine influenza viruses can compromise vaccine efficacy, pigs were vaccinated with experimental monovalent vaccines derived from different H1N1 strains or with a commercial bivalent vaccine (20). Clinical signs after challenge were negligible in all vaccinees, and all strains used for vaccination protected completely if antibody titers against the challenge virus were sufficiently high. The authors concluded that, apart from the vaccine strain, adjuvant and antigenic dose may play a crucial role in vaccine efficacy (20).
Epidemiologic data collected between 1999 and 2002 in U.S. swine populations indicate that cluster II viruses may not be prevalent (unpublished data). The apparent lack of significant numbers of circulating cluster II viruses is fortuitous for vaccination regimens. Our in vitro data suggest that cluster II viruses would be incompletely neutralized by a vaccine derived from the index H3N2 cluster I and III viruses. In contrast, our data would suggest that sufficient cross-protection is present between cluster I and cluster III viruses. It remains to be seen whether any single triple-reassortant strain will predominate in the U.S. swine population and whether the behavior of recent H3N2 viruses is similar to that of the viruses that we tested. What is known, however, is that swine influenza viruses in the United States are continuing to evolve and reassort. Surveillance studies in North American swine have revealed the presence of novel reassortant H1N2 (1, 2, 7) and H1N1 viruses carrying internal genes derived from the H3N2 viruses (R. J. Webby, unpublished data). Additionally, H4N6 viruses have been reported from an isolated outbreak of disease in a Canadian swine herd (8). The reassortment of swine viruses with each other and with circulating human strains has obvious implications for surveillance and vaccine efficacy. It is uncertain how broadly protective a vaccine produced with viruses from one clade of H3N2 isolates will be. Surveillance of the prevalent H3N2 strains will be necessary to determine what H3N2 viruses should be included in vaccine formulations. It will also be necessary to include newly emergent H3N2 viruses or antigens and antisera in swine surveillance, in addition to the index swine H1N1 and H3N2 viruses.
Swine influenza is a disease of the respiratory tract. Although the severity is affected by many factors, including viral strain, the onset of disease is typically sudden, and general signs include anorexia, inactivity, fever, respiratory distress, coughing, conjunctivitis, nasal discharge, and weight loss (4). The disease incubation period is between 1 and 3 days, with rapid recovery beginning 5 to 7 days after onset. Swine influenza is a herd disease characterized by high morbidity (approaching 100%) and generally low mortality (<1%) rates. However, the novel H3N2 viruses introduced into North American swine herds in the late 1990s induced dramatically severe disease in the naïve population, which resulted in abortion and an unusually high mortality rate in mature sows (B. H. Janke, unpublished data). Although infection of swine with these viruses caused severe disease, to our knowledge there have been no peer-reviewed reports describing the clinical and pathological effects. In our study, most of the symptoms typical of influenza virus infection were observed. Under experimental conditions, the 4-week-old pigs were more susceptible than the 12-week-old animals, as shown by clinical signs and by macroscopic and microscopic lesions in the respiratory tract. On day 5 p.i., microscopic examination showed changes in the bronchiolar epithelial layer, ranging from attenuation to marked proliferation. Epithelial cells were disorganized, and the epithelial layer was sometimes markedly disrupted, with necrotic cells observed in the lumen. All swine influenza viruses replicated in the lungs of pigs. Both gross and microscopic lesions were less pronounced in the 12-week-old group, although there was no difference between virus replication in the lungs of 4-week-old animals and that in 12-week-old animals infected with Sw/TX/98 or Sw/OK/99 virus. The difference in viral replication between the 4-week-old and 12-week-old animals infected with Sw/Co/99 virus was statistically significant only at the 95% confidence level (P = 0.05) and not at the 99% confidence level. These findings clearly demonstrate that there was no correlation between the levels of virus replication and clinical disease. The age-dependent susceptibility of pigs to swine influenza virus-induced signs and lesions may reflect a superior immune response to the virus or a lesser susceptibility to virus-mediated pulmonary damage in the older age group. Environmental and immunologic differences, as well as the presence of other pathogens, could play an important role in susceptibility. The fact that Sw/IL/99 virus showed higher replication titers than Sw/OK/99 in 4-week-old-pigs but significantly fewer macroscopic and microscopic lung lesions may be attributed to the individual growth curve of each virus in pigs and the fact that only one time point was analyzed (day 5 p.i.). The different pathogenicity of these two cluster III viruses also highlights the multigenic nature of influenza pathogenicity.
The presence of phylogenetically and antigenically distinct populations of H3N2 viruses in the U.S. swine population is of concern for human and animal health sectors. The antigenic variability demonstrated by the H3N2 viruses has potential implications for vaccination strategies within the swine industry, and the implications for human health are just as strong. The ability of swine viruses to sporadically enter the human population is well documented (17), and the increase in viral diversity at the swine-human interface translates to a more diverse group of viruses with this potential. Although the future composition of swine influenza in the United States is unclear, it is imperative that surveillance systems are implemented to monitor the present and future situation.
FIG. 3.
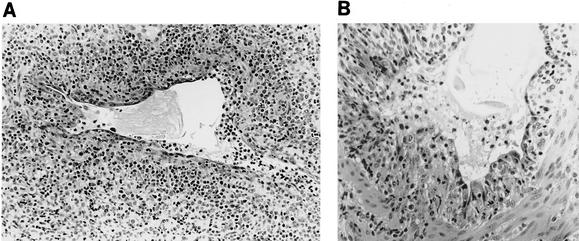
Lung from a 4-week-old pig 3 days p.i. with Sw/TX/98 virus (cluster I). Immunohistochemical staining techniques identify swine influenza virus antigen in epithelial cells in the bronchioles and in scattered pneumocytes in the alveoli. Light lymphocyte infiltration around infected airways is evident (magnification, ×149).
FIG. 4.
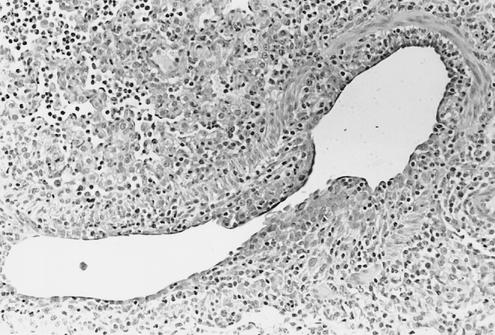
Medium-sized bronchiole from a 4-week-old pig 5 days p.i. with Sw/CO/99 virus (cluster II). The bronchiole is almost completely denuded with the remaining epithelial cells attenuated and irregularly distributed. Low numbers of infiltrating lymphocytes are around the bronchiole and are mixed with neutrophils in adjacent partially collapsed alveoli (magnification, ×149).
Acknowledgments
We thank Sagar Goyal and Sabrina Swenson for the kind gifts of viruses and reagents; Deb Adolphson, Randy Atchison, Deb Clouser, and Ann Vorwald for excellent technical assistance; and Sharon Naron for editorial assistance.
This work was funded in part (R.G.W. and R.J.W.) by NIH grants AI95357 and CA21765 and by the American Lebanese Syrian Associated Charities (ALSAC).
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Choi, Y. K., S. M. Goyal, M. W. Farnham, and H. S. Joo. 2002. Phylogenetic analysis of H1N2 isolates of influenza A virus from pigs in the United States. Virus Res. 87:173-179. [DOI] [PubMed] [Google Scholar]
- 2.Choi, Y. K., S. M. Goyal, S. W. Kang, M. W. Farnham, and H. S. Joo. 2002. Detection and subtyping of swine influenza H1N1, H1N2 and H3N2 viruses in clinical samples using two multiplex RT-PCR assays. J. Virol. Methods 102:53-59. [DOI] [PubMed] [Google Scholar]
- 3.Easterday, B. C. 1980. Animals in the influenza world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 288:433-437. [DOI] [PubMed] [Google Scholar]
- 4.Easterday, B. C., and V. S. Hinshaw. 1992. Swine influenza, p. 349-357. In A. D. Leman (ed.), Diseases of swine, 7th ed. Iowa State University Press, Ames.
- 5.Heinen, P. P., A. P. van Nieuwstadt, E. A. de Boer-Luijtze, and A. T. Bianchi. 2001. Analysis of the quality of protection induced by a porcine influenza A vaccine to challenge with an H3N2 virus. Vet. Immunol. Immunopathol. 82:39-56. [DOI] [PubMed] [Google Scholar]
- 6.Ito, T., Y. Kawaoka, A. Vines, H. Ishikawa, T. Asai, and H. Kida. 1998. Continued circulation of reassortant H1N2 influenza viruses in pigs in Japan. Arch. Virol. 143:1773-1782. [DOI] [PubMed] [Google Scholar]
- 7.Karasin, A. I., J. Landgraf, S. Swenson, G. Erickson, S. Goyal, M. Woodruff, G. Scherba, G. Anderson, and C. W. Olsen. 2002. Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J. Clin. Microbiol. 40:1073-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karasin, A. I., C. W. Olsen, I. H. Brown, S. Carman, M. Stalker, and G. Josephson. 2000. H4N6 influenza virus isolated from pigs in Ontario. Can. Vet. J. 41:938-939. [PMC free article] [PubMed] [Google Scholar]
- 9.Karasin, A. I., M. M. Schutten, L. A. Cooper, C. B. Smith, K. Subbarao, G. A. Anderson, S. Carman, and C. W. Olsen. 2000. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 68:71-85. [DOI] [PubMed] [Google Scholar]
- 10.Kida, H., T. Ito, J. Yasuda, Y. Shimizu, C. Itakura, K. F. Shortridge, Y. Kawaoka, and R. G. Webster. 1994. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 75:2183-2188. [DOI] [PubMed] [Google Scholar]
- 11.Mengeling, W. L., A. C. Vorwald, K. M. Lager, and S. L. Brockmeier. 1996. Comparison among strains of porcine reproductive and respiratory syndrome virus for their ability to cause reproductive failure. Am. J. Vet. Res. 57:834-839. [PubMed] [Google Scholar]
- 12.Olsen, C. W. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85:199-210. [DOI] [PubMed] [Google Scholar]
- 13.Olsen, C. W., S. Carey, L. Hinshaw, and A. I. Karasin. 2000. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol. 145:1399-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer, D. F., M. T. Coleman, W. R. Dowdle, and G. C. Schild. 1975. Advanced laboratory techniques for influenza diagnosis. Immunology series no. 6. U.S. Department of Health, Education, and Welfare, Washington, D.C.
- 15.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 16.Scholtissek, C. 1994. Source for influenza pandemics. Eur. J. Epidemiol. 10:455-458. [DOI] [PubMed] [Google Scholar]
- 17.Scholtissek, C., V. S. Hinshaw, and C. W. Olsen. 1998. Influenza in pigs and their role as the intermediate host, p. 137-145. In K. G. Nicholson, R. G. Webster, and A. J. Hay (ed.), Textbook of influenza. Blackwell Science, Oxford, United Kingdom.
- 18.Taubenberger, J. K., A. H. Reid, T. A. Janczewski, and T. G. Fanning. 2001. Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taubenberger, J. K., A. H. Reid, A. E. Krafft, K. E. Bijwaard, and T. G. Fanning. 1997. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science 275:1793-1796. [DOI] [PubMed] [Google Scholar]
- 20.Van Reeth, K., G. Labarque, S. De Clercq, and M. Pensaert. 2001. Efficacy of vaccination of pigs with different H1N1 swine influenza viruses using a recent challenge strain and different parameters of protection. Vaccine 19:4479-4486. [DOI] [PubMed] [Google Scholar]
- 21.Webby, R. J., S. L. Swenson, S. L. Krauss, P. J. Gerrish, S. M. Goyal, and R. G. Webster. 2000. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 74:8243-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webby, R. J., and R. G. Webster. 2001. Emergence of influenza A viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou, N. N., D. A. Senne, J. S. Landgraf, S. L. Swenson, G. Erickson, K. Rossow, L. Liu, K. J. Yoon, S. Krauss, and R. G. Webster. 2000. Emergence of H3N2 reassortant influenza A viruses in North American pigs. Vet. Microbiol. 74:47-58. [DOI] [PubMed] [Google Scholar]