Abstract
Seed formation in flowering plants requires meiosis of the megaspore mother cell (MMC) inside the ovule, selection of a megaspore that undergoes mitosis to form an embryo sac, and double fertilization to initiate embryo and endosperm formation. During apomixis, or asexual seed formation, in Hieracium ovules, a somatic aposporous initial (AI) cell divides to form a structurally variable aposporous embryo sac and embryo. This entire process, including endosperm development, is fertilization independent. Introduction of reproductive tissue marker genes into sexual and apomictic Hieracium showed that AI cells do not express a MMC marker. Spatial and temporal gene expression patterns of other introduced genes were conserved commencing with the first nuclear division of the AI cell in apomicts and the mitotic initiation of embryo sac formation in sexual plants. Conservation in expression patterns also occurred during embryo and endosperm development, indicating that sexuality and apomixis are interrelated pathways that share regulatory components. The induction of a modified sexual reproduction program in AI cells may enable the manifestation of apomixis in Hieracium.
INTRODUCTION
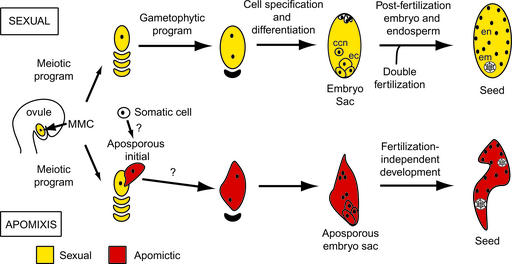
Seed formation in angiosperms begins with the formation of the flower and the male and female reproductive organs, the anther and ovule, respectively. Each contains cells that undergo meiotic reduction and then mitosis to form the male and female gametophytes. The entry of two male sperm cells into the female gametophyte (embryo sac) and their respective fusion with the egg and two central cell nuclei therein during double fertilization enables the growth of the embryo and the endosperm compartments of the seed (Figure 1).
Figure 1.
Mechanisms of Sexual and Apomictic Reproduction in Hieracium.
ccn, central cell nucleus; ec, egg cell; em, embryo; en, endosperm.
The events of sexual seed formation are characterized by spatial and temporal changes in the nuclear DNA content of cells with regard to both chromosome ploidy and the DNA contributed from maternal and paternal genomes. The gametophyte cell nuclei are reduced in DNA content, distinguishing them from the surrounding cells of the sporophyte. Fertilization restores the nuclear DNA content in the egg cell to that of the sporophytic generation, and each subsequent embryo cell contains maternal and paternal genomes present in equal ratio. By contrast, primary endosperm nuclei contain an unequal maternal-to-paternal genome ratio of 2:1, which is essential for seed viability in many species (Johnston et al., 1980; Lin, 1984; Chaudhury et al., 2001).
In some flowering plants, seeds also form asexually by apomixis. This term describes a highly variant suite of reproductive processes in which meiosis is avoided during embryo sac formation, embryo development is fertilization independent, and endosperm formation may or may not require fertilization (Nogler, 1984; Koltunow, 1993). These features result in unreduced embryo sac formation, embryos retaining the maternal genotype of the sporophyte, and endosperm that either retains the 2:1 maternal-to-paternal genome ratio or deviates significantly from it to the extent of completely lacking a paternal genome equivalent (for reviews, see Savidan, 2000; Grimanelli et al., 2001; Spillane et al., 2001).
The nature of the cells that initiate apomixis and the subsequent structures formed has been surmised from their positions within a tissue and their morphological appearance compared with those involved in sexual reproduction. Structures formed during apomixis can be similar to or can vary significantly in form and position relative to the surmised sexual counterpart (Nogler, 1984; Willemse and van Went, 1984; Koltunow, 1993; Naumova and Willemse, 1995; Koltunow et al., 1998; Araujo et al., 2000). In apomictic Beta (Jassem, 1990), Hieracium (Koltunow et al., 1998), Rosaceae species (Nybom, 1988; Izmailow, 1994), and Hypericum (Brutovska et al., 1998), variation in embryo sac, embryo, and endosperm development is evident among the ovules and seeds of an individual plant.
Plants that reproduce by apomixis also retain the capacity to reproduce sexually to varying degrees. Sexual and apomictic reproduction appear independent, but they are not mutually exclusive. For example, in some apomicts, such as Hieracium (Figure 1), the sexual process ceases if apomixis initiates in the ovule, whereas in others, both processes occur side by side in a competitive manner. The coexistence of apomixis and sexual reproduction in an apomictic plant and the polyploid nature of most apomicts complicate genetic analyses. Nevertheless, it has been demonstrated that different modes of apomixis are conferred by a few Mendelian loci in which apomixis usually is dominant over sexual reproduction (do Valle and Savidan, 1996; Barcaccia et al., 1998; Tas and Van Dijk, 1999; Bicknell et al., 2000). Models proposed to explain the manifestation of apomixis suggest that it may be a novel pathway distinct from sexual reproduction (Roche et al., 1999) or an aberrant form of sexual reproduction generated by hybridization, mutation, or epigenetic change (Peacock, 1992; Carman, 1997; Grossniklaus, 2001).
However, there is a complete absence of data concerning the identity of cells that initiate apomixis, the molecular processes that regulate apomixis, and the relationship between sexual and apomictic pathways. Relatively few apomicts have been studied at the molecular level. Differential and subtractive hybridization have been attempted to isolate genes expressed during apomictic reproduction (Vielle-Calzada et al., 1996; Guerin et al., 2000; Pessino et al., 2001). Some genes have been identified, but comparative spatial and temporal expression in sexual and apomictic plants and their functional relevance to apomixis have not been examined. Genes involved in and known to regulate gametophyte, endosperm, and embryo development have now been identified in sexually reproducing plant species, providing tools for the comparative analysis of gene expression programs during sexual and apomictic reproduction (Chaudhury et al., 1997; Christensen et al., 1998; Grini et al., 2002).
Many apomicts and their sexual relatives are not readily amenable to plant transformation. This is not the case for composite Hieracium species, which display a generation time from seed to seed of 4 months and are suited to the molecular analysis of apomixis (Koltunow et al., 1995; Bicknell et al., 2000). Apomixis is dominant in Hieracium, and a number of developmentally characterized lines that exhibit differences in the frequency and timing of apomictic initiation in addition to variable modes of embryo sac formation have been genotyped (Koltunow et al., 1998, 2000; Bicknell et al., 2000). Apomixis in Hieracium begins after the initiation of sexual reproduction. Somatic cells termed aposporous initial (AI) cells appear either before or after megaspore mother cell (MMC) meiosis, and sexual reproduction usually ceases (Figure 1). AI cells enlarge and undergo mitosis to form aposporous embryo sacs, and embryo and endosperm development is fertilization independent (Koltunow et al., 1998).
Here, we examine the molecular relationships between sexual and apomictic pathways in Hieracium. Developmental markers from other plant species representing genes with known and informative expression patterns in sexual reproduction were introduced into sexual and apomictic plants. Comparative analyses of gene expression patterns showed that sexual and apomictic reproduction are molecularly related pathways that share regulatory programs.
RESULTS
AtFIS:GUS Gene Expression Marks Mature Embryo Sacs and Initiating Seed Structures in Sexual Hieracium
Fertilization-independent (autonomous) endosperm development is one component of apomixis in Hieracium. In Arabidopsis thaliana (At), mutations in any of three members of the FERTILIZATION-INDEPENDENT SEED (FIS)–class genes MEDEA (MEA/FIS1), FIS2, and FERTILIZATION-INDEPENDENT ENDOSPERM (FIE/FIS3) result in the fertilization-independent initiation of endosperm development (Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998). Chimeric AtMEA:β-glucuronidase (GUS), AtFIS2:GUS, and AtFIE:GUS fusion constructs are coexpressed in comparable amounts during the late events of gametophyte development and early endosperm formation in transgenic Arabidopsis plants (Luo et al., 2000; Yadegari et al., 2000). However, the spatial expression patterns of the three chimeric genes vary slightly in developing seeds: AtFIS2:GUS activity is specific to the endosperm nuclei, AtMEA: GUS activity is restricted to the nuclear endosperm domains, and AtFIE:GUS activity is detected in both the endosperm nuclear domains and the embryo. The expression of AtFIS2:GUS was detected first around the two central cell nuclei before their fusion (Figure 2A) and continued in the dividing endosperm nuclei until the fifth or sixth nuclear division (Figure 2B) and thereafter only in the endosperm cyst (Figure 2C). AtFIE:GUS expression also is detected during pollen development (Luo et al., 2000).
Figure 2.
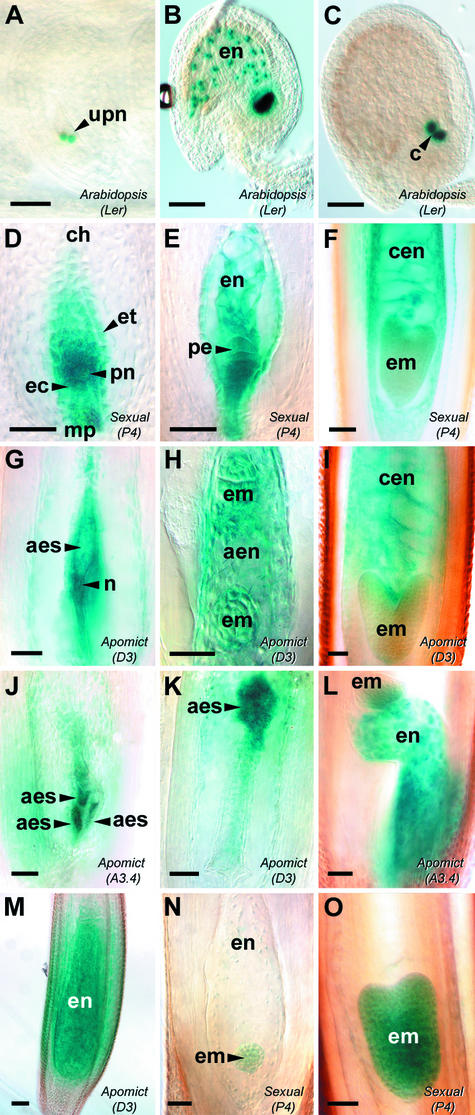
AtFIS2:GUS and AtFIE:GUS Expression during Early Seed Development.
Ovules from Arabidopsis Landsberg erecta AtFIS2:GUS ([A] to [C]), sexual Hieracium P4 ([D] to [F], [N], and [O]), and apomictic Hieracium D3 and A3.4 ([G] to [M]) were stained with GUS and viewed whole-mount using Nomarski differential interference contrast (DIC) microscopy. (A) to (M) show AtFIS2:GUS expression, and (N) and (O) show AtFIE:GUS expression. Bars = 50 μm.
(A) Anthesis ovule containing unfused polar nuclei (upn) within the mature embryo sac.
(B) Postfertilization ovule containing dividing endosperm nuclei (en).
(C) Postfertilization ovule after the sixth mitotic division of endosperm development, with GUS staining in the endosperm cyst (c).
(D) Mature embryo sac in the chalazal (ch)-to-micropylar (mp) orientation surrounded by the endothelium (et), containing a fused polar nucleus (pn) and egg cell (ec).
(E) Postfertilization embryo sac containing dividing endosperm nuclei and a dividing proembryo (pe).
(F) Developing fertilized seed containing an early-heart-stage embryo (em) and cellular endosperm (cen).
(G) Mature aposporous D3 embryo sac (aes) containing central cell–like nuclei (n).
(H) Aposporous D3 embryo sac showing dividing autonomous endosperm nuclei (aen) and two globular embryos.
(I) Developing D3 seed showing a heart-stage embryo and autonomous cellular endosperm.
(J) Multiple aposporous embryo sac structures developing within a single A3.4 ovule.
(K) Displaced aposporous embryo sac developing in the chalazal region of a D3 ovule.
(L) Developing A3.4 seed containing autonomous endosperm and showing parthenogenetic embryo development at the chalazal end of the embryo sac.
(M) Late D3 seed containing autonomous cellular endosperm and no embryo.
(N) Sexual P4 embryo sac containing a globular embryo and dividing endosperm nuclei.
(O) Developing P4 seed containing an early-heart-stage embryo.
The three chimeric AtFIS:GUS genes were introduced into Hieracium to compare their expression during fertilization-dependent and fertilization-independent seed development. The expression patterns of AtFIS2:GUS (Figures 2D to 2F) and AtMEA:GUS (data not shown) were identical at embryo sac maturity and during early seed development in sexual Hieracium P4. High levels of GUS activity were detected around the polar nuclei within the embryo sac and at a lower level throughout the central cell, the egg apparatus, and the endothelium (Figure 2D). After fertilization, AtFIS2:GUS expression in sexual Hieracium resembled that observed in Arabidopsis and was associated with the primary endosperm nucleus and the dividing endosperm nuclei (Figure 2E). However, AtFIS2:GUS expression was not localized tightly to endosperm nuclei, as observed in Arabidopsis (cf. Figures 2B and 2E), and continued to be expressed in endosperm cells after the formation of cell walls (Figure 2F). During embryogenesis, AtFIS2:GUS expression was detected in the dividing zygote and persisted until the heart stage of embryo development (Figures 2E and 2F). The same expression was observed for AtMEA:GUS in fertilized Hieracium embryos (data not shown), and this has not been observed in Arabidopsis.
The expression of AtFIE:GUS at embryo sac maturity and during early endosperm and early embryo development was barely detectable in the transgenic plants examined (Figure 2N) compared with the expression of the AtMEA:GUS and AtFIS2:GUS genes. However, the intensity of AtFIE:GUS expression increased during the later stages of embryo development in Hieracium (Figure 2O), attaining a level comparable to that of the other two genes.
The expression patterns of the three AtFIS:GUS genes at embryo sac maturity and seed initiation in sexual Hieracium were similar but not identical to those observed in Arabidopsis. However, the coordinated expression of the heterologous AtFIS:GUS genes in key structures formed during sexual reproduction underscored their suitability as markers for comparing gene expression patterns during aposporous embryo sac formation and fertilization-independent seed initiation in apomictic Hieracium.
Conservation of AtFIS:GUS Expression during Embryo Sac Maturation and Seed Initiation in Sexual and Apomictic Hieracium
Plants from two distinct apomictic Hieracium species, H. aurantiacum A3.4 and H. piloselloides D3, were selected to examine the expression patterns of introduced AtFIS:GUS genes. These plants are well characterized and vegetatively propagated and exhibit different modes of aposporous embryo sac formation (Koltunow et al., 1998). One to four aposporous initial cells differentiate in D3, of which only one usually gives rise to a mature embryo sac (Figure 1). A3.4 characteristically initiates multiple aposporous embryo sacs in different locations in an ovule, and these generally coalesce to form a single aposporous embryo sac of varying cellular structure with variable spatial locations. Mature ovules from both apomicts tend to contain a single mature aposporous embryo sac that is situated in the micropylar region of the ovule, the conserved position of embryo sacs in ovules of sexual plants. However, both apomicts also can develop an embryo sac in the chalazal region of the ovule.
Like that observed in sexual P4, the expression patterns of the AtFIS2:GUS (Figures 2G to 2M) and AtMEA:GUS (data not shown) chimeric genes were identical during embryo sac and fertilization-independent seed development in apomictic Hieracium. GUS activity arising from the introduced AtFIS2:GUS gene was evident in mature aposporous embryo sacs situated in the micropylar region (Figure 2G), multiple coalescing embryo sacs in A3.4 (Figure 2J), and aposporous embryo sacs found in the chalazal region of ovules from the transgenic apomictic Hieracium plants (Figure 2K). Similar to the expression in sexual P4, AtFIE:GUS expression was significantly lower than that of the other two genes during early embryo sac development (data not shown). Just before the initiation of fertilization-independent endosperm in both apomicts, a high level of AtFIS2:GUS activity was detected around nuclei positioned in the center of the aposporous embryo sac, and a lower level of GUS activity was observed consistently throughout the central cell (Figure 2G). This increase in staining around the polar nuclei suggests that, as in sexual reproduction, central cell nuclei are the likely progenitors of autonomous endosperm in apomictic Hieracium.
The nuclei formed during fertilization-independent endosperm divisions also were marked with AtFIS2:GUS (Figure 2H). After cell walls had formed between the syncytial nuclei, the endosperm cells continued to express the AtFIS2:GUS gene until the torpedo stage of embryo development (Figure 2I), consistent with the expression observed in the fertilized sexual plant (Figure 2F). Coexpression of AtFIS2:GUS and AtMEA:GUS also was observed in patches of endosperm that failed to cellularize (data not shown) and in the cellularized endosperm of seeds in which embryos failed to initiate (Figure 2M).
The AtFIS2:GUS (Figures 2H, 2I, and 2L), AtMEA:GUS, and AtFIE:GUS (data not shown) genes were coexpressed during fertilization-independent embryo formation when an embryo formed at the micropylar end of the aposporous embryo sac (Figure 2I) and also in multiple embryos that formed in different spatial locations (Figures 2H and 2L). GUS activity from the three chimeric genes in apomictic seeds diminished at the mid to late heart stage of embryo development (Figure 2I), comparable to the expression in the sexual plant (cf. with Figure 2F).
The three AtFIS:GUS genes were coexpressed in maturing aposporous embryo sacs and in the early embryo and endosperm of fertilization-independent seeds of both apomictic Hieracium species. The spatial and temporal expression patterns were comparable to those observed in embryo sacs, embryos, and endosperm in sexual Hieracium. This finding indicates that the spatial and temporal regulation of the introduced AtFIS:GUS genes is common to both sexual and apomictic pathways despite the absence of meiosis and fertilization in the apomictic pathway.
AtSERK1:GUS and SERK-Like Expression Confirm Shared Molecular Elements during Sexual and Apomictic Reproduction
Another component of apomixis is the fertilization-independent formation of embryos, a phenomenon that also occurs during somatic embryogenesis. The expression of the SOMATIC EMBRYO RECEPTOR KINASE (DcSERK) gene marks the vegetative-to-embryogenic transition in carrot, but its function remains unclear (Schmidt et al., 1997). The Arabidopsis SERK1 gene (AtSERK1) is expressed in germline cells during megagametogenesis as well as in pre-heart-stage developing embryos and is thought to play an important role in embryogenesis (Hecht et al., 2001).
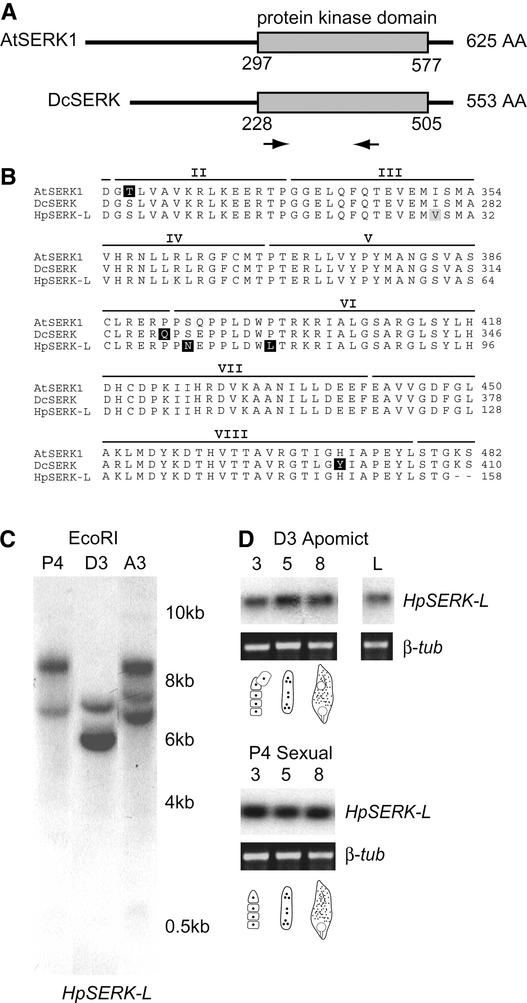
To determine if Hieracium contained SERK-like sequences, degenerate primers were designed to amplify part of the conserved kinase domain common to DcSERK and AtSERK1 (Figure 3A). Sequence analysis of the cloned reverse transcriptase–mediated (RT) PCR products obtained from early D3 Hieracium seeds revealed a partial 516-bp SERK-like clone (HpSERK-L) that was 82% identical to the DcSERK coding sequence. The cDNA fragment also showed >80% identity at the DNA level to the Arabidopsis SERK1, SERK2, and SERK3 genes. The HpSERK-L, DcSERK, and AtSERK1 putative amino acid sequences were almost identical across 7 of the 11 kinase domains (Figure 3B).
Figure 3.
SERK-Like Genes in Hieracium.
(A) Scheme of the AtSERK1 and DcSERK amino acid sequences showing the positions of the protein kinase domain and the degenerate primers (arrows). AA, amino acids.
(B) Alignment of the putative amino acid sequences for AtSERK1 and DcSERK over the length of the Hieracium SERK-like (HpSERK-L) fragment. Nonconserved residues are highlighted in black, and similar residues are highlighted in gray. The positions of the kinase domains (II to VIII) are indicated above the sequences (Schmidt et al., 1997).
(C) Genomic analysis of HpSERK-L. Genomic DNA samples were digested with EcoRI, which cuts external to the HpSERK-L fragment.
(D) RT-PCR expression analysis of HpSERK-L during ovary development. Total RNA was isolated from stages 3, 5, and 8 of ovary development, which coincide with meiosis, megagametogenesis, and early embryo development, respectively. The diagrams indicate the approximate stages of embryo sac development. RT-PCR samples were compared to a β-tubulin control (β-tub). L, leaves.
Hybridization of the HpSERK-L fragment to genomic DNA from sexual P4, apomictic D3, and apomictic A3.4 Hieracium revealed that few copies of the gene were likely to be present in these different polyploid plants (Figure 3C). Labeled AtSERK1 cDNA probe hybridized to the same genomic DNA fragments (data not shown). RT-PCR analysis using sequence-specific primers showed the presence of HpSERK-L mRNA transcripts from early ovule development until early seed development in both sexual and apomictic Hieracium (Figure 3D). Similar levels of expression were detected in both plants, and this temporal pattern of HpSERK-L gene expression corresponded to that observed for AtSERK1, except that HpSERK-L transcript also was detected in young, unexpanded Hieracium leaves. These data confirmed the presence of SERK-like sequences in Hieracium, and subsequent expression analyses used the AtSERK1:GUS and DcSERK probes with known expression patterns.
When the AtSERK1:GUS gene was introduced into Hieracium, the same spatial and temporal pattern of expression was observed in both sexual and apomictic plants. GUS activity was detected initially in developing florets in the region destined to give rise to the inferior ovary (data not shown). As the MMC and megaspores developed in the ovule, the expression of AtSERK1:GUS was not restricted to the nucellar region containing the megaspores, which is the case in Arabidopsis, but GUS activity was observed throughout the ovule (Figure 4A).
Figure 4.
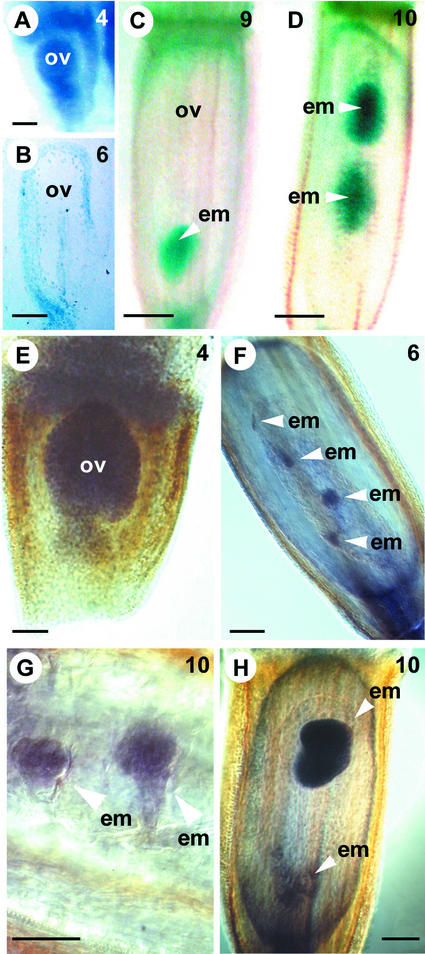
AtSERK1:GUS and HpSERK-L mRNA Expression during Seed Development in Hieracium.
(A) to (D) show cleared GUS-stained Hieracium ovaries viewed whole-mount or after sectioning (B) using DIC microscopy, and (E) to (H) show WISH samples viewed whole-mount or after sectioning (G) using Nomarski DIC microscopy. The numbers at top right indicate the ovary stage (Koltunow et al., 1998). Bars = 50 μm in (A), (B), (E), (F), and (H), 100 μm in (C) and (D), and 25 μm in (G).
(A) Early ovule (ov) from sexual P4.
(B) Thin section (2 μm) of a GUS-stained mature ovule from apomictic D3.
(C) Fertilized P4 seed containing a globular embryo (em).
(D) Seed from D3 containing two globular-to-heart stage embryos.
(E) Early ovule from apomictic D2.
(F) Anthesis ovule from apomictic D2 containing four small embryo structures, hybridized with antisense DcSERK probe.
(G) Early globular embryos in a D2 seed hybridized with antisense DcSERK probe.
(H) Seed from apomictic D2 containing a chalazal misshapen globular embryo and a micropylar heart-stage embryo hybridized with antisense DcSERK probe.
AtSERK1:GUS expression was not detected during megagametogenesis in sexual P4 or during aposporous embryo sac formation in apomictic D3, but expression was restricted to cells surrounding the vascular bundle of the ovule (Figure 4B), a pattern not evident in Arabidopsis. AtSERK1:GUS activity was not detected clearly in the egg cell of sexual plants or within the cells of the aposporous embryo sacs at seed initiation, and expression may have been below the sensitivity limit of detection. However, after fertilization in sexual Hieracium, embryos expressed GUS from the early four- to eight-cell stage until the heart stage of embryogenesis (Figure 4C). AtSERK1:GUS expression was not observed in developing endosperm. Similarly, in apomictic Hieracium, globular and also irregular clumps of cells morphologically resembling embryos were marked with GUS activity (Figure 4D), and embryo expression of AtSERK1:GUS began to decrease soon after the transition to the heart stage of development.
DcSERK was used as an antisense probe in whole-mount in situ hybridization (WISH) experiments. The same pattern of transcript localization was observed in both sexual and apomictic Hieracium and was essentially identical to that observed in transgenic plants expressing the AtSERK1:GUS gene. SERK-like mRNA sequences were detected in young ovules (Figure 4E) but not in the egg cell or the zygotes of sexual plants. WISH analysis of seeds from diploid apomictic Hieracium D2 that display a high frequency of multiple fertilization-independent embryos failed to detect SERK-like transcript until the embryos contained at least four to eight cells (Figure 4F). Similar to the AtSERK1:GUS expression, SERK-like mRNA sequences were detected clearly in small globular embryos (Figure 4G) and subsequently in embryos until the early heart stage in both sexual P4 (Figure 4C) and apomictic D2 (Figure 4H). These expression data reflect in part the patterns observed for AtSERK1 in Arabidopsis and suggest a role for endogenous Hieracium SERK-like sequences during ovule and seed development.
Both the AtSERK1:GUS gene and an endogenous HpSERK-L gene appear to be regulated spatially and temporally in a similar manner in sexual and apomictic Hieracium. The conserved patterns of SERK and AtFIS:GUS marker expression during the temporal events of sexual and apomictic reproduction in Hieracium collectively support the conclusion that both sexual and apomictic pathways share common molecular regulatory features and are not distinct pathways.
Aposporous Initial Cells Do Not Express an MMC Marker
Given the common spatial and temporal regulation of marker genes in sexual and apomictic pathways in Hieracium, we attempted to examine the identity of the AI cell to determine the point of molecular convergence between the two reproductive processes. We reasoned that the AI cell might differentiate with an identity resembling the MMC, whose fate is deregulated to avoid meiosis. Alternatively, the AI might differentiate with functional identity resembling a selected spore fated to undergo the mitotic events of embryo sac formation.
The marker genes available to test this are limited. The SPROCYTELESS (SPL) gene (Yang et al., 1999), also known as NOZZLE (Schiefthaler et al., 1999), is required for the initiation of male and female sporogenesis in Arabidopsis. SPL function is essential for MMC formation (Yang et al., 1999). Homozygous AtSPL:GUS Arabidopsis ovules show expression in the MMC, the megaspores after meiosis, the nonfunctional megaspores after spore selection, and intermittently in the surrounding nucellar epidermal layer but not in functional megaspores.
In sexual Hieracium (P4) plants, AtSPL:GUS expression was localized specifically to the male and female sporogenic tissues. GUS activity was detected clearly in developing anthers in pollen mother cells (Figures 5A and 5B) and microspore tetrads (Figure 5C), and also in the ovule, where it was restricted to the MMC (Figure 5D). In contrast to the expression of AtSPL:GUS in Arabidopsis ovules, GUS activity in sexual Hieracium was not evident in the nucellar epidermis or in any of the megaspores during functional megaspore selection (Figure 5E) or degeneration. The MMC-specific pattern was observed in four independent lines by dark-field microscopy and Nomarksi differential interference contrast microscopy and verified by serial sections (data not shown). AtSPL:GUS activity was detected clearly in MMCs within ovules from five apomictic (D3) Hieracium lines (Figure 5F) but not in the aposporous initial cells that differentiated soon afterward when the MMC underwent meiosis (Figure 5G).
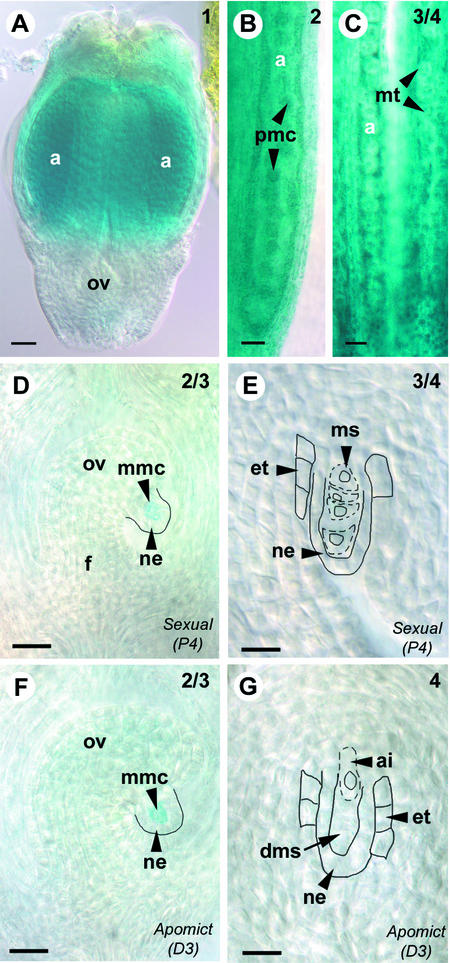
Figure 5.
AtSPL:GUS Expression during Floral Development in Hieracium.
Cleared GUS-stained Hieracium florets ([A] to [C]) and ovaries ([D] to [E]) viewed whole-mount using Nomarski DIC microscopy. The numbers at top right indicate the ovary stage (Koltunow et al., 1998). Bars = 50 μm in (A) to (C) and 25 μm in (D) to (G).
(A) Early floret from apomictic D3 containing a small ovule (ov) and immature anthers (a).
(B) Enlargement of an anther from apomictic D3 containing pollen mother cells (pmc).
(C) Anthers from sexual P4 containing microspore tetrads (mt).
(D) Ovule from sexual P4 containing a megaspore mother cell (mmc) showing GUS activity surrounded by the nucellar epidermis (ne). The funiculus (f) is indicated to aid orientation.
(E) Ovule from sexual P4 showing four megaspores (ms), outlined with dashed lines, surrounded by the nucellar epidermis and the developing endothelium (et), outlined with solid lines. No staining is detected in the indicated structures.
(F) Ovule from apomictic D3, showing the corresponding stage of apomictic development to (D), containing a megaspore mother cell showing GUS activity.
(G) Ovule from apomictic D3, showing the corresponding stage of apomictic development to (E), containing an expanding aposporous initial cell (ai) chalazal to the degenerating megaspores (dms). No staining is detected in the indicated structures.
These data show that the expression of the AtSPL:GUS chimeric gene clearly marks MMCs in Hieracium ovules but, unlike in Arabidopsis, it does not mark later temporal events of megasporogenesis. Nevertheless, the expression patterns in Hieracium show that at the time of cytologically discernible differentiation, aposporous initial cells do not express the AtSPL:GUS MMC marker gene; hence, they are unlikely to share identity with the MMC.
AtFIS2:GUS First Marks Megaspores and Is Downregulated in the Selected Megaspore
Surprisingly, AtFIS2:GUS expression also was detected early in Hieracium ovule development during megaspore selection. GUS activity was absent in the enlarging selected chalazal-most megaspore but was present in the three micropylar spores destined to degenerate (Figure 6A). These expression patterns were confirmed by serial sections (data not shown) and whole-mount analysis (Figure 6B). GUS activity remained absent in the functional selected megaspore as it expanded to fill the cavity left by the degenerating megaspores and the nucellar epidermis (Figure 6C). AtFIS2:GUS expression was then observed once the selected megaspore nucleus divided during the first division of embryo sac formation (Figure 6D), and expression continued during embryo sac maturation and fertilization-dependent seed initiation as described previously (Figure 2).
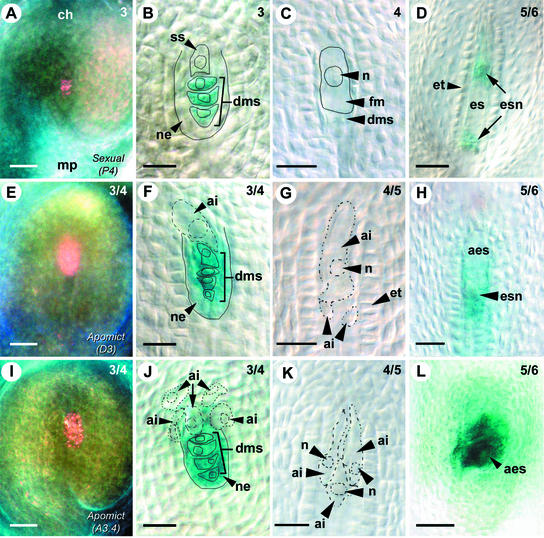
Figure 6.
AtFIS2:GUS Expression during Early Ovule Development in Hieracium.
Ovules from sexual Hieracium P4 ([A] to [D]), apomictic Hieracium D3 ([E] to [H]), and apomictic Hieracium A3.4 ([I] to [L]) were stained with GUS and viewed whole-mount using dark-field microscopy ([A], [E], and [I]) or Nomarski DIC microscopy. The numbers at top right indicate the ovary stage (Koltunow et al., 1998). Bars = 50 μm in (A), (E), (I), and (L) and 25 μm in (B) to (D), (F) to (H), (J), and (K).
(A) P4 ovule in the chalazal (ch)-to-micropylar (mp) orientation showing GUS stain as pink.
(B) Enlarging selected spore (ss) and blue GUS-stained degenerating megaspores (dms) surrounded by the nucellar epidermis (ne). Indicated structures are outlined with solid lines.
(C) Enlarging functional megaspore (fm) with a large nucleus (n) chalazal to degenerated megaspores. Indicated structures are outlined with solid lines.
(D) Ovule containing an early embryo sac (es) containing dividing embryo sac nuclei (esn) and surrounded by the endothelium (et).
(E) D3 ovule showing the corresponding stage of apomictic development to (A).
(F) Enlarging aposporous initial cell (ai) at the corresponding stage of apomictic development to (B). The aposporous initial cell, outlined with a broken line, forms in a slightly different plane than the other structures, which are outlined with solid lines.
(G) Enlarging aposporous initial cell above two smaller initial cells.
(H) D3 ovule containing a dividing aposporous embryo sac (aes) with embryo sac nuclei at the corresponding stage of apomictic development to (D).
(I) A3.4 ovule showing the corresponding stage of apomictic development to (A) and (E).
(J) Enlarging aposporous initial cells at the corresponding stage of apomictic development to (B) and (F). Multiple aposporous initial cells are indicated with broken lines, and some form in slightly different planes compared with the sexual structures, which are outlined with solid lines.
(K) Enlarging aposporous initial cells.
(L) Aposporous embryo sac structures at the corresponding stage of apomictic development to (D) and (H).
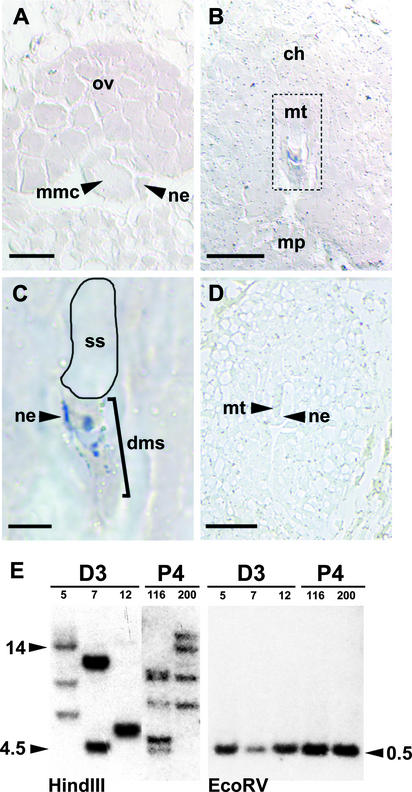
In situ hybridization using a probe to detect GUS mRNA transcripts was performed to determine if the megaspore expression pattern in transgenic plants containing AtFIS2:GUS was related to the selective distribution of mRNA synthesized in the MMC before meiosis or to regulatory events that occur during and/or after meiosis. AtFIS2:GUS mRNA was not detectable in the MMC of sexual plants (Figure 7A). Transcripts were not detected in the selected functional megaspore after meiosis but were evident in degenerating megaspores and some of the degenerating nucellar cells (Figures 7B and 7C). Once the nucleus of the functional megaspore divided, AtFIS2:GUS mRNA was detected in the early embryo sac (data not shown).
Figure 7.
In Situ Hybridization and Genomic Analysis of AtFIS2:GUS in Hieracium.
(A) to (D) show thin sections of AtFIS2:GUS sexual P4 ovules hybridized with antisense ([A] to [C]) or sense (D) labeled GUS RNA.
(A) Megaspore mother cell (mmc) surrounded by the nucellar epidermis (ne). ov, ovule. Bar = 15 μm.
(B) Ovule in the chalazal (ch)-to-micropylar (mp) orientation showing the megaspore tetrad (mt) outlined with a dashed line. Bar = 50 μm.
(C) Magnified outlined region from (B) showing staining in the degenerating megaspores (dms) and some nucellar epidermis cells. No staining is evident in the selected spore (ss). Bar = 12.5 μm.
(D) Ovule at a similar stage to that in (B), hybridized with sense GUS RNA. Bar = 50 μm.
(E) DNA gel blot analysis of the AtFIS2:GUS transgene in apomictic D3 (independent lines 5, 7, and 12) and sexual P4 (independent lines 116 and 200) Hieracium. Genomic DNA (15 μg each) was digested with HindIII, which cuts the transgene only 5′ of the probed sequence, and EcoRV, which cuts the transgene on both sides of the probed sequence. The expected size of the EcoRV fragment is 575 bp. The lengths of the DNA fragments (kilobases) are indicated for each blot.
The absence of both AtFIS2:GUS mRNA and protein activity in the enlarged functional megaspore was not a consequence of the absence of the AtFIS2:GUS gene from that cell as a result of meiotic segregation. Seven independent sexual AtFIS2:GUS transgenic lines were generated, and the copy number of the gene in these hemizygous plants varied from three to five insertions (Figure 7E). In AtFIS2:GUS plants containing a single copy of the transgene, 50% of the megaspores should contain the gene after meiosis, whereas all of the megaspores are likely to contain the transgene in plants with multiple unlinked copies of the gene.
Examination of 657 ovules from sexual P4 plants with different numbers of transgene insertions showed that the temporal and spatial patterns of AtFIS2:GUS expression were surprisingly conserved between MMC differentiation and meiosis. GUS activity was never evident in the MMC, but in ovules that contained early megaspores soon after meiosis (n = 148), GUS activity was detected in all four spores, with the activity in the selected chalazal-most spore weaker than that of the other megaspores. Ovules that contained an enlarged functional megaspore displayed GUS activity only in the three degenerating megaspores (n = 509). This finding suggests that in plants containing multiple copies of the transgene, in which all meiotic products should inherit a copy, processes associated with functional megaspore specification also are involved in switching off AtFIS2:GUS expression in that cell.
AtFIS2:GUS Is Expressed Differentially at Meiosis in Apomictic Hieracium
The AtFIS2:GUS gene was considered a suitable marker to further investigate AI cell identity. Differential expression of the AtFIS2:GUS gene was observed in two apomictic Hieracium species (D3 and A3.4) that possess different modes of aposporous embryo sac formation soon after meiosis by dark-field microscopy (Figures 6E and 6I), and the pattern was confirmed by serial sections (data not shown). In contrast to the pattern observed in sexual plants, AtFIS2:GUS was detected in all four megaspores soon after their formation and during their subsequent degeneration. AtFIS2:GUS also was expressed in the surrounding cells of the nucellar epidermis enveloping the tetrad of megaspores (Figures 6F and 6J), which degrades in both sexual and apomictic plants.
In both apomictic Hieracium species, AtFIS2:GUS expression was completely absent from aposporous initial cells as they enlarged and expanded toward the tetrad (Figures 6F and 6J), just as it was absent from the enlarging selected megaspore in sexual plants. In the apomict D3, a single aposporous initial usually expanded into the position vacated by the degenerating megaspores and nucellar epidermis (Figure 6F). GUS activity was absent from the aposporous initial cell as it expanded into the position vacated by the degenerated megaspores (Figure 6G), but it was detected in the aposporous structure as soon as the nucleus divided and aposporous embryo sac formation began (Figure 6H). In the apomict A3.4, multiple aposporous initials differentiated, and none of these initials expressed AtFIS2:GUS (Figure 6K) until each nucleus divided to initiate aposporous embryo sac development (Figure 6L). As in D3, the expression of AtFIS2: GUS in A3.4 ovules also was evident in the nucellar epidermis and in all four megaspores until these cells had degenerated.
The reasons for the expression of GUS in all four meiotic products of apomictic plants containing only a single insert of the AtFIS2:GUS transgene (Figure 7E) are not clear. Repeated transformation experiments have failed to identify a single-copy sexual P4 AtFIS2:GUS line to enable comparison. We speculate that postmeiotic movement of either the mRNA or the protein may occur during megaspore and/or nucellar epidermal cell degeneration.
The observation that AtFIS2:GUS expression is observed first in AI cells when the nucleus undergoes mitosis during early embryo sac formation suggests that the AI cell may acquire functional identity resembling that of a selected megaspore soon after differentiation. Alternatively, the AI cell may switch to an embryo sac program at the initiation of nuclear mitosis. These possibilities will require further clarification with appropriate markers as they become available.
DISCUSSION
Apomixis Reflects a Deregulated Sexual Program
In the past, apomixis and sexual reproduction were viewed as two distinct processes that have little in common. Ernst (1918) postulated that apomixis might result from the hybridization of related species based on the observation that the apomicts examined at that time were polyploid and highly heterozygous. The pioneering studies of Nogler (1984) and Savidan (1982) proved that apomixis is under genetic control, although genetic modifiers or environmental conditions may affect its expressivity (Nogler, 1984; Savidan, 2000). The subsequent confirmation that relatively few loci control different modes of apomixis and that mutagenesis can induce apparent components of apomixis in sexual plants has lent further support to the hypothesis that apomixis and sexual reproduction might share common elements.
The expression patterns of the AtSPL:GUS, AtSERK1:GUS, and various AtFIS-class:GUS chimeric constructs in sexual and apomictic Hieracium species described here have directly confirmed that apospory and the components of autonomous embryo and endosperm development share gene expression and regulatory components with sexual reproduction. This is in spite of the comparative structural differences in embryo sac, embryo, and endosperm development and also the temporal and spatial changes in cell and tissue ploidy status in the two reproductive pathways.
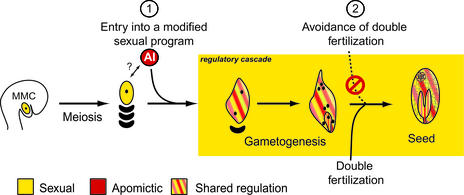
The spatial and temporal expression data of the marker genes were integrated to devise a model for the control of apomixis in Hieracium (Figure 8). Expression of the chimeric constructs was shared from the first nuclear division of the AI cell and the selected megaspore and continued during the mitotic events of embryo sac formation and also fertilization-dependent and fertilization-independent embryo and endosperm formation in sexual plants and apomictic plants, respectively. The markers used did not confirm the functional identity of the AI cell during differentiation from the nucellus and its subsequent enlargement but showed that the AI cell was unlikely to share identity with the MMC during this sequence of events.
Figure 8.
A Model for Apomixis in Hieracium.
The numerals 1 and 2 indicate the checkpoints at which the sexual process is modified in apomictic Hieracium.
The results of marker analyses indicate that aspects of meiosis are avoided during aposporous embryo sac formation in Hieracium, because the AI cell that differentiates from the nucellus (or nucellar epidermis) is directed onto a mitotic embryo sac formation pathway combined with fertilization-independent embryo and endosperm development. This finding, coupled with the shared nature of gene expression programs and regulators in sexual and apomictic pathways, suggests that apomixis is manifested by the induction of a sexual developmental program deregulated in both time and space that leads to cell fate changes and the omission of critical steps in the sexual process (Figure 8).
Interactions between Sporophytic Tissues and Sexual and Apomictic Pathways
Genetic analysis of sexual plants has shown that the events leading to female gametophyte formation and seed development are both independent of and interdependent on the events and signals from surrounding sporophytic ovule tissues (Gasser et al., 1998). The importance of sporophytic ovule signals in governing the progression of the apomictic process is not entirely clear. Earlier studies in Hieracium species with apospory show that induced alterations in ovule development correlate with changes in the apomictic process with regard to the frequency of initiating cells (Koltunow et al., 2000, 2001). Continuation of studies targeting different ovule cell types in Hieracium and similar examinations in other apomicts should further clarify the dependence of sporophytic ovule signals in apomictic development.
Differentiation of the AI cell in apomictic Hieracium coincides with the demise of the sexual process in the ovule. Previous studies have shown that this does not simply occur by physical displacement (Tucker et al., 2001). The demise of the sexual pathway in apomictic Hieracium was shown to correlate with a change in the spatial pattern of AtFIS2:GUS marker expression. A shift in GUS activity from the three nonselected megaspores in the sexual plant to all four megaspores and also the surrounding nucellar epidermal cells in two apomictic species suggested that the presence of the AI might directly influence the demise of the sexual program by altering gene expression programs.
Molecular studies in Arabidopsis have suggested that positional signals involving the functional megaspore control cell fate and degeneration and result in the death of the three nonselected megaspores during megaspore selection in sexual plants (Wu and Cheung, 2000; Yang and Sundaresan, 2000). These mechanisms ensure that only a single cell is specified to initiate megagametogenesis and form an embryo sac. The use of such signals by the AI might influence the demise of all four sexual megaspores in Hieracium, a notion that could be tested with the appropriate markers as they become available.
Other aposporous species form initials at different times of ovule development compared with Hieracium species, and in some of these, such as Brachiaria species (Araujo et al., 2000), the demise of the sexual pathway does not occur. Marker studies to examine cell fate in other aposporous species should determine whether there are differences in the identities of cells that initiate apospory. They also should lead to an understanding of the factors that favor the development of one type at the expense of the other or that enable their coexistence.
Roles of FIS-Class Genes and Genes That Stimulate Somatic Embryogenesis in Apomictic Reproduction
Overexpression of AtSERK1 increases the embryogenic potential of cultured Arabidopsis cells (Hecht et al., 2001). In this study, AtSERK1:GUS and HpSERK-L expression was not observed clearly in the egg of sexual Hieracium and it was detected in embryos only once they contained four to eight cells. Our study does not exclude the possibility that other previously characterized genes may be involved in the parthenogenetic induction of embryogenesis in Hieracium. Several other genes that may be involved in embryo initiation have been described. The overexpression of the unrelated transcription factors BABY BOOM (BBM; Boutilier et al., 2002), LEAFY COTYLEDON1 (LEC1; Lotan et al., 1998) and LEC2 (Stone et al., 2001), and WUSCHEL (WUS; Zuo et al., 2002) leads to somatic embryo formation in transgenic plants. Examination of the expression of LEC1, LEC2, BBM, and WUS markers or their homologs in apomictic species may be instructive regarding their relevance to the induction of parthenogenesis and adventitious embryony.
The early expression of AtFIS2:GUS in Hieracium and the conservation of synchronized expression of the AtMEA:GUS, AtFIS2:GUS, and AtFIE:GUS genes during the late events of embryo sac formation and early seed development raise questions concerning the role of Hieracium FIS-class genes during sexual and apomictic reproduction. The FIS-class genes are postulated to form complexes that alter chromatin structure to inhibit or promote the expression of seed development genes and to regulate endosperm polarity in Arabidopsis (Sorensen et al., 2001; Chaudhury et al., 2001). Recently, homologs of the MEA and FIE genes were cloned from maize, but the functional roles of these genes during maize ovule and seed development have not been ascertained (Springer et al., 2002; Danilevskaya et al., 2003).
A plausible hypothesis is that the altered regulation of Hieracium FIS genes may enable autonomous seed development in apomictic plants. We have isolated homologs of the Arabidopsis FIS genes from sexual and apomictic Hieracium species. Analysis of transgenic plants in which these genes have been upregulated and downregulated may determine their function during fertilized and autonomous seed development.
METHODS
Plant Material
All Hieracium plants examined were maintained by vegetative propagation under the growth conditions described by Koltunow et al. (1998). Four different genetically characterized Hieracium lines that have been described previously (Koltunow et al., 1998, 2000; Tucker et al., 2001) were used in this study. P4 is a self-incompatible tetraploid sexual accession of H. pilosella, D3 is an apomictic triploid accession of H. piloselloides, D2 is derived from D3 and is an apomictic diploid derived from parthenogenetic development of a reduced egg cell, and A3.4 is an apomictic aneuploid accession of H. aurantiacum. In all three apomictic plants, >93% of the viable seeds are apomictic (Koltunow et al., 1998, 2000). Tissues for GUS staining, histology, and in situ hybridization were staged and collected as described by Koltunow et al. (1998).
Arabidopsis thaliana ecotype Landsberg erecta seeds homozygous for AtFIS2:GUS were provided by Abed Chaudhury (Commonwealth Scientific and Industrial Research Organization, Canberra, Australia) and grown at 20°C with a 16-h daylength. Ovules and early seeds were staged as described by Schneitz et al. (1995).
Arabidopsis SPL, MEA, FIS2, FIE, and SERK1 Chimeric Genes and Hieracium Transformants
The AtFIS:GUS constructs used in this study were provided by Abed Chaudhury. The AtMEA:GUS fusion contains 2070 bp of nucleotide sequence upstream of the predicted translation site, exons 1 and 2 and 305 bp of exon 3, in the vector pBI101-3 (Clontech, Palo Alto, CA). The AtFIS2:GUS and AtFIE:GUS fusions are identical to those used by Luo et al. (2000). The AtSPL:GUS fusion was provided by Venkatesan Sundaresan (National University of Singapore) and is identical to that used by Yang et al. (1999). The AtSERK1:GUS promoter fusion is identical to that used by Hecht et al. (2001). All binary constructs were mobilized in Agrobacterium tumefaciens AGL1, and the T-DNA was transformed into Hieracium leaf pieces as described by Bicknell and Borst (1994). Kanamycin-resistant progeny were screened for the presence of the desired construct using the GUS forward primer 5′-CTGTAGAAACCCCAACCCGTG-3′ and the GUS reverse primer 5′-CATTACGCTGCGATGGATCCC-3′, which produce a 515-bp product after PCR. The number of independent, kanamycin-resistant, and GUS PCR-positive transformants obtained was 24 (7 P4, 11 D3, and 6 A3.4) for AtFIS2:GUS, 28 (11 P4 and 17 D3) for AtMEA:GUS, 19 (5 P4 and 14 D3) for AtFIE:GUS, 9 (4 P4 and 5 D3) for AtSPL:GUS, and 5 (5 D3) for AtSERK1:GUS. Two D3 AtSERK1:GUS plants (numbers 6 and 7) were crossed to sexual P4 plants, and resulting progeny were identified that contained the AtSERK1:GUS gene and required fertilization for embryogenesis. These plants were assayed for the expression of the marker gene during zygotic embryogenesis.
To determine transgene copy number, genomic DNA was isolated from the leaves of transformed plants as described by Tucker et al. (2001), and DNA samples were digested with HindIII and EcoRV. Digested DNA was transferred to Hybond-N membranes (Amersham) and hybridized with an α-32P-dCTP–labeled GUS probe (515-bp PCR fragment; see above). Hybridization solutions and washes were as described by Tucker et al. (2001).
Isolation, Reverse Transcriptase–Mediated PCR Analysis, and Genomic Organization of a Hieracium SERK Homolog
The SERK Leu-rich repeat–containing kinase is highly conserved in a number of species (Hecht et al., 2001). The degenerate primers 5′ld forward (5′-GGNGGATTTGGTAARGTNAY-3′) and 3′ld reverse (5′-DATNCCRTAACCAAANACATC-3′) were designed to part of the conserved protein kinase domain from DcSERK and AtSERK1. Stage-8 D3 ovary total RNA was extracted as described by Tucker et al. (2001), and 500 ng was used in reverse transcriptase–mediated (RT) PCR with reverse transcriptase from Avian myeloblastosis virus (Promega). The resulting PCR products were cloned into pGEM T-Easy (Promega) and sequenced to identify a 516-bp HpSERK-L fragment.
RNA samples were harvested from various stages of leaf and ovary development (ovule initiation and heart stage of embryogenesis) (Koltunow et al., 1998), and single-stranded cDNA was generated using 500 ng of total RNA template, oligo(dT) primer, and the Thermoscript cDNA synthesis kit (Invitrogen, Carlsbad, CA). PCR was performed in a 20-μL total volume containing 1 μL (∼25 ng) of cDNA, 0.5 μM of each gene-specific primer, 1 unit of RedTaq polymerase (Sigma), 2 μL of 10× PCR buffer (100 mM Tris-HCI, pH 8.3, 500 mM KCI, 11 mM MgCI2, and 0.1% gelatin) and 50 μM of each deoxynucleotide triphosphate. Primers used for the RT-PCR procedure were HpSERKF (5′-GGCTGTCAAAACGGGTTAAA-3′) and HpSERKR (5′-CATTTGGTGGTCGCTCTCTTAA-3′). PCR samples were transferred to Hybond-N membranes (Amersham) and hybridized with α-32P-dCTP–labeled HpSERK-L probe (516-bp cDNA fragment). The β-tubulin primers BtubRTfwd (5′-GGGTGCTGGAAACAATTGGGCTAA-3′) and BtubRTrev (5′-ACTGCTCACTCACGCGCCTAA-3′) were used to amplify cDNA as a loading control.
Genomic DNA was isolated from the leaves of P4, D3, and A3.4 plants as described by Tucker et al. (2001), and DNA samples were digested with EcoRI. Digested DNA was transferred to Hybond-N membranes (Amersham) and hybridized with α-32P-dCTP–labeled HpSERK-L probe. Membranes were washed three times for 15 min at 60°C in 0.2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS. After exposure to Biomax autoradiography film (Kodak), membranes were stripped in 0.4 M NaOH at 45°C for 30 min, washed in 0.1× SSC, 0.1% SDS, and 0.2 M Tris-HCl, pH 7.5, at 45°C for 15 min, and then rehybridized with a labeled 1455-bp BamHI fragment from AtSERK1. Membranes were washed three times for 15 min at 60°C in 2× SSC and 0.1% SDS and exposed to film.
Whole-Mount in Situ Hybridization of a Hieracium SERK Homolog
Based on the high similarity of the Hieracium, Arabidopsis, and carrot SERK sequences (>80% identical), digoxigenin (DIG)-labeled DcSERK RNA probe was used to detect homologous HpSERK mRNA in Hieracium ovules and seeds by whole-mount in situ hybridization. The procedure was performed as described by Schmidt et al. (1997). Whole-mount expression patterns were verified by thin sections and then stained with 0.1% toluidine blue in 0.02% sodium carbonate to determine cell identity.
In Situ Localization of GUS mRNA
In situ hybridization was performed as described by Guerin et al. (2000) with modifications. To verify ovule staging, capitula collected for in situ analysis were cut in two, and one-half was submitted to GUS stain analysis (see below) and the other half was fixed in 4% paraformaldehyde and 0.25% glutaraldehyde for in situ analysis. Fixed tissues were embedded in butyl-methyl methacrylate and then sectioned to 2 μM.
A 515-bp fragment of GUS (for primers, see above) was amplified by PCR from pBI101-3 and cloned into pGEM T-Easy (Promega). The clone was linearized with SalI (sense) and NcoI (antisense), and DIG-labeled RNA probe was produced using the DIG-RNA labeling kit (Boehringer Mannheim) and SP6 or T7 RNA polymerase to generate sense and antisense probes, respectively.
Microscopy and GUS Staining
Histological analysis of GUS enzyme activity in Hieracium tissues was performed as described by Koltunow et al. (2001). Stained tissues were fixed in formalin:acetic acid:alcohol overnight and then cleared in methyl salicylate as described by Stelly et al. (1984). Arabidopsis AtFIS2:GUS ovule tissues were stained for GUS activity as described by Koltunow et al. (2001). Ovules were cleared in a solution of 20% lactic acid and 20% glycerol in 1× PBS. Whole-mount tissue samples were placed on concave slides (ProSciTech, Thuringowa, Australia) in clearing solution and viewed by Nomarski differential interference contrast microscopy or by dark-field microscopy with a Zeiss Axioplan microscope (Jena, Germany). Selected GUS-stained samples were embedded in butyl-methyl methacrylate, sectioned to 2 μM, and counterstained with Fuschin red. Digital images were captured using a Spot II camera (Diagnostic Instruments, Sterling Heights, MI).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
This article is dedicated to the memory of our friend, Nicholas Paech, a student who contributed much to our work on apomixis and who passed away on September 25, 2002. We thank Venkatesan Sundaresan for the AtSPL:GUS construct, Abed Chaudhury and Ming Luo for the AtFIS: GUS constructs and Landsberg erecta AtFIS2:GUS seeds, Melissa Pickering for Arabidopsis GUS stains, and Anne Tassie for generating Hieracium transformants. We also thank Ueli Grossniklaus, Ross Bicknell, members of the European Plant Embryogenesis Network funded by European Union Framework 4, and members of the CSIRO Plant Industry Molecular Basis of Seed and Fruit Formation laboratory for many helpful discussions. This study was supported by an Adelaide University Premier Scholarship to M.R.T., an Australian Center for International Agricultural Research grant to A.M.G.K., a Brazilian Agricultural Research Corporation grant to A.-C.G.A., and a University of Amsterdam travel fellowship to J.-B.R.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011742.
References
- Araujo, A.C.G., Mukhambetzhanov, S., Pozzobon, M.T., Santana, E.F., and Carneiro, V.T.C. (2000). Female gametophyte development in apomictic and sexual Brachiaria brizantha (Poaceae). Rev. Cytol. Biol. Veg. Botan. 23, 13–28. [Google Scholar]
- Barcaccia, G., Mazzucato, A., Albertini, E., Zethof, J., Gerats, A., Pezzotti, M., and Falcinelli, M. (1998). Inheritance of parthenogenesis in Poa pratensis L.: Auxin test and AFLP linkage analyses support monogenic control. Theor. Appl. Genet. 97, 74–82. [Google Scholar]
- Bicknell, R.A., and Borst, N.K. (1994). Agrobacterium-mediated transformation of Hieracium aurantiacum. Int. J. Plant Sci. 155, 467–470. [Google Scholar]
- Bicknell, R.A., Borst, N.K., and Koltunow, A.M. (2000). Monogenic inheritance of apomixis in two Hieracium species with distinct developmental mechanisms. Heredity 84, 228–237. [DOI] [PubMed] [Google Scholar]
- Boutilier, K., Offringa, R., Sharma, V.K., Kieft, H., Ouellet, T., Zhang, L., Hattori, J., Liu, C.M., van Lammeren, A.A., Miki, B.L., Custers, J.B., and van Lookeren Campagne, M.M. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutovska, R., Cellarova, E., and Dolezel, L. (1998). Cytogenetic variability of in vitro regenerated Hypericum perforatum L. plants and their seed progenies. Plant Sci. 133, 221–229. [Google Scholar]
- Carman, J.G. (1997). Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 61, 51–94. [Google Scholar]
- Chaudhury, A.M., Koltunow, A., Payne, T., Luo, M., Tucker, M.R., Dennis, E.S., and Peacock, W.J. (2001). Control of early seed development. Annu. Rev. Cell Dev. Biol. 17, 677–699. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202, 136–151. [DOI] [PubMed] [Google Scholar]
- Danilevskaya, O.N., Hermon, P., Hantke, S., Muszynski, M.G., Kollipara, K., and Ananiev, E.V. (2003). Duplicated fie genes in maize: Expression pattern and imprinting suggest distinct functions. Plant Cell 15, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Valle, C.B., and Savidan, Y.H. (1996). Genetics, Cytogenetics and Reproductive Biology of Brachiaria. In Brachiaria: Biology, Agronomy and Improvement, J.W. Miles, B.L. Maass, and C.B. do Valle, eds (Cali, Colombia: International Center for Tropical Agriculture), pp. 164–177.
- Ernst, A. (1918). Die Bastardierung als Ursache der Apogamie im Pflanzenreiche. (Jena, Germany: Fischer).
- Gasser, C.S., Broadhvest, J., and Hauser, B.A. (1998). Genetic analysis of ovule development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 1–24. [DOI] [PubMed] [Google Scholar]
- Grimanelli, D., Leblanc, O., Perotti, E., and Grossniklaus, U. (2001). Developmental genetics of gametophytic apomixis. Trends Genet. 17, 597–604. [DOI] [PubMed] [Google Scholar]
- Grini, P.E., Jurgens, G., and Hulskamp, M. (2002). Embryo and endosperm development is disrupted in the female gametophytic capulet mutants of Arabidopsis. Genetics 162, 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U. (2001). From sexuality to apomixis: Molecular and genetic approaches. In The Flowering of Apomixis: From Mechanisms to Genetic Engineering, Y. Savidan, J.G. Carman, and T. Dresselhaus, eds (Texcoco, Mexico: CIMMYT Press), pp. 168–211.
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Guerin, J., Rossel, J.B., Robert, S., Tsuchiya, T., and Koltunow, A. (2000). A DEFICIENS homologue is down-regulated during apomictic initiation in ovules of Hieracium. Planta 210, 914–920. [DOI] [PubMed] [Google Scholar]
- Hecht, V., Vielle-Calzada, J.-P., Hartog, M.V., Schmidt, E.D., Boutilier, K., Grossniklaus, U., and de Vries, S.C. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816. [PMC free article] [PubMed] [Google Scholar]
- Izmailow, R. (1994). Further observations in embryo sac development in Alchemilla L. (subsection Heliodrosium rothm). Acta Biol. Cracoviensca Ser. Bot. 36, 37–41. [Google Scholar]
- Jassem, B. (1990). Apomixis in the genus Beta. Apomixis Newsl. 2, 7–23. [Google Scholar]
- Johnston, S.A., den Nijs, T.P.M., Peloquin, S.J., and Hanneman, R.E., Jr. (1980). The significance of genic balance to endosperm development in interspecific crosses. Theor. Appl. Genet. 57, 5–9. [DOI] [PubMed] [Google Scholar]
- Koltunow, A.M. (1993). Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5, 1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow, A.M., Bicknell, R.A., and Chaudhury, A.M. (1995). Apomixis: Molecular strategies for the generation of genetically identical seeds without fertilization. Plant Physiol. 108, 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow, A.M., Johnson, S.D., and Bicknell, R.A. (1998). Sexual and apomictic development in Hieracium. Sex. Plant Reprod. 11, 213–230. [Google Scholar]
- Koltunow, A.M., Johnson, S.D., and Bicknell, R.A. (2000). Apomixis is not developmentally conserved in related, genetically characterized Hieracium plants of varying ploidy. Sex. Plant Reprod. 12, 253–266. [Google Scholar]
- Koltunow, A.M., Johnson, S.D., Lynch, M., Yoshihara, T., and Costantino, P. (2001). Expression of rolB in apomictic Hieracium piloselloides Vill. causes ectopic meristems in planta and changes in ovule formation, where apomixis initiates at higher frequency. Planta 214, 196–205. [DOI] [PubMed] [Google Scholar]
- Lin, B.-Y. (1984). Ploidy barrier to endosperm development in maize. Genetics 107, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97, 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova, T.N., and Willemse, M.T.M. (1995). Ultrastructural characterization of apospory in Panicum maximum. Sex. Plant Reprod. 8, 197–204. [Google Scholar]
- Nogler, G. (1984). Gametophytic apomixis. In Embryology of An-giosperms, B.M. Johri, ed (Berlin: Springer-Verlag), pp. 475–518.
- Nybom, H. (1988). Apomixis versus sexuality in blackberries (Rubus subgen. Rubus, Rosaceae). Plant Syst. Evol. 160, 207–218. [Google Scholar]
- Ohad, N., Margossian, L., Hsu, Y.C., Williams, C., Repetti, P., and Fischer, R.L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 93, 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock, J.W. (1992). Genetic engineering and mutagenesis for apomixis in rice. In Proceedings of the International Workshop on Apomixis in Rice, Changsha, China, K.J. Wilson, ed (New York: The Rockefeller Foundation), pp. 11–21.
- Pessino, S.C., Espinoza, F., Martinez, E.J., Ortiz, J.P.A., Valle, E.M., and Quarin, C.L. (2001). Isolation of cDNA clones differentially expressed in flowers of apomictic and sexual Paspalum notatum. Hereditas 134, 35–42. [DOI] [PubMed] [Google Scholar]
- Roche, D., Cong, P.S., Chen, Z.B., Hanna, W.W., Gustine, D.L., Sherwood, R.T., and Ozias-Akins, P. (1999). An apospory-specific genomic region is conserved between buffelgrass (Cenchrus ciliaris L.) and Pennisetum squamulatum Fresen. Plant J. 19, 203–208. [DOI] [PubMed] [Google Scholar]
- Savidan, Y. (2000). Apomixis: Genetics and breeding. In Plant Breeding Reviews, J. Janick, ed (New York: Wiley), pp. 13–86.
- Savidan, Y.H. (1982). Nature et hérédité de l' apomixie chez Panicum maximum Jacq. Trav. Doc. ORSTOM 153, 1–159. [Google Scholar]
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, E.D., Guzzo, F., Toonen, M.A., and de Vries, S.C. (1997). A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124, 2049–2062. [DOI] [PubMed] [Google Scholar]
- Schneitz, K., Hulskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light-microscope study of cleared whole-mount tissue. Plant J. 7, 731–749. [Google Scholar]
- Sorensen, M.B., Chaudhury, A.M., Robert, H., Bancharel, E., and Berger, F. (2001). Polycomb group genes control pattern formation in plant seed. Curr. Biol. 11, 277–281. [DOI] [PubMed] [Google Scholar]
- Spillane, C., Steimer, A., and Grossniklaus, U. (2001). Apomixis in agriculture: The quest for clonal seeds. Sex. Plant Reprod. 14, 179–187. [DOI] [PubMed] [Google Scholar]
- Springer, N.M., Danilevskaya, O.N., Hermon, P., Helentjaris, T.G., Phillips, R.L., Kaeppler, H.F., and Kaeppler, S.M. (2002). Sequence relationships, conserved domains and expression patterns for maize homologs of the Polycomb group genes E(z), esc, and E(Pc). Plant Physiol. 128, 1332–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelly, D.M., Peloquin, S.J., Palmer, R.G., and Crane, C.F. (1984). Mayer's hemalum-methyl salicylate: A stain clearing technique for observations within whole ovules. Stain Technol. 59, 155–161. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Kwong, L.W., Yee, K.M., Pelletier, J., Lepiniec, L., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98, 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas, I.C., and Van Dijk, P.J. (1999). Crosses between sexual and apomictic dandelions (Taraxacum). I. The inheritance of apomixis. Heredity 83, 707–714. [DOI] [PubMed] [Google Scholar]
- Tucker, M.R., Paech, N.A., Willemse, M.T.M., and Koltunow, A.M.G. (2001). Dynamics of callose deposition and β-1,3-glucanase expression during reproductive events in sexual and apomictic Hieracium. Planta 212, 487–498. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Nuccio, M.L., Budiman, M.A., Thomas, T.L., Burson, B.L., Hussey, M.A., and Wing, R.A. (1996). Comparative gene expression in sexual and apomictic ovaries of Pennisetum ciliare (L) Link. Plant Mol. Biol. 32, 1085–1092. [DOI] [PubMed] [Google Scholar]
- Willemse, M.T.M., and van Went, J.L. (1984). The female gametophyte. In Embryology of Angiosperms, B.M. Johri, ed (Berlin: Springer-Verlag), pp. 159–191.
- Wu, H.M., and Cheung, A.Y. (2000). Programmed cell death in plant reproduction. Plant Mol. Biol. 44, 267–281. [DOI] [PubMed] [Google Scholar]
- Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y., Nakashima, K., Harada, J.J., Goldberg, R.B., Fischer, R.L., and Ohad, N. (2000). Mutations in the FIE and MEA genes that encode interacting Polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12, 2367–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.C., and Sundaresan, V. (2000). Genetics of gametophyte biogenesis in Arabidopsis. Curr. Opin. Plant Biol. 3, 53–57. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., Frugis, G., and Chua, N.H. (2002). The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 30, 349–359. [DOI] [PubMed] [Google Scholar]