Abstract
Sequencing-based human immunodeficiency virus type 1 (HIV-1) genotyping assays require subjective interpretation (editing) of sequence data from multiple primers to form consensus sequences and identify antiretroviral drug resistance mutations. We assessed interlaboratory variations in editing and their impact on the recognition of resistance mutations. Six samples were analyzed in a central laboratory by using a research-use-only HIV-1 genotyping system previously produced by Applied Biosystems. The electronic files of individual primer sequences from the samples were sent to 10 laboratories to compare sequence editing strategies. Each sequence data set included sequences from seven primers spanning protease codons 1 to 99 and reverse transcriptase codons 1 to 320. Each laboratory generated a consensus sequence for each sample and completed a questionnaire about editing strategy. The amount of editing performed, the concordance of consensus sequences among the laboratories, and the identification of resistance mutations were evaluated. Sequence agreement was high among the laboratories despite wide variations in editing strategies. All laboratories identified 66 (88%) of 75 resistance mutations in the samples. Nonconcordant identifications were made for 9 (12%) of the 75 mutations, all of which required editing for identification. These results indicate a need for standardized editing guidelines in genotyping assays. Proficiency in editing should be assessed in training and included in quality control programs for HIV-1 genotyping.
The use of sequencing-based human immunodeficiency virus (HIV) type 1 (HIV-1) genotyping assays to identify antiretroviral drug resistance mutations for the clinical management of HIV-infected patients is increasing rapidly (1-4, 6, 8, 13). These assays typically involve HIV RNA extraction, reverse transcription, PCR amplification, population sequencing of PCR products, electrophoresis of sequencing products, and evaluation of sequence data. In HIV genotyping, multiple primers are used to provide overlapping sequence data spanning the region of interest. Data from two or more sequencing primers spanning a given region are then compared to determine the correct consensus sequence. This subjective process is referred to as editing. Although HIV genotyping assays can be powerful tools for patient management, the data produced can be strongly affected by the editing process.
The need to edit sequence data can be influenced by several factors. Editing is typically required when there is some discrepancy between the data obtained from individual sequencing primers covering the same sequence region. This process is complicated by the fact that HIV drug resistance mutations are often present in clinical samples as mixtures (e.g., mutant plus wild type), reflecting either emergence or fading of the resistant viral variants in a genetically heterogeneous viral population. Visual inspection of sequence data from bidirectional primers is required to confirm the presence of nucleotide mixtures in clinical samples. Different laboratories use different criteria to confirm the presence of mixtures. A variety of technical problems can also produce peaks in sequencing electropherograms that suggest the presence of nucleotide mixtures. These artifacts can usually be identified by visual inspection of the sequence data from individual primers. Once identified, such artifactual mixtures can be removed from a consensus sequence before a resistance report is generated. In some instances, the generation of suboptimal sequence data is related to inherent characteristics of the HIV template or assay performance. For instance, poor sample preparation, inefficient reverse transcription or PCR amplification, nonspecific binding of a sequencing primer to an alternate region of the sequencing template, inadequate purification of sequencing products prior to electrophoresis, and problems in gel preparation, sample loading, or electrophoresis of sequencing gels can all contribute to poor-quality data (from Comparative PCR Sequencing: a Guide to Sequencing-Based Mutation Detection, 1995; Perkin-Elmer Corporation, Applied Biosystems, Foster City, Calif.). When the quality of data is poor, nucleotide mixtures may be introduced into a sequence, requiring the sequence editor to decide whether the observed mixtures are artifactual or real. The number of bases edited in a given sequence can also be influenced by the amount of experience a person has in evaluating electropherograms, as well as the strategy chosen to assemble and evaluate sequence data from individual primers to generate a consensus sequence. It is important that the data generated from the laboratories performing these assays accurately reflect the presence of true mixtures, especially for base positions linked to antiretroviral drug resistance.
In a previous study, two panels, each containing individual plasma samples from three HIV-1-infected persons, were sent to 10 laboratories participating in the Pediatric AIDS Clinical Trials Group Sequencing Working Group. The laboratories used a research-use-only genotyping system to genotype the samples. The consensus sequences generated for each plasma sample by the individual laboratories showed a very high concordance to a group consensus sequence, ranging from 98.0 to 100.0% for protease (PR) (297 bases) and 97.3 to 100.0% for reverse transcriptase (RT) (960 bases). However, the laboratories varied widely in the percentages of codons edited: 2.0 to 87.8% of PR codons and 4.7 to 63.6% of RT codons were edited (D. Huang, J. Bremer, D. Brambilla, S. Eshleman, R. Nutter, S. Hart, M. Wantman, and P. Palumbo, Abstr. 7th Conf. Retrovir. Opportunistic Infect., abstr. 792, 2000).
In this study, the influence of the editing process was assessed independently by requesting laboratories to produce consensus sequences directly from sequence data sets provided to them from the Virology Quality Assurance (VQA) Laboratory (Chicago, Ill.). The amount of editing used to form a consensus sequence for each sequence data set, the concordance of the consensus sequences among the laboratories, and successful identification of resistance mutations were examined.
MATERIALS AND METHODS
Production of sequence data sets.
At the VQA Laboratory, plasma HIV-1 from five infected donors was genotyped with a research-use-only genotyping system (HIV-1 Genotyping System, version 1 [HGS], with software version 2.1; Applied Biosystems). One cloned insert in pGEM T (Promega, Madison, Wis.) derived from plasma HIV-1 virions was also sequenced by using the sequencing module of the HGS. The HGS uses seven primers to provide continuous, overlapping, bidirectional sequences that encode PR and the first 320 codons of RT (Fig. 1). Six sequence data sets (each containing the sequence files for the seven primers in the HGS) were selected to contain representative problems in sequence interpretation and were distributed electronically to 10 laboratories. No information about the origin of the samples was provided to the laboratories to reduce bias in interpretation. The laboratories had various levels of experience with the HGS. All had received training from Applied Biosystems prior to analysis of the sequence data sets.
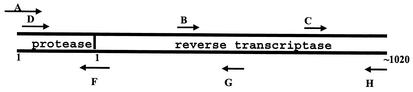
FIG. 1.
Diagram of the positions of sequencing primers in the HGS. The positions of selected nucleotides in PR and RT are shown. The blunt end of the arrow is positioned at the approximate nucleotide start site of the primer.
Analysis of sequence data sets.
Each of the 10 testing laboratories was provided with the following guidelines for editing the sequence data. (i) Manually “trim” further areas of poor sequence data from the ends of the individual segments if needed to resolve ambiguities in the consensus sequence. Trimmed areas become shaded and are not used to edit the base(s) in question. (ii) Each base reported in the consensus sequence should be seen in both the forward and the reverse directions (i.e., sense and antisense). (iii) For verifying the presence of mixtures, (a) mixed bases should be seen in both the forward and the reverse directions with peaks comigrating; (b) in one direction, the smaller peak should be at least 30% the maximum peak height; (c) in the other direction, the smaller peak should be clearly visible but does not have to reach 30% the maximum peak height; and (d) positions should not be identified as containing mixed bases if data are present only for a single direction.
Using these guidelines, as well as in-house guidelines, each laboratory aligned and edited the sequence data to generate a single consensus sequence for each sample. The HGS software documents the editing of individual nucleotides in the consensus sequence by using a lowercase letter. Unedited bases in the consensus sequence remain in uppercase letters. Positions of mixed nucleotides are indicated with the appropriate International Union of Biochemistry (IUB) codes. Both the lowercase and the IUB designations for mixed bases are preserved in the FASTA (a standardized text format for nucleic acid sequences) text file when saved. Each laboratory submitted the following files to a central laboratory for analysis: (i) the edited “project” file, showing the alignment and editing of individual sequences; (ii) the consensus sequence for each sequence data set, saved in FASTA format; and (iii) a list of the mutations (variations from the reference sequence) identified by the software after editing.
Analysis of data from 10 testing laboratories.
The 10 testing laboratories submitted their data to Frontier Science and Technology Research Foundation, Buffalo, N.Y., where the data were collated for analysis. The combined data set was then sent to the VQA Statistical Center at New England Research Institute, Watertown, Mass., for further analyses. These analyses included (i) identification of codons that differed among the 10 testing laboratories (discrepant codons) and (ii) determination of the percentage of codons edited by each laboratory. The consensus sequences submitted for each sequence data set were aligned by using Align Plus (Scientific Educational Software, Durham, N.C.) to form a group consensus sequence. The overall concordance of each laboratory's consensus sequence with the group consensus sequence (homology) was then determined. Mutations associated with antiretroviral drug resistance were identified by using the HGS software. Mutations identified by the 10 testing laboratories were tabulated.
Analysis of data from a questionnaire.
After completing the analysis of the sequence data sets, participants from each of the 10 testing laboratories completed a questionnaire about their editing strategy. Each person who analyzed the sequence data sets completed the questionnaire. Answers to the questionnaire were tabulated.
RESULTS
The relative positions of the forward and reverse sequencing primers in the HGS are shown in Fig. 1. Successful sequencing with this system yields overlapping sequences in both the forward and the reverse directions over a contiguous template from codon 1 of PR through codon 320 of RT. The regions of sequence ambiguity in each of the sequence data sets are described in Table 1. None of these ambiguities prohibited the testing laboratories from aligning the sequencing files for each data set to form a project file by using the HGS software. Each laboratory used its own strategy to edit the consensus sequences and resolve the sequence ambiguities. Antiretroviral drug resistance mutations in the edited consensus sequences were identified by the HGS software.
TABLE 1.
Regions of sequence ambiguity in sequence data sets
| Sample | Primer | Region to be resolveda | Gene affected |
|---|---|---|---|
| 03rg01 | A | 1-120b | PR and RT |
| 03rg02 | D | 1-50 | PR |
| F | 1-400 | ||
| 03rg03 | H | 1-600 | RT |
| 03rg04 | A | 1-600 | PR and RT |
| B | 1-40 | ||
| C | 1-70 | ||
| D | 1-600 | ||
| F | 1-30 | ||
| H | 1-50 | ||
| 03rg05 | A | 1-600 | PR |
| F | 350-410 | ||
| 03rg06 | H | 170-320 | RT |
The numbers refer to nucleotides starting with the first 5′ nucleotide of each primer sequence file.
Consensus sequence, bases 1027 to 1038. An insertion or deletion is present in the region from RT codons 244 to 247.
Homology of consensus sequences generated in 10 testing laboratories.
All 10 testing laboratories generated consensus sequences with very high concordance to the group consensus sequences for PR and RT (Tables 2 and 3). The mean nucleotide homologies among the 10 consensus sequences for the six sequence data sets ranged from 98.5 to 100% for PR and 99.5 to 99.8% for RT. The data indicate that all 10 laboratories produced similar consensus sequences, regardless of their experience with the HGS.
TABLE 2.
Percent PR gene homology among laboratoriesa
| Laboratory | % Homology in the following sequence data set:
|
|||||
|---|---|---|---|---|---|---|
| 03rg01 | 03rg02 | 03rg03 | 03rg04 | 03rg05 | 03rg06 | |
| 1 | 99.3 | 100.0 | 100.0 | 99.0 | 99.7 | 99.7 |
| 2 | 99.6 | 100.0 | 100.0 | 98.3 | 99.7 | 100.0 |
| 3 | 99.3 | 100.0 | 100.0 | 99.0 | 99.0 | 100.0 |
| 4 | 99.6 | 100.0 | 100.0 | 98.6 | 99.3 | 100.0 |
| 5 | 99.3 | 98.7 | 100.0 | 99.0 | 98.6 | 94.3 |
| 6 | 99.3 | 99.7 | 100.0 | 99.0 | 99.7 | 98.3 |
| 7 | 98.9 | 100.0 | 100.0 | 99.3 | 99.7 | 99.0 |
| 8 | 100.0 | 100.0 | 100.0 | 98.3 | 100.0 | 100.0 |
| 9 | 98.9 | 99.7 | 99.7 | 95.3 | 100.0 | 99.3 |
| 10 | 99.3 | 100.0 | 100.0 | 99.0 | 99.7 | 100.0 |
| Mean | 99.4 | 99.8 | 100.0 | 98.5 | 99.5 | 99.5 |
The reference sequence is the group consensus sequence from 10 laboratories. Percent homology is the number of bases in agreement/the number of bases submitted.
TABLE 3.
Percent RT gene homology among laboratoriesa
| Laboratory | % Homology in the following sequence data set:
|
|||||
|---|---|---|---|---|---|---|
| 03rg01 | 03rg02 | 03rg03 | 03rg04 | 03rg05 | 03rg06 | |
| 1 | 99.6 | 99.8 | 100.0 | 99.9 | 100.0 | 99.8 |
| 2 | 99.6 | 99.9 | 99.9 | 99.7 | 99.4 | 100.0 |
| 3 | 99.7 | 100.0 | 100.0 | 99.9 | 99.7 | 99.9 |
| 4 | 99.3 | 99.5 | 99.9 | 99.6 | 99.4 | 99.8 |
| 5 | 99.5 | 99.1 | 99.6 | 99.0 | 99.1 | 96.0 |
| 6 | 99.9 | 99.7 | 99.9 | 99.5 | 99.9 | 99.6 |
| 7 | 99.9 | 99.9 | 99.7 | 99.8 | 100.0 | 99.9 |
| 8 | 99.8 | 99.7 | 99.6 | 99.8 | 99.3 | 99.9 |
| 9 | 98.8 | 99.9 | 99.7 | 99.1 | 99.9 | 99.8 |
| 10 | 99.6 | 99.5 | 100.0 | 99.8 | 100.0 | 99.9 |
| Mean | 99.6 | 99.7 | 99.8 | 99.6 | 99.7 | 99.5 |
The reference sequence is the group consensus sequence from 10 laboratories. Percent homology is the number of bases in agreement/the number of bases submitted.
Analysis of percentages of codons edited by testing laboratories.
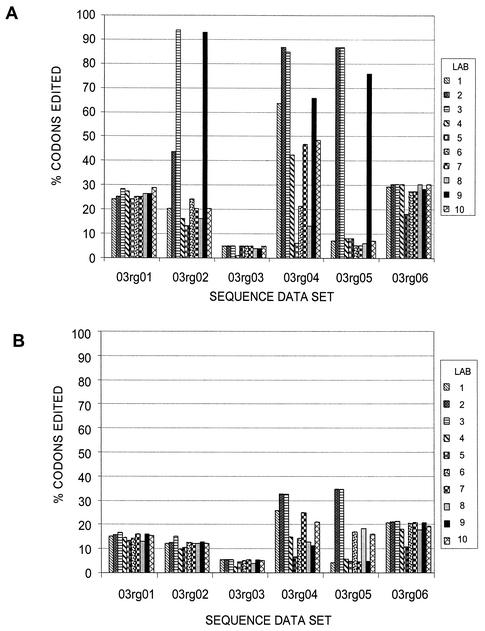
Surprisingly, the amounts of editing used to generate the individual consensus sequences varied widely for the individual sequence data sets (Fig. 2). The sequence data set from the plasmid-derived sample, 03rg03, was edited the least across the 10 laboratories for both PR and RT. Also, there was little variation in the percentages of codons edited by the laboratories for this sample. For the other sequence data sets, some of the testing laboratories (e.g., laboratories 2 and 3) tended to edit a higher percentage of codons regardless of the location of the sequence ambiguity in the sequence data set, whereas other laboratories (e.g., laboratories 5 and 8) tended to edit fewer codons.
FIG. 2.
Frequency of editing of PR (99 codons) (A) and RT (320 codons) (B) sequences. The percentages of the edited codons for the sequence data sets were compared. Codons that contained any bases designated by lowercase letters, indicating editing, were tabulated for each of the 10 laboratories. The percentages of edited codons for the samples were determined for each laboratory.
Identification of antiretroviral drug resistance mutations.
Table 4 shows the number of PR and RT mutations associated with antiretroviral drug resistance for each sequence data set. These mutations were identified in the edited consensus sequences by using the HGS software. A total of 75 mutations were distributed among the six sequence data sets. All of the laboratories identified 66 (88%) of the 75 mutations. Twenty-five of these 66 mutations required editing of the corresponding codons for identification. The remaining 9 (12%) of the 75 mutations, all of which required editing for identification, were identified by some but not all of the testing laboratories. For these, results from one to four laboratories differed from the rest of the results. Discrepant interpretations of resistance occurred for 9 (26.4%) of the 34 codons that were edited.
TABLE 4.
Identification of antiretroviral drug resistance mutations
| Data set | No. of resistance mutations:
|
||
|---|---|---|---|
| Noted in the sequence (PR + RT) | Identified by:
|
||
| All laboratories | Some laboratories | ||
| 03rg01 | 3 | 2 | 1 |
| 03rg02 | 16 | 15 | 1 |
| 03rg03 | 18 | 18 | 0 |
| 03rg04 | 14 | 11 | 3 |
| 03rg05 | 6 | 4 | 2 |
| 03rg06 | 18 | 16 | 2 |
| Total | 75 (100%) | 66 (88%) | 9 (12%) |
Analysis of representative sequence ambiguities.
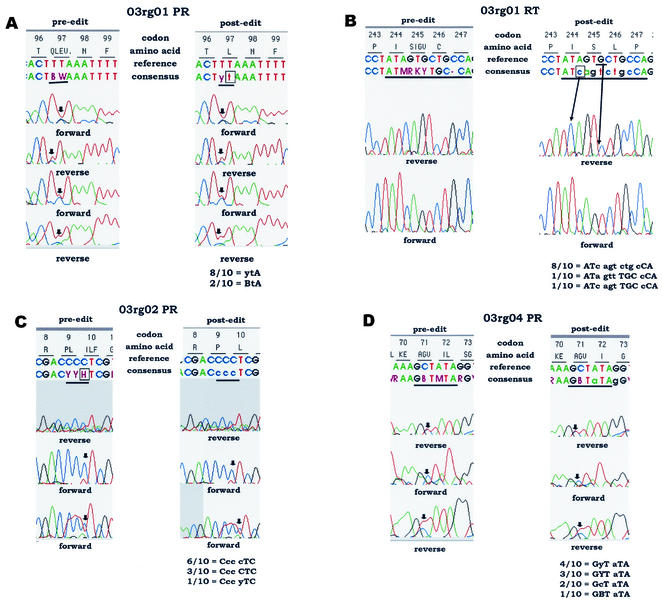
Figure 3 shows representative electropherograms from sequence data sets, with pre- and postediting sequence interpretations. Individual panels are described below.
FIG. 3.
Examples of sequence ambiguity for which discordant results were obtained after editing in the test laboratories. (A to D) Examples of sequence ambiguity requiring different editing strategies (see the text). The data on the left in each panel show the unedited sequence interpretation (preediting), including any nucleotide mixtures identified by the HGS software. The data on the right in each panel show an example of the same sequence file after editing. Note that different editing strategies were used by different laboratories; a single example is shown for each panel. The positions of PR and RT codons are indicated at the top of each panel (e.g., codons 96 to 99 for PR in panel A). Immediately below the codon positions are the amino acid interpretations for the positions in the consensus sequence produced. Two nucleotide sequences are shown below the codon sequences above each set of electropherograms. The upper nucleotide sequence is a reference sequence provided in the software for comparison. The lower nucleotide sequence is the consensus sequence derived from analysis of data from individual electropherograms (before or after editing). Unedited bases are shown in uppercase letters. Edited bases are shown in lowercase letters. Nucleotide mixtures are indicated with standard IUB codes (e.g., C + T = Y). The orientation of each electropherogram is indicated. Sequences that were trimmed by either the software or the user during editing are shaded; trimmed sequences are not used in the base-calling process. Ambiguous nucleotide positions are underlined in the consensus sequences. In panels A, C, and D, arrows indicate positions in the electropherograms that were interpreted differently by the testing laboratories (discordant results). In panel B, arrows indicate the positions of bases inserted and deleted during editing. The nucleotides reported by the testing laboratories at the ambiguous positions are shown below each set of electropherograms. The number of laboratories that provided each interpretation is indicated. Interpretations of the edited sequences are described in the text.
(i) Panel A: sequence data set 03rg01, PR codon 97.
The unedited consensus sequence at PR codon 97 was BWA (B = C + G + T; W = A + T). This sequence encodes a mixture of four amino acids: glutamine (Q), leucine (L), glutamate (E), and valine (V). Eight of the 10 laboratories edited this codon to ytA, which encodes the wild-type amino acid leucine. The remaining two laboratories edited the W at position 2 in the codon to t but did not edit the B at position 1 in the codon. Their interpretation, BtA, encodes a mixture of leucine (CTA or TTA) and valine (GTA). The substitution of valine at position 97 in PR is associated with in vitro resistance to DMP-323 (9, 10).
(ii) Panel B: sequence data set 03rg01, RT codons 244 to 247.
The unedited consensus sequence at RT codons 244 to 247 was ATM RKY TGC-CA. This sequence encodes isoleucine (I) at codon 244; a mixture of serine (S), isoleucine (I), glycine (G), and valine (V) at position 245; cysteine (C) at position 246; and a frameshift at position 247 (single base deletion). Eight of the 10 laboratories edited the sequence to ATc agt ctg cCA, which encodes the amino acid sequence isoleucine (I), serine (S), leucine (L), and proline (P) at codons 244 to 247 and corrects the reading frame. The postediting panel indicates the position of a c, shown in the consensus line, that was added in editing and the position of a G, shown in the reference line, that was deleted in editing. The other two laboratories edited the sequence incorrectly. One laboratory edited codons 244 to 247 to ATa gtt TGC cCA. This sequence encodes isoleucine (I) at codon 244, valine (V) at position 245, cysteine (C) at position 246, and proline (P) at position 247. For this sequence, the editor did not include the inserted base, c, in codon 244 but placed an extra c, not present in the electropherogram, in codon 247, which corrected the frameshift. The other laboratory edited the codons to ATc agt TGC cCA. This sequence encodes isoleucine (I), serine (S), cysteine (C), and proline (P) at codons 244 to 247. This laboratory correctly inserted a c in codon 244 but also placed an extra base, c, in codon 247.
(iii) Panel C: sequence data set 03rg02, PR codon 10.
The unedited consensus sequence at PR codons 9 and 10 was CYY HTC (Y = C + T; H = A + C + T). This sequence encodes a mixture of proline (P) and leucine (L) at codon 9 and a mixture of isoleucine (I), leucine (L), and phenylalanine (F) at codon 10. All 10 laboratories edited codon 9 from CYY to Ccc, which encodes proline (P). Six of the 10 laboratories edited codon 10 from HTC to cTC, which encodes the wild-type amino acid leucine (L). This strategy was consistent with the editing guidelines (see Materials and Methods), which require that mixed bases be observed in both forward and reverse directions. Three laboratories first modified the sequence by trimming in that region, generating the consensus sequence CTC. This alternative editing strategy produced the same result as the strategy used by the other six laboratories mentioned above. The last laboratory edited codon 10 from HTC to yTC, which encodes a mixture of the wild-type amino acid leucine (L) and the mutant amino acid phenylalnine (F). The mutation L10F is associated with lopinavir resistance (10).
(iv) Panel D: sequence data set 03rg04, PR codon 71.
The unedited consensus sequence at PR codons 71 and 72 was GBT MTA (B = C + G + T; M = A + C). This sequence encodes a mixture of alanine (A), glycine (G), and valine (V) at codon 71 and a mixture of isoleucine (I) and leucine (L) at codon 72. All 10 laboratories edited MTA at codon 72 to aTA, which encodes the wild-type amino acid isoleucine (I). Four laboratories edited GBT at codon 71 to GyT, which encodes a mixture of the wild-type amino acid alanine (A) and the mutant amino acid valine (V). The mutation A71V is associated with resistance to lopinavir, nelfinavir, and indinavir (10). Three laboratories trimmed the sequence, generating the same result (GYT = alanine + valine) at codon 71. Two laboratories edited GBT to GcT, which encodes alanine only, and one laboratory did not edit the codon, leaving the sequence GBT, which encodes the A71V mutation as well as the A71G mutation. The A71G mutation has not been reported to be associated with resistance.
Some of the variability in the editing and interpretation of this sequence data set resulted because the laboratories differed in which primer sequences were used to generate the consensus sequence prior to editing individual base positions. In this genotyping system (Fig. 1), the two forward primers, A and D, serve as alternate primers for the PR gene. Either primer may be used in sequence interpretation. The editor has the option to use only one of these two primer sequences for interpretation. This choice may simplify the editing of individual bases if one of the sequences is of lesser quality. In editing this sequence data set, three laboratories used both primer A and primer D sequences, four laboratories used only the primer A sequence, and three laboratories deleted both primer A and primer D sequences, leaving only the reverse primer F sequence for interpretation in this region.
When both primer A and primer D sequences were removed, the unedited consensus sequence generated for this codon was GYT (rather than GBT, as shown in Fig. 3D); none of the three laboratories edited this codon. The codon GYT encodes a mixture of the wild-type amino acid alanine and the mutant amino acid valine. The mutation A71V is associated with resistance to lopinavir, ritonavir, nelfinavir, and indinavir. The decision not to edit the GYT codon by these three laboratories was not consistent with the editing guidelines (see Materials and Methods), which state that a nucleotide mixture (e.g., Y) can be confirmed only if it is present in both forward and reverse sequences. Since only the reverse primer F sequence was used to form the consensus sequence, this sequence should have been edited to the wild-type codon GcT, which encodes alanine.
Analysis of questionnaire on editing strategies.
A questionnaire on editing practices was distributed to the 10 participating laboratories (Fig. 4). Analysis of the questionnaires revealed variability in editing strategies among the laboratories. The majority of the respondents indicated that they prescreened sequences prior to editing and trimmed sequences further after editing had begun if this was deemed necessary. In general, the quality of data from an individual primer was considered to be more important than the direction of the sequence (question 4). The majority of the respondents also would not edit a single base to a mixture unless the mixture was clearly present in both directions (question 5a). These strategies seemed to be applied by most of the respondents in most of those instances (e.g., in the examples shown in Fig. 3). Questions 6a and 6b describe examples similar to that shown in Fig. 3D. From their answers, 7 (58%) of 12 respondents would have changed the mixed base B or Y to c in codon 71, but only 3 (25%) of 12 would have made this change all of the time. However, in practice, only 2 (20%) of 10 respondents changed the mixed base to a pure base. In question 6b, the first and third situations are applicable to the example shown in Fig. 3D. The majority of laboratories indicated that they would have changed codon 71 to GcT; however, in practice, most did not.
FIG. 4.
Questions regarding evaluation of mixtures and tabulated responses. A questionnaire regarding editing strategy was sent with the sequence data sets. Every person (n = 12) who submitted edited data was asked to complete a questionnaire postediting. Only questions relevant to the examples shown in Fig. 3 are shown; the numbers of responses are summarized.
DISCUSSION
True nucleotide mixtures are often present in plasma samples from HIV-1-infected patients, representing the genetic variation among quasispecies in the viral population. In addition, erroneous “apparent” mixtures may be detected in sequencing data due to technical artifacts. For this reason, persons performing HIV-1 genotyping assays must often make subjective decisions to define a mixture as real or artifactual. Artifactual mixtures should be edited from sequence data before algorithms to identify antiretroviral drug resistance mutations are applied.
We examined the editing step performed in a research-use-only HIV-1 genotyping assay previously produced by Applied Biosystems, the HGS. This system provided software for analysis and editing of sequence data. By providing the laboratories with identical electronic sequence data sets, we were able to exclude any variations in interpretation due to technical issues associated with other steps in the assay (e.g., HIV-1 RNA isolation, reverse transcription, PCR amplification, cycle sequencing, or electrophoresis of sequencing products). We found a high level of concordance among the laboratories with regard to their final genotyping interpretations of sequence data sets from six selected samples. Interestingly, we found significant variability in the strategies used by the laboratories for editing sequencing data and in the percentages of codons edited. In some cases, differences in sequence editing influenced the identification of mutations associated with antiretroviral drug resistance. This finding may have implications for the clinical application of genotypic resistance data in the management of HIV-1-infected patients.
It was interesting to observe that relatively little editing was performed for the sequence data set generated from the plasmid-derived sample compared to the data sets generated from clinical plasma samples. The mixtures in the plasmid-derived sample were presumably all artifactual, since the template used for sequencing was clonal. In some cases, laboratories prepare quality-controlled reagents for analysis of mixtures by mixing defined proportions of homogeneous templates containing genetically engineered mutations at specific sites (7, 12). While this approach may provide some useful information, our data suggest that simple plasmid-derived mixtures may not be complex enough to thoroughly test proficiency in the performance of HIV-1 genotyping assays.
The variability among laboratories in the perception of what constitutes an “acceptable” sequence (prior to editing) was unexpected. Our data suggest that individuals within and between laboratories may have different perceptions of data quality. In order to evaluate a laboratory's genotyping performance, it will be necessary to define what decision-making factors are used in sequence analysis and the extent to which they need to be or can be monitored. It may be difficult to establish methods that account for an individual's perception of data quality.
In addition, evaluation of a questionnaire about editing was enlightening. The individuals performing the assay clearly knew the guidelines for editing that were presented during their training and apparently used these guidelines most of the time. However, among the 12 respondents from 10 laboratories, 9 developed their own set of guidelines for editing. The questionnaire also indicated that the guidelines developed for editing by the manufacturer were used inconsistently. The overall concordance of the sequence data indicates that the application of multiple sets of in-house editing guidelines, overlaid on those learned during training, was not generally detrimental to the consistency and quality of the data.
Editing decisions are inherently made during the use of all commercially available genotyping systems. The genotyping system described here has been modified in the Celera Diagnostics ViroSeq HIV-1 Genotyping System, recently granted 510K approval by the U.S. Food and Drug Administration for clinical use. Editing still must be performed despite the incorporation of a more simplified editing process and improved algorithms for base calling. Generally, the discrepancies noted in data interpretation indicate the need to form more specific editing guidelines that can be applied consistently to sequence-based genotyping assays currently in use. Future studies are needed to evaluate the impact of editing on each available sequence-based HIV genotyping system.
Our study suggests that global guidelines should be designed to help make editing practices consistent for all laboratories, regardless of the assay used. Editing strategies and evaluation of data quality are an integral part of any commercial or in-house sequence-based genotyping assay (5, 11, 12). The technical performance of genotyping assays, as well as inherent sample variability, is also likely to influence the amount of editing required for data interpretation. Editing procedures and practices should be evaluated as part of any proficiency testing, quality control, or quality assurance program for HIV-1 genotyping.
Acknowledgments
This work was supported by Public Health Service contract NO1-AI-85354 from the National Institute of Allergy and Infectious Diseases.
We thank Applied Biosystems for providing the HGS with software version 2.1.
Members of the Pediatric AIDS Clinical Trials Group Sequencing Working Group include Grace Aldrovandi, University of Alabama at Birmingham; Donald J. Brambilla, New England Research Institute, Inc.; Clark Brown, Applied Biosystems; Susan H. Eshleman, Johns Hopkins Medical Institutions; Susan Fiscus, University of North Carolina; Lisa Frenkel, University of Washington; Hasnah Hamdan, Nichols Institute; Stephen Hart, Frontier Science and Technology Research Foundation; Diana D. Huang, Rush Medical College; Andrea Kovacs, University of Southern California; Paul Krogstad, University of California at Los Angeles; Phillip LaRussa, Columbia University; Paul E. Palumbo, University of Medicine and Dentristry of New Jersey; Walter Scott, University of Miami; Stephen Spector, University of California at San Diego; John Sullivan, University of Massachusetts; Adriana Weinberg, University of Colorado Health Sciences Center; and Yu Qi Zhao, Northwestern University.
REFERENCES
- 1.Arens, M. 2001. Clinically relevant sequence-based genotyping of HBV, HCV, CMV, and HIV. J. Clin. Virol. 22:11-29. [DOI] [PubMed] [Google Scholar]
- 2.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. I. Abrams, B. J. Brizz, J. P. Ioannidis, T. C. Merigan, et al. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. AIDS 14:F83-F93. [DOI] [PubMed] [Google Scholar]
- 3.Clevenbergh, P., J. Durant, P. Halfon, P. del Giudice, V. Mondain, N. Montagne, J. M. Schapiro, C. A. Boucher, and P. Dellamonica. 2000. Persisting long-term benefit of genotype-guided treatment for HIV-infected patients failing HAART. The Viradapt Study: week 48 follow-up. Antiviral Ther. 5:65-70. [PubMed] [Google Scholar]
- 4.DeGruttola, V., L. Dix, R. D'Aquila, D. Holder, A. Phillips, M. Ait-Khaled, J. Baxter, P. Clevenbergh, S. Hammer, R. Harrigan, D. Katzenstein, R. Lanier, M. Miller, M. Para, S. Yerly, A. Zolopa, J. Murray, A. Patick, V. Miller, S. Castillo, L. Pedneault, and J. Mellors. 2000. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antiviral Ther. 5:41-48. [DOI] [PubMed] [Google Scholar]
- 5.Erali, M., S. Page, L. G. Reimer, and D. R. Hillyard. 2001. Human immunodeficiency virus type 1 drug resistance testing: a comparison of three sequence-based methods. J. Clin. Microbiol. 39:2157-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanna, G. J., and R. T. D'Aquila. 2001. Clinical use of genotypic and phenotypic drug resistance testing to monitor antiretroviral chemotherapy. Clin. Infect. Dis. 32:774-782. [DOI] [PubMed] [Google Scholar]
- 7.Hanna, G. J., V. A. Johnson, D. R. Kuritzkes, D. D. Richman, J. Martinez-Picado, L. Sutton, J. D. Hazelwood, and R. T. D'Aquila. 2000. Comparison of sequencing by hybridization and cycle sequencing for genotyping of human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 38:2715-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA Panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 9.King, R. W., S. Garber, D. L. Winslow, C. Reid, B. L. T., E. Anton, and M. J. Otto. 1995. Multiple mutations in the human immunodeficiency virus protease gene are responsible for decreased susceptibility to protease inhibitors. Antiviral Chem. Chemother. 669:80-88. [Google Scholar]
- 10.Parikh, U., C. Calef, B. Larder, R. Schnazi, and J. W. Mellors. 2001. Mutations in retroviral genes associated with drug resistance, p. 191-277. In C. Kuiken, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. W. Mellors, S. Wolinsky, and B. Korber (ed.), HIV sequence compendium 2001. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 11.Romanelli, F., and C. Pomeroy. 2000. Human immunodeficiency virus drug resistance testing: state of the art in genotypic and phenotypic testing of antiretrovirals. Pharmacotherapy 20:151-157. [DOI] [PubMed] [Google Scholar]
- 12.Schuurman, R., L. Demeter, P. Reichelderfer, J. Tijnagel, T. de Groot, and C. Boucher. 1999. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 37:2291-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinstein, M. C., S. J. Goldie, E. Losina, C. J. Cohen, J. D. Baxter, H. Zhang, A. D. Kimmel, and K. A. Freedberg. 2001. Use of genotypic resistance testing to guide hiv therapy: clinical impact and cost-effectiveness. Ann. Intern. Med. 134:440-450. [DOI] [PubMed] [Google Scholar]