Abstract
The tumor suppressor gene p53 in mammalian cells plays a critical role in safeguarding the integrity of genome. It functions as a sequence-specific transcription factor. Upon activation by a variety of cellular stresses, p53 transactivates downstream target genes, through which it regulates cell cycle and apoptosis. However, little is known about p53 in invertebrates. Here we report the identification and characterization of a Drosophila p53 homologue gene, dp53. dp53 encodes a 385-amino acid protein with significant homology to human p53 (hp53) in the region of the DNA-binding domain, and to a lesser extent the tetramerization domain. Purified dp53 DNA-binding domain protein was shown to bind to the consensus hp53-binding site by gel mobility analysis. In transient transfection assays, expression of dp53 in Schneider cells transcriptionally activated promoters that contained consensus hp53-responsive elements. Moreover, a mutant dp53 (Arg-155 to His-155), like its hp53 counterpart mutant, exerted a dominant-negative effect on transactivation. Ectopic expression of dp53 in Drosophila eye disk caused cell death and led to a rough eye phenotype. dp53 is expressed throughout the development of Drosophila with highest expression levels in early embryogenesis, which has a maternal component. Consistent with this, dp53 RNA levels were high in the nurse cells of the ovary. It appears that p53 is structurally and functionally conserved from flies to mammals. Drosophila will provide a useful genetic system to the further study of the p53 network.
The tumor suppressor gene p53 plays a pivotal role in safeguarding the integrity of the genome (1). Most human tumors (2, 3) have a mutation in the p53 gene or a functional defect in the p53 pathway, highlighting its importance for preventing tumorigenesis. p53 is a sequence-specific transcription factor. The human p53 contains 393 amino acids and has been divided structurally and functionally into several domains (1). The transcription activation domain of p53 is localized in the N-terminal 42 amino acids. A region within this domain (amino acids 13–29) also interacts with the human MDM2 protein (4), which regulates p53 exporting to the cytoplasm and its degradation (5). The sequence-specific DNA-binding domain is localized between amino acids 94 and 292. This domain folds into a β-sheet sandwich that forms a scaffold for a loop-sheet-helix motif and a large loop, which interacts directly with DNA (6). More than 90% of the missense mutations of p53 found in cancers are found in this region (2, 7). These mutations change the conserved amino acids that contact DNA, or which maintain the three-dimensional conformation of p53. They are defective in DNA binding, and consequently, are incapable of transactivation. The tetramerization domain of p53 is located in the C-terminal region, from amino acid 324 to 355 (8). The native and functional p53 protein is a tetramer in solution.
Normally, the amount of p53 protein in a cell is kept at a low level by its relatively short half-life. Cellular stresses, such as DNA damage, hypoxia, or abnormal oncogene activation, signal to p53 and stabilize it. The p53 protein levels rapidly increase, and it is activated as a transcription factor. The transcriptional activation of downstream target genes, such as p21, cyclin G, and Bax, appears to account for most of the mechanism by which p53 regulates cell cycle progression and apoptosis.
In addition to mammals, p53 cDNAs have been cloned in a number of other vertebrate species, including rainbow trout, Xenopus, and chicken (9). The p53 gene from these different vertebrate species share high homology at five different regions, which reside in the DNA-binding domain and the MDM2 interaction domain (9). Biochemical analysis of Xenopus p53 (Xp53) indicates that Xp53 binds to the consensus hp53-binding site, activates transcription, and forms a tetramer in solution (10). Moreover, MDM2 is also present in Xenopus and has a similar function as human MDM2 in regulating p53 stability (11). These data suggest that these key biochemical properties of p53, i.e., sequence-specific binding, transactivation, tetramerization, and MDM2-targeted degradation, are conserved among all vertebrates.
Despite intensive efforts, the complex p53 network in mammalian cells is far from fully understood. Moreover, the majority of this information was obtained from cultured cells and may not fully reflect the physiological function of p53 in the context of the whole organism. Therefore, it would be of great use to study p53 in a simpler organism such as Drosophila to better elucidate the fundamental components of the p53 network and their functions. Moreover such a system would provide a convenient genetic tool to study gene–gene interaction within the p53 network in the context of an organism. Despite the fact that Drosophila is a very well-characterized genetic model system, no p53 homologue gene has been identified to date. Nevertheless, studies have shown that ectopic expression of the human p53 protein in Drosophila eye disk induced cell cycle arrest and apoptosis (12), suggesting that the cell cycle and apoptosis regulatory machinery be conserved between Drosophila and mammal. Here we report the identification and characterization of a p53 homologue in Drosophila (dp53). We demonstrate that dp53 is structurally and functionally homologous to the vertebrate p53. This may provide a new simple model system, as well as a powerful genetic tool, for the study of the p53 network.
Materials and Methods
Cloning, Sequencing, and Plasmid Constructing.
Drosophila expressed sequence tag (EST) clones were purchased from Research Genetics (Huntsville, AL) and sequenced by Rockefeller University Sequencing Facility (New York, NY). The Drosophila expression vector of dp53, pAKS-dp53(F), was constructed by excising the EcoRI–XhoI fragment (full cDNA of dp53) from the expressed sequence tag clone GH11591. The fragment was end-blunted and inserted into EcoRV-digested pAKSII-pBS SK(−) vector (13). hp53 Drosophila expression vector pAKS-hp53 was constructed by cloning the hp53 full cDNA (BamHI–BamHI fragment from pC53-SN3) into BamHI-digested pAKSII-pBS SK(−). pAKS-dp53 (mut155) was generated by site-directed mutagenesis using QuikChange kit (Stratagene). The template for the mutagenesis is pAKS-dp53 (F) and the sequence of the sense strand of the mutation oligonucleotides is 5′-GTGCTCCCGTGGTCCaCTGTCAAAATCACCTTAGC-3′, with the mutation base shown in lowercase [from CGC (Arg) to CaC (His)]. The mutation was confirmed by direct sequencing. The dp53-coding region was amplified by PCR, subcloned into EcoRI–BglII-digested pUAST (14) or EcoRI–BamHI-digested pBluescript SK(−) to generate pUAST-dp53 or pSK-BX, respectively.
Cell Lines, Transfection, and Luciferase Assay.
Schneider Cell Line 2 (S2) was maintained at 22°C–23°C in Shields and Sang M3 insect medium supplemented with 10% FCS, 10 units/ml penicillin, and 10 μg/ml streptomycin. Cells were transfected by using the calcium phosphate precipitation method essentially as described (15). In brief, S2 cells were seeded in six-well plates at a density of 1.5 × 106 cells per well. The following day, cells were cotransfected with 0.2 μg per well of pG13 reporter construct (16) and 0.2 μg per well (or as indicated) of different effector plasmids [pAKS-dp53(F) or pAKS-dp53(mut155)]. The total amount of DNA transfected was adjusted to 2 μg per well by the addition of pBluescript SK(−) plasmid DNA. The precipitate was removed and fresh medium was added 24 h after transfection. Cells were allowed to grow for another 48 h before harvesting for luciferase assay (Promega). Luciferase activity was normalized relative to protein concentration as determined by Bradford method (Bio-Rad).
Protein Purification and Gel-Mobility Shift Assay.
The gene fragments for dp53-N + Core (amino acids 1–297) was amplified from clone GH11591 and subcloned into pGEX-4T1 vector (Pharmacia). The dp53-N + Core was expressed in Escherichia coli BL21(DE3) as glutathione S-transferase fusion proteins, purified by glutathione-Sepharose affinity chromatography, and cleaved with thrombin. The cleaved proteins were purified by cation-exchange chromatography, followed by Superdex-75 gel filtration chromatography. The hp53 DNA-binding domain (amino acids 94–292) was purified by methods as described before (6). The hp53 consensus oligonucleotide was end-labeled with [γ-32P]ATP. Gel-mobility shift analysis was performed as described (17). The sequences of the upper strands of the double-stranded oligonucleotide used as probe and competitors are as follows: probe and specific competitor (SP): 5′-TACAGAACATGTCTAAGCATGCTGGG-3′; nonspecific competitor (NS): 5′-GTGCTCCCGTGGTCCACTGTCAAAATCACCTTAGC-3′.
Fly Culture and Generation of Transgenic Animals.
Drosophila melanogaster were reared on standard cornmeal medium at 25°C. The pUAST-dp53 constructs were injected into Drosophila embryos by standard procedures.
In Situ Hybridization.
In situ hybridizations were performed as described earlier for embryos (18) and for egg chambers (19). In brief, embryos were dechorionized, fixed in 4% paraformaldehyde in PBS, devitellinized, and proteinase K digested (2 min, 50 μg/ml). The StuI–BamHI fragment of the dp53 cDNA (361–1158) from pSK-BX served as template for the transcription of digoxigenin-labeled RNA (Boehringer Mannheim). The hybridization was performed at 55°C in hybridization buffer (50% formamide/5× SSC/100 μg/ml salmon sperm DNA/50 μg/ml heparin/0.1% Tween-20/100 μg/ml total yeast RNA). The probe was detected by using an alkaline phosphatase-conjugated antibody, and the staining procedure was carried out according to the manufacturer's instructions (Boehringer Mannheim).
RNase Protection Assay.
Total RNA was isolated from heads as described in the manual of Tel-Test (Friendswood, TX). The RNase protection assay was performed as described in the manufacturer's manual for the RPAII kit (Ambion, Austin, TX). Ten micrograms of total RNA was used for each sample. The EcoRV–BamHI fragment (dp53 cDNA 741-1158) from pSK-BX was used as template to generate dp53 riboprobe.
Acridine Orange Staining.
Eye imaginal discs from wandering third instar larvae were dissected in PBS and stained in 1.6 μM acridine orange (Sigma) in PBS for 5 min. After three brief washes in PBS, the discs were mounted in PBS and viewed immediately by confocal microscopy.
Results
Cloning and Modeling of a p53 Homologue in Drosophila.
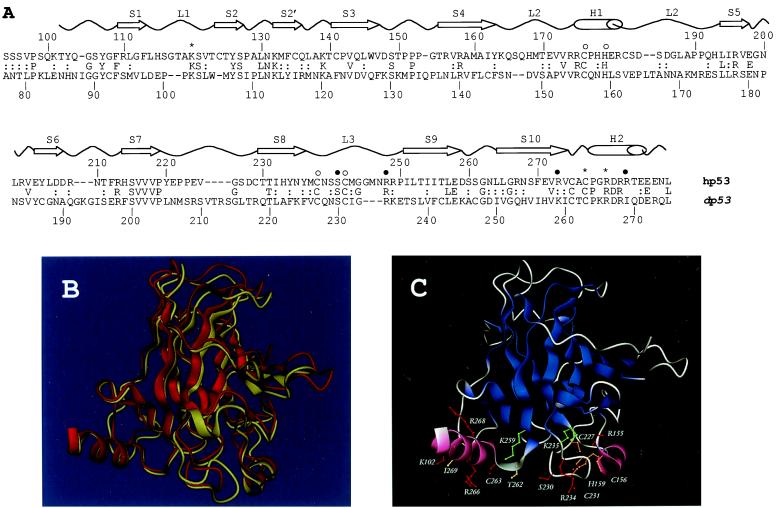
Despite the fact that Drosophila is a very well-characterized genetic organism, a p53 homologue has not been reported in Drosophila to date. Recent advances in the sequencing of the Drosophila genome by a joint effort of Celera Genomics and the Berkeley Drosophila Genome Project provided a new opportunity to search for a p53 homolog. Homology search was performed using the blast program (www.fruitfly.org) with amino acid sequence of human DNA-binding domain (from 100 to 300) against the Drosophila Gene Bank (www.fruitfly.org). Genomic sequences of a bacterial artificial chromosome (BAC) (GenBank accession no. AC008200) was obtained. By blast search with sequence 5′ to the homologous region in the BAC, several overlapping expressed sequence tag clones were obtained and further sequenced. The sequence of the GH11591 clone suggested some sequence homology to human p53. It predicted an ORF of 385 amino acids (GenBank accession no. AF250918), which was termed Drosophila p53 (dp53). Alignment of the protein sequences of Drosophila and human p53 DNA-binding domains was obtained by the profile-based program psi-blast (20). The alignment of the DNA-binding domains was between Drosophila residues 77 and 275 and human residues 94 and 289 (Fig. 1A). Even though the match is statistically significant, the two domains share only 24% sequence identity and 44% similarity over 207 amino acids (Fig. 1A). This low similarity explains for previous unsuccessful attempts to directly clone the Drosophila p53 using vertebrate sequences as molecular probes (9). The alignment was used to generate a comparative protein structure model for the Drosophila sequence based on the crystallographic structure of the hp53 DNA-binding domain (6), using the program modeller (21). The elements of secondary structure are conserved between the Drosophila and human proteins, with bigger differences observed in loop regions (Fig. 1 A and B). As shown in Fig. 1 A and C, the amino acid residues that are known to be important for DNA–protein interactions as determined by x-ray crystallography and mutagenesis (6) are well conserved between hp53 and dp53.
Figure 1.
(A) Sequence and structure comparison of the Drosophila and human p53 DNA-binding domains. Sequence alignment of the dp53 and hp53 DNA-binding domains as produced by psi-blast. The secondary structure elements of hp53 are shown above (S, β-strand; L, loop; H, α-helix). Residues involved in DNA binding (*, contacting bases; ●, contacting phosphate backbone) and zinc binding (○) also are indicated (6). (B) Superimposition of the crystal structure of hp53 (yellow cartoon) DNA-binding domain and the model of the dp53 domain (red cartoon) predicted by program modeller. (C) Protein structure model of the dp53 DNA-binding domain. Color scheme: red, residues preserved between the human and Drosophila sequences; green, conservative substitutions; orange, preserved Zn-coordinating residues; and yellow, nonconservative substitutions. B and C were rendered by program dino (http://www.biozentrum.unibas.ch/∼x-ray/dino).
Another region of similarity was indicated by the three-dimensional structure of the tetramerization domain of hp53 (8). The β-strand and α-helix structural elements that constitute this domain in hp53 would be consistent with the sequence of dp53 spanning residues from 316 to 360 (data not shown). These findings suggest that the dp53 might have a tetramerization domain similar to that of the human p53.
dp53 Binds to the Mammalian p53 Consensus Binding Site.
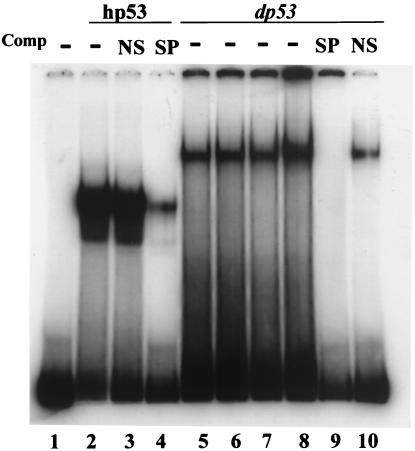
The similarity between predicted structure of dp53 and the crystal structure of the hp53 DNA-binding domain prompted us to ask whether dp53 is able to bind to the hp53 consensus binding site. Previous studies have shown that the DNA-binding domain plus N terminus and the DNA-binding domain alone of hp53 had similar affinities to the consensus DNA site as did the full-length protein (10, 22). Therefore, we purified the fragment of hp53 and dp53 containing the DNA-binding domains to test their DNA-binding ability. The consensus site for human p53 is PuPuPuCA/T-T/AGPyPyPy-N0–13-PuPuPuCA/T-T/AGPyPyPy (23). A double-stranded oligonucleotide matching the consensus sequence (5′-TACAGAACATGTCTAAGCATGCTGGG-3′) was end labeled and used for gel mobility-shift assay. As shown in Fig. 2, dp53 formed a DNA–protein complex (lanes 5–8) as did hp53 (lane 2). The specificity of the DNA–protein interaction was demonstrated by competition assays, in which the unlabeled specific oligonucleotide (SP, the consensus oligonucleotide itself) effectively competes with the labeled probe (lane 4 for hp53; lane 9 for dp53), whereas a nonspecific oligonucleotide (NS, lane 3 for hp53; lane 10 for dp53) had little effect in inhibiting binding. The specificity of interaction was further demonstrated by using a mutated p53 consensus oligonucleotide (5′-TACAGAAaATtTCTAAGaATtCTGGG-3′, mutation in consensus sequence shown in lowercase) as a probe in a similar gel shift assay. Both dp53 and hp53 proteins failed to form a complex with the mutated oligonucleotide (data not shown). The gel mobility-shift analysis demonstrated that the binding affinity of dp53 to the oligonucleotide sequence was lower than that of hp53. It is not clear whether this was due to the specific oligonucleotide sequence chosen or that dp53 prefers a similar but a slightly different consensus site than that of hp53.
Figure 2.
dp53 binds to consensus hp53-binding sequence. Gel mobility-shift assay was performed by using 32P-end-labeled consensus p53-binding oligonucleotide. hp53 DNA-binding domain (amino acids 94–292) or dp53 N terminus plus DNA-binding domain (amino acids 1–297) were purified from bacteria. hp53 protein (100 ng; lanes 2, 3, and 4) or different amount of dp53 protein (lane 5, 50 ng; lane 6, 100 ng; lane 7, 150 ng; lane 8, 200 ng; and lanes 9 and 10, 100 ng) were added into the reactions without competitor, with nonspecific competitor (NS, 500 ng) or with specific competitor (SP, 500 ng) as indicated. Lane 1 contained the probe only.
dp53 Displays Transcriptional Activation Activity.
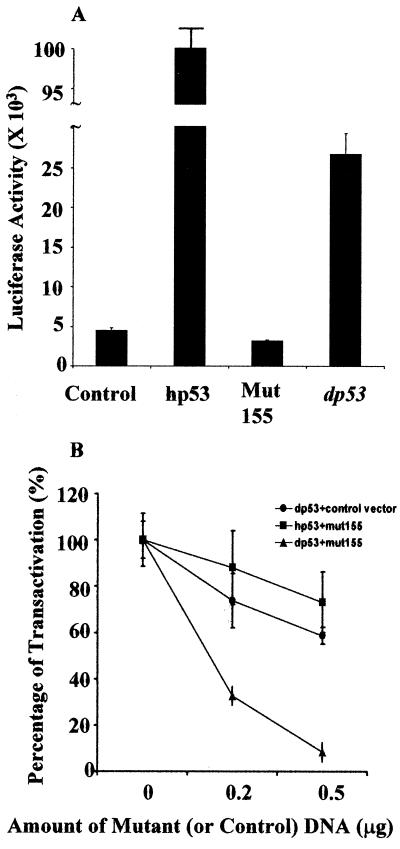
Upon activation, hp53 binds to consensus sites and activates transcription of its target genes. The N terminus of hp53 is essential for this transactivation. Although there is less homology in the N terminus region between dp53 and hp53, dp53 contains a small acidic (aspartate-glutamate rich) region (KESTDSEDDSTEVDIKEDIPKTVEVSGSEL, amino acids 11–40). Acidic amino acid-rich regions are commonly found in transactivation domains of transcription factors, suggesting dp53 might have transcriptional activation activity. To test whether dp53 is able to activate transcription in Drosophila cells, we cloned the dp53 cDNA into the Drosophila expression vector pAKSII-pBS SK(−) (13), in which transcription of dp53 gene is under control of a constitutively active Drosophila actin promoter (14). The construct, pAKS-dp53(F), was cotransfected into Schneider cells with a luciferase reporter construct pG13, which contains 13 tandem repeats of hp53-binding sites within its promoter (16). As shown in Fig. 3A, expression of dp53 activated transcription of the promoter by 6- to 7-fold, compared to the control. The transcriptional activation was dependent on DNA binding of dp53. A mutant form of dp53 (Arg-155 to His-155) was unable to transactivate the promoter (Fig. 3A). dp53 Arg-155 corresponds to Arg-175 in hp53 (Fig. 1A), and the substitution of histidine for Arg-175 abolishes the DNA-binding activity of hp53 (6). Moreover, expression of dp53 cannot activate promoters or vectors without hp53-binding sites (the thymidine kinase promoter, the simian virus 40 promoter (pGL2P, Promega), and a promoterless construct pGL2B (Promega) were tested, data not shown).
Figure 3.
dp53 transcriptionally activates promoter with hp53 responsive elements in Schneider cells. (A) Expression of dp53 activates pG13. (B) a mutant dp53 (Arg-155 to His-155) has a dominant-negative effect. In A, 0.2 μg of pG13 was cotransfected into Schneider cells with 0.2 μg of control vector (control) or with expression vectors of hp53, mutant dp53 (Mut155), dp53 as indicated. In B, 0.2 μg of pG13 and 0.2 μg of expression vectors of wild-type p53 (dp53 or hp53, as indicated) were cotransfected into Schneider cells with the indicated amount (0, 0.2 μg, 0.5 μg) of mutant dp53 (mut155) expression vector or control vector. In each case, transcription activation by wild-type p53 without the cotransfection of mutant dp53 (or control vector) is arbitrarily set as 100%. Percentage of activation is the ratio of the fold activation with the cotransfection of mutant dp53 (or control vector) divided by the fold activation without the cotransfection of mutant dp53.
Next, dp53 was used to transactivate a natural promoter with a single p53-binding site. When using the MDM2-promoter–luciferase construct, pBL100GL2, which contains a p53-responsive element within the promoter, as the reporter construct, the results obtained were essentially identical to those of pG13 (data not shown).
Many cancer-derived hp53 mutants, including the hp53 (Arg-175 to His-175), have a dominant-negative effect on wild-type hp53. To test whether similar mutations in dp53 have dominant-negative effects, we examined the effect of the dp53 mutant (Arg-155 to His-155). Expression of this mutant form of dp53 abrogated the transcriptional activation by the wild-type dp53 (Fig. 3B). On the other hand, coexpression of the same amount of the dp53 mutant appeared to have little effect (similar to control vector) on the transactivation by hp53 (Fig. 3B).
The transactivation activity (6- to 7-fold) of dp53 was lower compared to that of hp53 (>20-fold, Fig. 3A). This may reflect a difference in protein stability, a difference in the intrinsic transactivation activity of the proteins, or the relative affinities for the human consensus site. It will be of interest to examine the exact mechanisms and their implications.
Overexpression of dp53 Induces Cell Death.
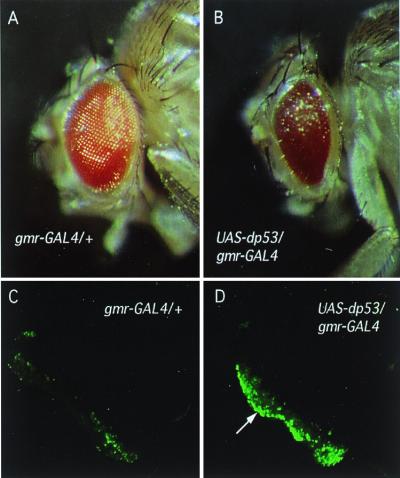
The similarity between dp53 and hp53 prompted us to explore the role dp53 plays in vivo. It is known that p53 exerts its role as a tumor suppressor partially through initiation of apoptosis. Consistently, expression of human p53 in the fly eye initiates apoptosis (12). We reasoned that overexpression of wild-type dp53 might trigger an ectopic cellular response, thereby revealing some of its in vivo function. Using the UAS/GAL4 binary expression system (14), we overexpressed dp53 under the control of a photoreceptor specific promoter, gmr-GAL4 (flybase.bio.indiana.edu). One of five transgenic fly lines tested showed a rough eye phenotype (Fig. 4 A and B). At least 2-fold higher dp53 RNA levels were observed in the line that shows the phenotype than in any of the other lines (data not shown); we therefore believe that the phenotype is indeed caused by dp53 overexpression. Because the eyes of flies overexpressing dp53 are smaller than those of wild-type controls, it seemed likely that the observed phenotype is partially caused by ectopic apoptosis. To test this possibility, we subjected third-instar eye imaginal discs from animals overexpressing dp53 to an acridine orange staining to visualize cell death. As shown in Fig. 4 C and D, overexpression of dp53 in the developing retina causes an increase in cell death. This observation is in accordance with an apoptosis-inducing function of dp53. Moreover, ubiquitous expression in transgenic Drosophila resulted in a high percentage of lethality (data not shown). This observation is consistent with an induction of extensive apoptosis in essential tissues of the fly, thereby reducing viability.
Figure 4.
Increased cell death in the fly retina induced by dp53. dp53 was overexpressed in the fly retina under the control of a photoreceptor-specific promoter (gmr) using the UAS/GAL4 binary expression system (B and D). gmr-GAL4/+ served as wild-type control (A and C). Note the reduced size and the roughness of the eye caused by the overexpression of dp53 (B). (C and D) Third instar eye imaginal discs were subjected to acridine orange staining. Note the increased amount of cell death in the discs from animals overexpressing dp53 (D, arrow).
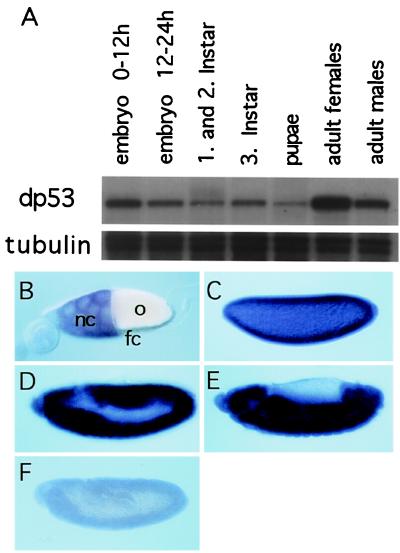
dp53 Is Expressed Throughout Development.
We further examined the expression pattern of dp53. A developmental profile of dp53 RNA levels shows that dp53 is present throughout development (Fig. 5A). dp53 RNA levels seem to be highest during early embryogenesis and in females, suggesting a maternal contribution of dp53 RNA. Consistent with this notion, in those cells of the egg chamber that provide the maternal contribution, the nurse cells, dp53 RNA was detected (Fig. 5B), but dp53 RNA was undetectable in the somatic follicle cells of the egg chamber. Additionally, dp53 RNA was found expressed ubiquitously in early embryogenesis (Fig. 5 C–F). The staining inside the blastoderm embryo (Fig. 5C) probably stems from the maternal contribution. The predominant expression of dp53 in the germ line and during embryogenesis indicates a role for dp53 as a teratogenesis suppressor, as was previously suggested for mammalian p53 (24).
Figure 5.
(A) dp53 RNA is expressed throughout development. Total RNA from the indicated developmental stages was subjected to an RNase-protection assay using a dp53-specific probe. Note the elevated levels of dp53 RNA in females and early embryos. Tubulin RNA was used as loading control. (B–F) In situ hybridization was performed on wild-type embryos using a dp53-specific RNA probe. (B) Stage 10 egg chamber. Note presence of dp53 RNA in the nurse cells (nc) of the egg chamber. No signal was observed in the somatic follicle cells (fc). o, oocyte. (C) Blastoderm embryo probed with antisense RNA; the signal inside the egg indicates a maternal component of dp53 RNA. (D and E) Embryos before and after germ band retraction, showing a ubiquitous expression of dp53. (E) Embryo probed with sense RNA as a control for the specificity of the probe.
Discussion
Drosophila is one of the best-characterized model organisms for genetic study. Recently, the whole Drosophila genome has been sequenced (25) through the joint effort of Celera Genomics and the Berkeley Drosophila Genome Project. This database will make Drosophila a more appealing system in which to study gene functions and interactions. Here we report the identification a gene in Drosophila that is homologous to the mammalian p53 gene. We believe that dp53 is the Drosophila counterpart of mammalian p53 for the following reasons: first, dp53 has sequence homology to hp53 in the DNA-binding domain (the residues of hp53 that contact the DNA are overall conserved in dp53); second, dp53 biochemically resembles hp53 in that it recognizes and binds to the hp53 consensus binding site and activates transcription; third, expression of dp53 has a similar cellular effect, resulting in cell death, probably through apoptosis; finally, dp53 has an expression pattern similar to that of vertebrate p53, with high expression in the adult gonads and in early embryonic development.
Insects and mammals diverged ≈150 million years ago in evolution. The striking conservation of p53 in the two systems suggests that p53 is an early-evolved gene and its functions were under strong selection pressure. In Drosophila, dp53 RNA levels are high in the nurse cells of the ovary. Subsequently, dp53 RNA seems to be transported into the eggs, because the early embryo contains high levels of dp53 RNA. As the embryo differentiates dp53 RNA levels decrease. This expression pattern exactly mirrors that of Xenopus (26) and is also very similar to that of mice in early embryonic development. The expression pattern in mice may reflect the function of p53 as teratogenesis suppressor, as shown by the observation that p53-null mice had a higher teratogenesis rate and lower abortion rate upon γ-irradiation than wild-type mice (24). This conserved expression pattern in all species examined to date suggests that one major function of p53 might be protecting the genomic integrity of early embryos and that of the germ-line cells. This would of course be critical to ensure proper development of an individual organism, eliminating embryos with DNA damage and genetic defects. Therefore this function is strongly selected and maintained during evolution. The tumor suppressor function of p53 in differentiated somatic cells might be a more recently evolved adaptation. As organisms appeared with a long lifespan and activating dividing cells in the adult, the selection pressure to eliminate somatic mutation concomitantly increased. Although p53−/− mice appear to develop normally, it would be interesting to see if the dp53−/− Drosophila has an elevated degree of developmental defect and germ-line instability.
dp53 has been shown in the present study to be a sequence-specific transcription factor. Because the whole Drosophila genome has been sequenced, this provides an opportunity for a genome-wide search for all possible dp53 downstream targets and even computer modeling the dp53 network in Drosophila. If indeed the cellular effect of dp53 is to induce apoptosis, then pro-apoptotic genes are good candidate targets of dp53. The Reaper (27) and Hid (28) genes trigger apoptosis when activated. Interestingly, transcription of Reaper is rapidly activated by ionizing irradiation (29), which also strongly activates p53 in mammalian cells. Likewise, there are several p53 consensus sites near the transcription initiation site of Hid (data not shown). It would be interesting to study whether Reaper and Hid are, in fact, dp53 downstream target genes.
Although dp53 and hp53 share much sequence and biochemical homology, one major difference between dp53 and hp53 is that dp53 lacks the consensus box I sequence found in all vertebrate p53 proteins (9), which is located in the p53–MDM2 interaction region (4). Moreover, genome-wide searches in Drosophila have failed to identify an MDM2 homolog. Therefore, MDM2-mediated p53 degradation could be a later evolutionary event. Interestingly, there is a putative PEST region at the N terminus of dp53 but not hp53. These are P- (proline), E- (glutamate), S- (serine), and T- (threonine) rich sequences flanked by K (lysine) or R (arginine) but not interrupted by any basic amino acids (30), which act as protein degradation signals. It seems possible that dp53 protein stability is regulated through this PEST sequence instead of the more specific MDM2-p53 autoregulating loop in vertebrates (1).
In summary, a p53 homologue in Drosophila has been identified based on the sequence homology, the biochemical properties, and the conserved cellular function. It thus appears that p53 is conserved from insects to mammals. It is hoped that this finding will help us to better understand the function of p53 by providing a convenient and simpler genetic model system in which to study the p53 network.
Acknowledgments
We thank Dr. Ken Onel for reading the manuscript and advice, Dr. Toby Lieber for helpful suggestions, Dr. Simon Kidd for the pAKSII-pBS SK(−) vector, and Ms. Angelika Teresky for various help. S.M. was supported by a Beckman fellowship.
Abbreviations
- dp53
Drosophila p53
- hp53
human p53
Note Added in Proof
Recently, C. Kopczynski (31) and J. M. Abrams (32) had similar results; J. M. Abrams showed Reaper is a dp53 downstream target.
Footnotes
Data deposition: The dp53 cDNA sequence reported in this paper has been deposited in the GenBank database (accession no. AF250918).
References
- 1.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 3.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Nature (London) 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 4.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 5.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 7.Levine A J, Perry M E, Chang A, Silver A, Dittmer D, Wu M, Welsh D. Br J Cancer. 1994;69:409–416. doi: 10.1038/bjc.1994.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffrey P D, Gorina S, Pavletich N P. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 9.Soussi T, Caron de Fromentel C, May P. Oncogene. 1990;5:945–952. [PubMed] [Google Scholar]
- 10.Wang Y, Farmer G, Soussi T, Prives C. Oncogene. 1995;10:779–784. [PubMed] [Google Scholar]
- 11.Marechal V, Elenbaas B, Taneyhill L, Piette J, Mechali M, Nicolas J C, Levine A J, Moreau J. Oncogene. 1997;14:1427–1433. doi: 10.1038/sj.onc.1200967. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi M, Hirose F, Inoue Y H, Shiraki M, Hayashi Y, Nishi Y, Matsukage A. Oncogene. 1999;18:6767–6775. doi: 10.1038/sj.onc.1203113. [DOI] [PubMed] [Google Scholar]
- 13.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 14.Brand A H, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 15.Kindston R E. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1997. pp. 9.1.5–9.1.7. [Google Scholar]
- 16.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 18.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 19.Buszczak M, Freeman M R, Carlson J R, Bender M, Cooley L, Segraves W A. Development (Cambridge, UK) 1999;126:4581–4589. doi: 10.1242/dev.126.20.4581. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sali A, Blundell T. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 22.Pavletich N P, Chambers K A, Pabo C O. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 23.el-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 24.Norimura T, Nomoto S, Katsuki M, Gondo Y, Kondo S. Nat Med. 1996;2:577–580. doi: 10.1038/nm0596-577. [DOI] [PubMed] [Google Scholar]
- 25.Myers E W, Sutton G G, Delcher A L, Dew I M, Fasulo D P, Flanigan M J, Kravitz S A, Mobarry C M, Reinert K H, Remington K A, et al. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 26.Tchang F, Gusse M, Soussi T, Mechali M. Dev Biol. 1993;159:163–172. doi: 10.1006/dbio.1993.1230. [DOI] [PubMed] [Google Scholar]
- 27.White K, Tahaoglu E, Steller H. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- 28.Grether M E, Abrams J M, Agapite J, White K, Steller H. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 29.Nordstrom W, Chen P, Steller H, Abrams J M. Dev Biol. 1996;180:213–226. doi: 10.1006/dbio.1996.0296. [DOI] [PubMed] [Google Scholar]
- 30.Rechsteiner M, Rogers S W. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 31.Kopczynski C. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- 32.Abrams J M. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]