Abstract
v-Crk induces cellular tyrosine phosphorylation and transformation of chicken embryo fibroblasts (CEF). We studied the molecular mechanism of the v-Crk-induced transformation. Experiments with Src homology (SH)2 and SH3 domain mutants revealed that the induction of tyrosine phosphorylation of cellular proteins requires only the SH2 domain, but both the SH2 and SH3 domains are required for complete transformation. Analysis of three well defined signaling pathways, the mitogen-activated protein kinase (MAPK) pathway, the Jun N-terminal kinase (JNK) pathway, and the phosphoinositide 3-kinase (PI3K)/AKT pathway, demonstrated that only the PI3K/AKT pathway is constitutively activated in v-Crk-transformed CEF. Both the SH2 and SH3 domains are required for this activation of the PI3K/AKT pathway in CEF. We also found that the colony formation of CEF is strongly induced by a constitutively active PI3K mutant, and that a PI3K inhibitor, LY294002, suppresses the v-Crk-induced transformation. These results strongly suggest that constitutive activation of the PI3K/AKT pathway plays an essential role in v-Crk-induced transformation of CEF.
v-Crk, an oncogene product of avian sarcoma virus CT10, efficiently transforms chicken embryo fibroblasts (CEF) (1). It is comprised of a viral Gag sequence fused to Src homology (SH)2 and one of the two SH3 domains of c-Crk (2, 3). v-Crk is the earliest identified member of the adaptor protein family having primarily SH2 and SH3 domains, and it is now well established that these domains mediate many protein–protein interactions and play pivotal roles in signal transduction (4, 5). The v-Crk SH2 domain binds phosphorylated tyrosine residues of proteins such as p130Cas and Paxillin, and the v-Crk SH3 domain binds proline-rich sequences of several proteins, including c-Abl, C3G, SOS, and DOCK180 (5, 6). Although the basic nature of the protein–protein interactions mediated through these domains has been well characterized, the precise mechanism by which v-Crk transmits oncogenic signals is not understood.
One of the most interesting features in v-Crk-induced cell transformation is the elevated level of tyrosine phosphorylation of several signaling molecules, such as p130cas and Paxillin, although v-Crk itself has no catalytic tyrosine kinase domain (5, 6). It has been hypothesized that v-Crk somehow regulates endogenous tyrosine kinases and induces tyrosine phosphorylation of signaling molecules and thereby leads to transformation (1). Our earlier studies with SH2 and SH3 domain mutants demonstrated that both domains were required for the induction of tyrosine phosphorylation, and that there was an absolute correlation between the increased tyrosine phosphorylation level and the cell transformation (7). However, this conclusion seemed to be somewhat premature because the levels of expression of these mutants were lower than those of the wild type.
In this study, we reexamined the activity of v-Crk SH2 and SH3 mutants in CEF to clarify the role of each domain in cell transformation by using a newly developed mouse-avian chimeric retroviral vector system in which the expression levels of each mutant were comparable to those of wild type. Moreover, we analyzed three well defined signaling pathways: the Jun N-terminal kinase (JNK) pathway, the mitogen-activated kinase (MAPK) pathway, and the phosphoinositide 3-kinase (PI3K)/AKT pathway, for the elucidation of the downstream signal(s) critical in the cell transformation induced by v-Crk.
Materials and Methods
Construction of Retroviral Vectors Expressing v-Crk Mutants And Other Expression Vectors.
pCXbsr, a Moloney murine leukemia virus (MuLV)-based retroviral vector plasmid, was constructed with a chimeric 5′ long terminal repeat in which the Moloney MuLV enhancer was replaced by a human cytomegalovirus immediate-early enhancer, and the Blasticidin S resistance gene (bsr) was inserted after the internal ribosome entry site of encephalomyocarditis virus as a selectable marker (Fig. 1A). pCXneo had a neomycin resistance gene (neo) instead of bsr. The complete sequences of pCXbsr and pCXneo have been submitted to the GenBank database under the accession nos. AB041927 and AB041928, respectively. Wild-type (WT) and mutated v-crk were subcloned into the BamHI-NotI site of pCXbsr by attaching appropriate linkers at both ends. v-Crk mutants used are as follows (Fig. 1A). R273N is an SH2 point mutant in which the Arg at position 273 was changed to Asn (7). The Arg 273 is highly conserved within all known SH2 domains. BSP is an SH3 insertional mutant with a four amino acid insertion at position 386 in the N-terminal part of SH3 (7). W405K is an SH3 point mutant in which the Trp at position 405 was changed to Lys. The Trp 405 is absolutely conserved among all known SH3 domains (8). N273/K405 is an SH2 and SH3 double mutant having both R273N and W405K mutations. CCSv is an SH3-swapped mutant in which the v-crk-SH3 domain was swapped with the v-src-SH3 domain (9). We confirmed that all of the mutations indeed disrupted the function of each respective domain of v-Crk. For the tetracycline-inducible expression system, pCXneo/TR-2 was constructed by inserting the tetR gene from pcDNA6/TR (Invitrogen) into the BamHI-HpaI site of pCXneo, and pCXbsrR(TO/v-Crk WT) was constructed by inserting the WT v-crk cDNA linked after the T-Rex promoter from pcDNA4/TO (Invitrogen) into the HpaI-NotI site of pCXbsr in an antisense orientation to the vector.
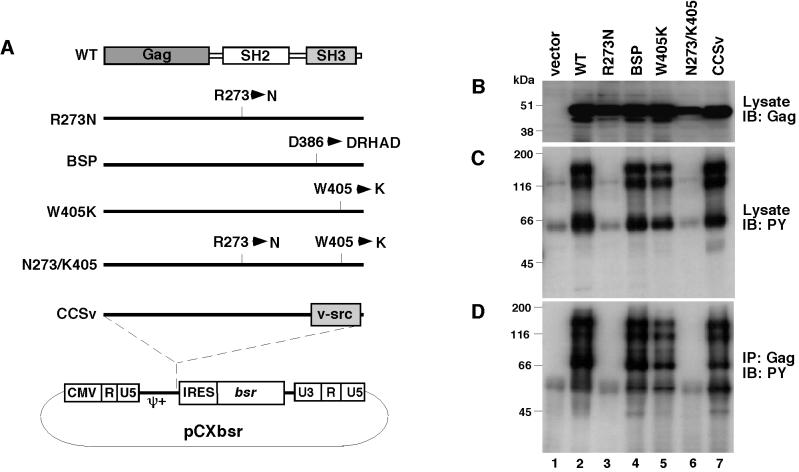
Figure 1.
Characterization of CEF expressing v-Crk mutants. (A) Schematic structure of the v-Crk mutants used in this study. Amino acid changes in each mutant are indicated in the single-letter amino acid code. Detailed descriptions about each mutant are given in Materials and Methods. pCXbsr is a Moloney MuLV-based retroviral vector. CMV, human cytomegalovirus; ψ+, extended retroviral packaging signal; IRES, internal ribose entry site; bsr, Blasticidin S resistance gene. (B) Expression of v-Crk mutants. v-Crk proteins were detected by immunoblotting with anti-Gag antibody 3C2 by using total cell lysates from CEF expressing v-Crk mutants as indicated above each lane. (C) Protein tyrosine phosphorylation in CEF expressing v-Crk mutants. Tyrosine phosphorylated proteins were detected by immunoblotting with anti-phosphotyrosine mouse monoclonal antibody 4G10 by using total cell lysates from CEF expressing v-Crk mutants as indicated above each lane. (D) Association of tyrosine-phosphorylated proteins with v-Crk mutants. Tyrosine phosphorylated proteins associated with v-Crk mutants were detected by immunoprecipitation with the anti-Gag antibody 3C2 followed by immunoblotting with the anti-phosphotyrosine rabbit polyclonal antibody.
An expression vector for Rous sarcoma virus (RSV) envelope glycoprotein (env), pcDNA3/RSV-env, was constructed by inserting the 1.8-kb SacI fragment from pLC32 (10) containing the RSV env cDNA into the EcoRV-XbaI site of pcDNA3 (Invitrogen) after attaching appropriate linkers at both ends.
A retroviral vector expressing a constitutively active mutant of PI3K BD110 (11), which was provided by Y. Fukui (University of Tokyo), was constructed by inserting the entire coding region of BD110 into pCXneo.
Production of Mouse-Avian Chimeric Retroviruses.
Retroviral vector plasmids made with pCXbsr were cotransfected with pcDNA3/RSV-env into BOSC23 (12), an ecotropic MuLV packaging cell line, by using Fugene6 (Boehringer Mannheim) according to the manufacturer's directions. At 2 days after the transfection, culture supernatants were collected and used as viral stocks.
Culture, Infection, and Soft-Agar Colony Assay of CEF.
CEF were prepared and maintained essentially as described previously (13). Infection of CEF was carried out by adding the filtered culture supernatants from transfected BOSC23 cells supplemented with 6.25 μg/ml DEAE-dextran. At 2 days after the infection, drug selection of infected CEF was started, and the selected drug-resistant populations were used for all of the experiments. For tetracycline-inducible expression of v-Crk, CEF were dually infected with CXneo/TR-2 and CXbsrR(TO/v-Crk WT). For the soft-agar colony formation assay, 5 × 104 infected CEF were plated per 6-cm culture dish as a suspension in 3 ml of DMEM containing 10% calf serum and 0.4% agar on a layer of 5 ml of the same medium containing 0.7% agar. Plates were incubated at 37°C for 3–4 weeks until colonies were formed.
Protein Analysis.
Immunoblotting and immunoprecipitation were performed as described previously (14). Anti-p19gag mouse monoclonal antibody 3C2 (15) was provided by D. Boettiger (University of Pennsylvania, Philadelphia). Anti-phosphotyrosine mouse monoclonal antibody 4G10 was purchased from Upstate Biotechnology. Anti-phosphotyrosine rabbit polyclonal antibody was purchased from Zymed. Anti-JNK1 rabbit polyclonal antibody was purchased from Santa Cruz Biotechnology. Anti-phospho-SAPK/JNK (Thr183/Tyr185) mouse monoclonal antibody, anti-Akt rabbit polyclonal antibody, and anti-phospho-Akt(Ser473) rabbit polyclonal antibody were purchased from New England Biolabs. Anti-c-Jun mouse monoclonal antibody was purchased from Transduction Laboratories (Lexington, KY). Anti-phospho-c-Jun (Ser63) rabbit polyclonal antibody was purchased from Calbiochem. Anti-MAPK rabbit polyclonal antibody was purchased from Promega. Anti-activated MAPK (diphosphorylated ERK1 and -2) mouse monoclonal antibody was purchased from Sigma.
Results
Establishment of an Efficient Retroviral Vector System for CEF.
Because v-Crk efficiently transforms CEF but not rodent fibroblasts (1, 16, 17), CEF seemed to be the most suitable cell system for the analysis of v-Crk-induced transformation. Therefore, we first attempted to establish an efficient helper-free retroviral vector system for CEF by modifying the well established murine retroviral vector system. We cotransfected the MuLV-based retroviral vector with the RSV env expression vector into the murine retrovirus packaging cell line BOSC23, and successfully produced chimeric viruses with both MuLV-env and RSV-env, which can infect not only mouse cells but also chicken cells. After optimizing the infection conditions, we finally obtained a greater than 50% infection efficiency for CEF (data not shown). By using this system, we infected CEF with various v-crk mutants to analyze the roles of the SH2 and SH3 domains in transformation. We used one SH2-deficient mutant (R273N), two SH3-deficient mutants (W405K and BSP), one SH2 and SH3 double mutant (N273/K405), and one mutant in which Crk SH3 was replaced by Src SH3 (CCSv) (Fig. 1A). Cells infected with these mutants were subjected to drug selection, and the selected populations then were used for all of the subsequent experiments.
Only the SH2 Domain Is Essential for the Induction of Protein Tyrosine Phosphorylation by v-Crk.
First, we checked the expression of v-Crk protein in CEF infected with various mutants by immunoblotting and confirmed that most of the mutants were expressed at levels comparable to the WT levels (Fig. 1B). Next, we examined the state of tyrosine phosphorylation of cellular proteins by anti-phosphotyrosine immunoblotting (Fig. 1C), and found that whereas the three SH3 mutants (BSP, W405K, and CCSv) induced tyrosine phosphorylation at almost the same level as WT, the SH2 mutant (R273N) and the double mutant (N273/W405K) did not. The association of tyrosine-phosphorylated proteins with v-Crk was analyzed by anti-Gag immunoprecipitation followed by anti-phosphotyrosine immunoblotting. All of the three SH3 mutants were shown to be similar to WT with respect to its association with most of the tyrosine-phosphorylated proteins detected in these cells (Fig. 1D, lanes 4, 5, and 7). We confirmed that the 130-kDa band corresponded to p130Cas and the 66-kDa band corresponded to Paxillin by immunoblotting with each specific antibody (data not shown). These results clearly show that although the SH2 domain is essential for the induction of tyrosine-phosphorylation of some signaling molecules, and for the association with these proteins, the SH3 domain is not necessary.
Both the SH2 and SH3 Domains Are Required for Cell Transformation by v-Crk.
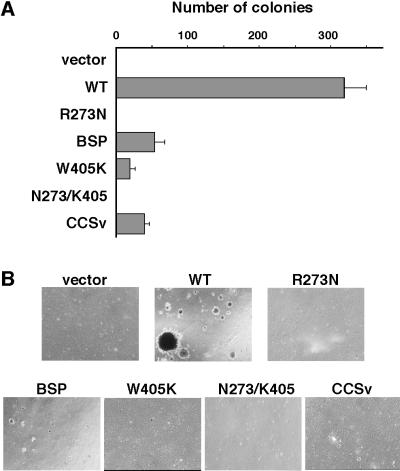
Next, we compared the transforming activities of these mutants by soft-agar colony formation assay. The transforming activities of SH3 mutants were severely impaired. The numbers of colonies formed by CEF infected with SH3 mutants were about 1/6 to 1/10 of those formed by CEF infected with WT (Fig. 2A), and also the size of the colonies formed by SH3 mutant-infected cells were much smaller than those formed by WT-infected cells (Fig. 2B). No colony was found in CEF expressing the SH2 mutant or double mutant. These results indicate that both the SH2 and SH3 domains are essential for the complete transformation induced by v-Crk.
Figure 2.
Soft-agar colony formation of CEF expressing v-Crk mutants. (A) The numbers of colonies in soft-agar were counted 3 weeks after plating. Bars represent the averages (±SD) of three independent experiments. (B) Photomicrographs of colonies were taken 3 weeks after plating. (×40.)
The PI3K/AKT Pathway Is Activated in v-Crk-Transformed CEF.
To clarify the downstream signal(s) critical for the transformation by v-Crk, we investigated three well defined signaling pathways: the JNK pathway, the MAPK pathway, and the PI3K/AKT pathway.
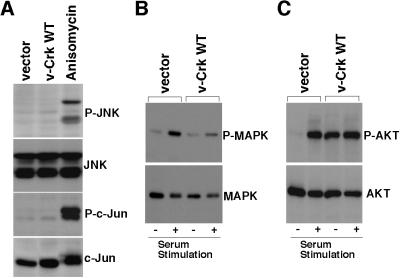
First, we examined the JNK pathway, because there are several lines of evidence that indicate that Crk activates JNK (18, 19). We analyzed the activation state of endogenous JNK proteins by immunoblotting with antibody specific for JNK phosphorylated both at Thr183 and Tyr185 (P-JNK), which represents the activated state of JNK (20). Although phosphorylated JNKs clearly were detected in the positive control anisomycin-treated cells, there was no significant difference between v-Crk-transformed CEF and vector control cells (Fig. 3A, first panel). We further analyzed the phosphorylation state of endogenous c-Jun by immunoblotting with the antibody specific for c-Jun phosphorylated at Ser63 (P-c-Jun). Again, although the strongly phosphorylated c-Jun was detected in the positive control anisomycin-treated cells, there was no significant difference between v-Crk-transformed CEF and vector control cells (Fig. 3A, third panel). Thus, we concluded that there was no evidence of endogenous JNK activation in the v-Crk-transformed CEF.
Figure 3.
Investigation of the downstream signals of v-Crk. (A) Analysis of the JNK pathway. Total cell lysates were prepared from CEF transduced with control vector, CEF transduced with v-crk WT, and CEF treated with anisomycin (10 μg/ml for 20 min). Cell lysates then were subjected to immunoblot analysis with antibodies specific for the phosphorylated forms of either JNK (P-JNK) (first panel) or c-Jun (P-c-Jun) (third panel). The same blots also were probed with antibodies to JNK (second panel) or to c-Jun (fourth panel) that react with these proteins irrespective of phosphorylation state to confirm that equal amounts of each protein were present in each lane. (B) Analysis of the MAPK pathway. CEF transduced with vector or v-crk WT were serum-starved for 24 h and then stimulated with 20% calf serum for 20 min (+), or left unstimulated (−). Total cell lysates from these cells were subjected to immunoblot analysis with antibody specific for the phosphorylated form of MAPK (P-MAPK) (Upper). The same blots were also probed with antibody to MAPK that reacts with MAPK irrespective of phosphorylation state to confirm that equal amounts of this protein were present in each lane (Lower). (C) Analysis of the PI3K/AKT pathway. CEF transduced with vector and CEF transduced with v-crk WT were serum-starved for 24 h and then stimulated with 20% calf serum for 20 min (+), or left unstimulated (−). Total cell lysates from these cells were subjected to immunoblot analysis with antibody specific for the phosphorylated form of AKT (P-AKT) (Upper). The same blots also were probed with antibody to AKT that reacts with AKT irrespective of phosphorylation state to confirm that equal amounts of this protein were present in each lane (Lower).
By using the same strategy as that for the study of JNK, we examined the MAPK pathway by immunoblotting with antibody specific for MAPK phosphorylated at Thr202 and Tyr204 (P-MAPK), which represents the activated state of MAPK (21). As expected, MAPK was activated on serum stimulation in both control and v-Crk-transformed CEF. However, we could not find any evidence of such activation in v-Crk-transformed CEF (Fig. 3B).
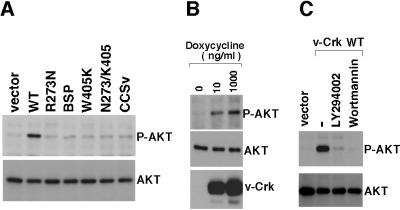
Next, to examine the PI3K/AKT pathway, we analyzed the activation state of endogenous AKT protein in v-Crk-transformed CEF by immunoblotting with antibody specific for AKT phosphorylated at Ser473 (P-AKT), which represents the activated state of AKT (22). There was a significant difference between the results in v-Crk-transformed CEF and vector control cells. Although phosphorylated AKT was detected only after serum stimulation in vector control cells, it was detected readily even before serum stimulation in v-Crk-transformed CEF (Fig. 3C). This marked difference in AKT phosphorylation was detected even in cells maintained in the medium containing serum (data not shown). These results clearly show that AKT is constitutively activated in v-Crk-transformed CEF. This constitutive activation of AKT was detected only in WT v-Crk-expressing cells and not in SH2- or SH3-mutant-expressing cells (Fig. 4A), indicating that both the domains are required and that AKT phosphorylation is closely correlated with transformation.
Figure 4.
Analysis of v-Crk-induced AKT phosphorylation. (A) AKT phosphorylation in CEF expressing v-Crk mutants. Total cell lysates from CEF expressing v-Crk mutants (indicated above each lane) were subjected to immunoblot analysis with antibody specific for the phosphorylated form of AKT (P-AKT) (Upper) or with antibody to AKT that reacts with AKT irrespective of phosphorylation state (Lower). (B) AKT phosphorylation in an inducible v-Crk expression system. CEF dually infected with CXneo/TR-2 and CXbsrR(TO/v-Crk WT) were cultured with the indicated concentrations of doxycycline for 24 h. Total cell lysates from these cells then were subjected to immunoblot analysis with antibody specific for the phosphorylated form of AKT (P-AKT) (Top), with antibody to AKT that reacts with AKT irrespective of phosphorylation state (Middle), or with anti-Gag antibody (Bottom). (C) Effects of PI3K inhibitors. CEF expressing WT v-Crk were treated for 2 h with LY294002 (Calbiochem) at 10 μM, wortmannin (Calbiochem) at 200 nM, or with the solvent DMSO (−). Then, total cell lysates from these cells as well as from vector-transduced cells were subjected to immunoblot analysis with antibody specific for the phosphorylated form of AKT (P-AKT) (Upper) or with antibody to AKT that reacts with AKT irrespective of phosphorylation state (Lower)
To further confirm their direct relationship, we established a tetracycline-inducible expression system of v-Crk in CEF (Fig. 4B). In this system, 24 h after the addition of doxycycline, a tetracycline derivative, v-Crk protein expression was strongly induced (Fig. 4B, Bottom). Accompanying this induction, phosphorylation of AKT also was induced (Fig. 4B, Top). These results strongly suggest the assumption that AKT phosphorylation is a direct effect of v-Crk expression.
Although it is well established that AKT phosphorylation is a downstream effect of PI3K activation, there are several instances of PI3K-independent activation of AKT (23). Therefore, we examined whether v-Crk-induced AKT phosphorylation actually is mediated by PI3K by testing the effects of PI3K inhibitors. As shown in Fig. 4C, LY294002 and wortmannin, two well known PI3K inhibitors, at concentrations supposed to be specific for PI3K (24, 25), almost completely suppressed the v-Crk-induced AKT phosphorylation. This finding clearly shows that v-Crk-induced constitutive phosphorylation of AKT is mediated by PI3K and that PI3K is constitutively activated in v-Crk-transformed CEF, which is consistent with our previous finding of the association of active PI3K with v-Crk (26).
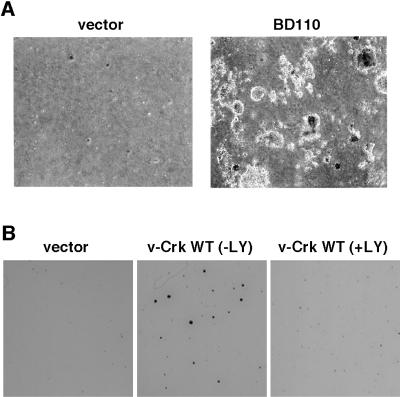
To test whether the constitutive activation of PI3K is sufficient for the transformation of CEF, we introduced a constitutively active mutant of PI3K, BD110 (11), into CEF and examined its transforming activity by soft-agar colony formation assay. As shown in Fig. 5A, BD110 strongly transformed CEF, which is consistent with the recent identification of a p110 PI3K catalytic subunit as an oncogene of ASV16 avian sarcoma virus (27, 28). Moreover, as shown in Fig. 5B, we found that the soft-agar colony formation of CEF induced by v-Crk was significantly suppressed by the addition of a PI3K inhibitor LY294002. All of these results strongly suggest that constitutive activation of the PI3K/AKT pathway plays a major role in the v-Crk-induced transformation of CEF.
Figure 5.
Transformation by a constitutively active mutant of PI3K and suppression of v-Crk-induced transformation by a PI3K inhibitor. (A) Soft-agar colony formation by the PI3K constitutively active mutant BD110. CEF transduced with BD110 or with vector were subjected to soft-agar colony formation assay. Photomicrographs of colonies were taken 3 weeks after plating. (×40.) (B) Suppression of v-Crk-induced soft-agar colony formation by a PI3K inhibitor. CEF expressing v-Crk WT were subjected to soft-agar colony formation assay with (+LY) or without (−LY) LY294002. LY294002 was included in top agar layer at 10 μM final concentration, and 0.5 ml of medium containing the same final concentration of LY294002 was added onto the top agar every 4 days. We confirmed that long-term treatment with this concentration of LY294002 showed no apparent toxicity to CEF cultured in monolayer (data not shown). CEF transduced with vector without such treatment also were examined. At 3 weeks after plating, colonies were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) as described (37) and photographs of the stained colonies were taken. (×1.)
Discussion
Enhanced Cellular Tyrosine Phosphorylation Is Not Sufficient for Transformation by v-Crk.
Cells transformed by v-Crk exhibit elevated levels of tyrosine phosphorylation of several cellular proteins (1). In a previous study, we concluded that both SH2 and SH3 mutants were unable either to activate the tyrosine phosphorylation of cellular proteins or to induce cell transformation (7). In the present study, however, we found that v-Crk SH3 mutants are able to induce tyrosine phosphorylation to almost the same extent as found in WT, yet the cell transformation was severely impaired. This apparent discrepancy may be attributable to the difference in the degree of expression of the v-Crk mutants. In a previous study, we used an avian replicating retroviral system requiring helper viruses, which lead to a lower percentage of cells expressing nontransforming mutants than the WT, and the apparent expression levels of such mutants were decreased (7). In the present study, we used a helper-free murine retroviral vector system by which one can easily obtain 100% infected cell populations expressing v-Crk mutants. This system enabled us to better compare the biological activities of each mutant at comparable expression levels. We found that only the v-Crk SH2 domain is responsible for the induction of tyrosine phosphorylation of cellular proteins. It also became evident that this phosphorylation is necessary but not sufficient for the complete cell transformation and that signal(s) from the SH3 domain also are required.
Activation of the PI3K/AKT Pathway by v-Crk and Its Implication in Transformation.
Comparative analysis of the three well defined kinase (JNK, MAPK, and PI3K/AKT) pathways highlighted the importance of the PI3K/AKT pathway in v-Crk-induced transformation. Only the PI3K/AKT pathway was constitutively activated in CEF transformed by WT v-Crk but not in CEF expressing the v-Crk SH2 or SH3 mutants. Moreover, we confirmed that a constitutively active mutant of PI3K transforms CEF and that LY294002, a PI3K inhibitor, significantly suppresses the v-Crk-induced transformation. Activation of the PI3K/AKT pathway also was observed in the v-Src-induced transformation of CEF, however, this transformation was suppressed only slightly by the treatment with LY294002 at the same concentration as that used in this study (29). Therefore, v-Crk seems to be more dependent on the PI3K/AKT pathway for its transforming activity than is v-Src. Taken together with the identification of the p110 PI3K catalytic subunit as an oncogene of ASV16 avian sarcoma virus (27) and the recent demonstration of the transforming activity of a constitutively active Akt mutant in CEF (30), our results strongly suggest that the PI3K/AKT pathway is a critical mediator of v-Crk-induced transformation of CEF. It is well established that the activation of the PI3K/AKT pathway provides a survival signal in many circumstances such as growth factor stimulation and attachment to the extracellular matrix (23). Thus, it is conceivable that v-Crk facilitates anchorage-independent growth by generating a survival signal through the constitutive activation of the PI3K/AKT pathway. Indeed, we detected AKT phosphorylation in v-Crk-transformed CEF even in suspension cultures (T.A. and H.H., unpublished data).
Although we previously reported the activation of the JNK pathway by v-Crk (18), we could not find any evidence for such activation in the present study. Because v-Crk-induced JNK activation was very subtle in CEF as compared with that in NIH 3T3 cells, even in the previous study, the difference in species may explain this discrepancy. However, the precise reason for the more prominent JNK activation reported in NIH 3T3 cells than that in CEF is not clear. This difference in JNK activation may bear some relation to the difference in the efficiency of v-Crk-induced transformation between these cell types. Whereas CEF can be very efficiently transformed by v-Crk (1), rodent fibroblasts are rather refractory and only a small portion of the rodent cells expressing v-Crk are transformed (16, 17). By using the same retroviral vector system as that used in this study, we confirmed the inability of v-Crk to induce the transformation of rodent fibroblasts efficiently (data not shown). Therefore, it is possible that some unknown events in addition to v-Crk expression are involved in the transformation, of which the activation of the JNK pathway may be one.
A Possible Mechanism of PI3K Activation by v-Crk.
Although we previously demonstrated that v-Crk is associated with active PI3K (26), the precise mechanism by which v-Crk activates the PI3K/AKT pathway is not clear at present. Considering that activation of PI3K requires binding of the SH2 domains of the p85 regulatory subunit to tyrosine-phosphorylated motifs on growth factor receptors and other signaling molecules (31), it is conceivable that the SH2 domains of p85 bind to some signaling molecule that is tyrosine-phosphorylated by v-Crk. An interesting candidate for such a molecule is FAK. FAK binds through its phosphorylated Tyr-397 to the SH2 domains of the PI3K p85 subunit on integrin stimulation, and this interaction is supposed to play an important role in PI3K activation after integrin engagement (32). We have evidence demonstrating that v-Crk induces phosphorylation of Tyr-397 of FAK (T.A. and H.H., unpublished data). Therefore, v-Crk might mimic the integrin-stimulated PI3K activation by inducing an interaction between the PI3K p85 subunit and FAK. However, this binding of the p85 subunit to a tyrosine-phosphorylated signaling molecule seems to be insufficient for the activation of PI3K, because v-Crk SH3 mutants that are totally competent in inducing tyrosine phosphorylation still failed to activate the PI3K/AKT pathway (Fig. 4A). Some signal(s) from the SH3 domain also are likely to be required. In this context, it is intriguing that v-Crk SH3 binds to guanine nucleotide-releasing factors for some Ras family proteins such as C3G and mSOS (33, 34), because Ras and R-Ras are reported to activate PI3K by binding to the p110 catalytic subunit (35). PI3K might be cooperatively activated by the binding of a tyrosine-phosphorylated signaling molecule to the p85 regulatory subunit for which the v-Crk SH2 domain is responsible, and by the binding of active Ras or R-Ras to the p110 catalytic subunit for which the v-Crk SH3 domain is responsible. Consistent with this finding, we previously demonstrated that a dominant-negative mutant of Ras suppressed v-Crk-induced cell transformation (36). Experiments to verify this hypothesis further remain to be performed.
Acknowledgments
We thank D. Boettiger, J. Fujisawa, Y. Fukui, and C. M. Stoltzfus for providing the reagents, and M. Fukuda for the technical assistance. This work was supported by the National Institutes of Health Grant R35CA44356 and by a grant-in-aid for Specially Promoted Research from the Ministry of Education, Science, Sports, and Culture of Japan, as well as a grant from The Mitsubishi Foundation.
Abbreviations
- CEF
chicken embryo fibroblasts
- SHn
Src homology n domain
- PI3K
phosphoinositide 3-kinase
- JNK
Jun N-terminal kinase
- MAPK
mitogen activated protein kinase
- MuLV
murine leukemia virus
- WT
wild type
- RSV
Rous sarcoma virus
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AB041927 (pCXbsr) and AB041928 (pCXneo)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140210297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140210297
References
- 1.Mayer B J, Hamaguchi M, Hanafusa H. Nature (London) 1988;332:272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda M, Tanaka S, Nagata S, Kojima A, Kurata T, Shibuya M. Mol Cell Biol. 1992;12:3482–3489. doi: 10.1128/mcb.12.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichman C T, Mayer B J, Keshav S, Hanafusa H. Cell Growth Differ. 1992;3:451–460. [PubMed] [Google Scholar]
- 4.Pawson T, Gish G D. Cell. 1992;71:359–362. doi: 10.1016/0092-8674(92)90504-6. [DOI] [PubMed] [Google Scholar]
- 5.Birge R B, Knudsen B S, Besser D, Hanafusa H. Genes Cells. 1996;1:595–613. doi: 10.1046/j.1365-2443.1996.00258.x. [DOI] [PubMed] [Google Scholar]
- 6.Feller S M, Posern G, Voss J, Kardinal C, Sakkab D, Zheng J, Knudsen B S. J Cell Physiol. 1998;177:535–552. doi: 10.1002/(SICI)1097-4652(199812)177:4<535::AID-JCP5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 7.Mayer B J, Hanafusa H. J Virol. 1990;64:3581–3589. doi: 10.1128/jvi.64.8.3581-3589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka M, Gupta R, Mayer B J. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuda M, Reichman C T, Hanafusa H. J Virol. 1992;66:115–121. doi: 10.1128/jvi.66.1.115-121.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang L J, Stoltzfus C M. Mol Cell Biol. 1985;5:2341–2348. doi: 10.1128/mcb.5.9.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi M, Nagata S, Kita Y, Nakatsu N, Ihara S, Kaibuchi K, Kuroda S, Ui M, Iba H, Konishi H, et al. J Biol Chem. 1997;272:16089–16092. doi: 10.1074/jbc.272.26.16089. [DOI] [PubMed] [Google Scholar]
- 12.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanafusa H. Proc Natl Acad Sci USA. 1969;63:318–325. doi: 10.1073/pnas.63.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akagi T, Ono H, Shimotohno K. Oncogene. 1996;12:1645–1652. [PubMed] [Google Scholar]
- 15.Potts W M, Olsen M, Boettiger D, Vogt V M. J Gen Virol. 1987;68:3177–3182. doi: 10.1099/0022-1317-68-12-3177. [DOI] [PubMed] [Google Scholar]
- 16.Sabe H, Okada M, Nakagawa H, Hanafusa H. Mol Cell Biol. 1992;12:4706–4713. doi: 10.1128/mcb.12.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa S, Toyoshima H, Kozutsumi H, Hagiwara K, Sakai R, Tanaka T, Hirano N, Mano H, Yazaki Y, Hirai H. Oncogene. 1994;9:1669–1678. [PubMed] [Google Scholar]
- 18.Tanaka S, Ouchi T, Hanafusa H. Proc Natl Acad Sci USA. 1997;94:2356–2361. doi: 10.1073/pnas.94.6.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolfi F, Garcia-Guzman M, Ojaniemi M, Nakamura H, Matsuda M, Vuori K. Proc Natl Acad Sci USA. 1998;95:15394–15399. doi: 10.1073/pnas.95.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minden A, Karin M. Biochim Biophys Acta. 1997;1333:F85–F104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 21.Cobb M H, Goldsmith E J. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 22.Downward J. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 23.Datta S R, Brunet A, Greenberg M E. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 24.Vlahos C J, Matter W F, Hui K Y, Brown R F. J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 25.Okada T, Kawano Y, Sakakibara T, Hazeki O, Ui M. J Biol Chem. 1994;269:3568–3573. [PubMed] [Google Scholar]
- 26.Fukui Y, Kornbluth S, Jong S M, Wang L H, Hanafusa H. Oncogene Res. 1989;4:283–292. [PubMed] [Google Scholar]
- 27.Chang H W, Aoki M, Fruman D, Auger K R, Bellacosa A, Tsichlis P N, Cantley L C, Roberts T M, Vogt P K. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 28.Aoki M, Schetter C, Himly M, Batista O, Chang H W, Vogt P K. J Biol Chem. 2000;275:6267–6275. doi: 10.1074/jbc.275.9.6267. [DOI] [PubMed] [Google Scholar]
- 29.Penuel E, Martin G S. Mol Biol Cell. 1999;10:1693–1703. doi: 10.1091/mbc.10.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aoki M, Batista O, Bellacosa A, Tsichlis P, Vogt P K. Proc Natl Acad Sci USA. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanhaesebroeck B, Leevers S J, Panayotou G, Waterfield M D. Trends Biochem Sci. 1997;22:267–272. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- 32.Chen H C, Appeddu P A, Isoda H, Guan J L. J Biol Chem. 1996;271:26329–26334. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nagashima K, et al. Proc Natl Acad Sci USA. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuda M, Hashimoto Y, Muroya K, Hasegawa H, Kurata T, Tanaka S, Nakamura S, Hattori S. Mol Cell Biol. 1994;14:5495–5500. doi: 10.1128/mcb.14.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marte B M, Rodriguez-Viciana P, Wennstrom S, Warne P H, Downward J. Curr Biol. 1997;7:63–70. doi: 10.1016/s0960-9822(06)00028-5. [DOI] [PubMed] [Google Scholar]
- 36.Greulich H, Hanafusa H. Cell Growth Differ. 1996;7:1443–1451. [PubMed] [Google Scholar]
- 37.Alley M C, Pacula-Cox C M, Hursey M L, Rubinstein L R, Boyd M R. Cancer Res. 1991;51:1247–1256. [PubMed] [Google Scholar]