Abstract
The timing of flowering initiation depends on the balanced expression of a complex network of genes that are regulated by both endogenous and environmental factors. We showed previously that mutations at the EARLY BOLTING IN SHORT DAYS (EBS) locus of Arabidopsis result in an acceleration of flowering, especially in noninductive photoperiods (short days), and other phenotypic anomalies. We have identified the EBS gene and demonstrate that it encodes a nuclear protein that contains a bromoadjacent homology domain and a plant homeodomain Zn finger. Both types of motif are thought to mediate protein–protein interactions and occur in transcriptional regulators involved in chromatin remodeling, suggesting that EBS is part of a transcriptional repressor complex that modulates chromatin structure and is required to repress the initiation of flowering in short days. Overexpression of EBS has phenotypic effects similar to those of recessive ebs mutations, suggesting that both might disrupt the formation of protein complexes that contain EBS. Analysis of the expression of flowering-time genes in ebs mutants and in EBS-overexpressing plants indicates that EBS participates in the regulation of flowering time by specifically repressing the expression of FT, a key gene in the integration of floral promotion pathways in Arabidopsis.
INTRODUCTION
During the embryogenesis and organogenesis of all organisms, groups of genes are specifically activated or silenced to establish the spatial and temporal patterns of expression that allow proper development. In higher plants, the production of organs continues during postembryonic growth, and changes in gene expression initiate progressive phases of development that are characterized by the identity of the lateral primordia produced on the flanks of the shoot apical meristem. Initially, vegetative organs are formed, but after the floral transition, the shoot apical meristem initiates the production of inflorescences and flowers. The time of the initiation of flowering is crucial for the reproductive success of plants; therefore, they have developed mechanisms to integrate both environmental and endogenous cues to regulate flowering time precisely.
In Arabidopsis, the floral transition is promoted by exposure to low temperatures (vernalization) and growth under long-day (LD) conditions; by contrast, growth under short-day (SD) conditions delays the initiation of flowering. As a result of physiological, genetic, and molecular analyses of Arabidopsis mutants altered in flowering time, the existence of four floral promotion pathways was proposed in Arabidopsis (for reviews, see Mouradov et al., 2002; Simpson and Dean, 2002). The LD and vernalization pathways promote flowering in response to photoperiod and low temperature, respectively, whereas the autonomous and gibberellin (GA) pathways act independently of environmental signals, although the latter is required most strongly under SD conditions (Wilson et al., 1992). Recently, significant progress was made in understanding how these floral promotion pathways are integrated at the molecular level to regulate the time of flowering (Mouradov et al., 2002; Simpson and Dean, 2002). The pathways that regulate vernalization requirement and response converge on FLOWERING LOCUS C (FLC), a gene that encodes a MADS-box transcription factor that represses flowering. The analysis of FLC expression revealed that both the vernalization (in vernalization-responsive accessions) and autonomous pathways promote flowering by repressing FLC expression (Michaels and Amasino, 1999; Sheldon et al., 1999). FLC, in turn, appears to repress the expression of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Lee et al., 2000), a gene that encodes another MADS-box protein, and FT (Samach et al., 2000), which encodes a protein with similarity to RAF kinase inhibitor proteins (Kardailsky et al., 1999; Kobayashi et al., 1999). However, the expression of SOC1 and FT is not regulated exclusively by FLC, because both genes also are direct targets of CONSTANS (CO), a gene that mediates the flowering response to LD conditions (Suárez-López et al., 2001). These observations suggest that FT and SOC1 integrate signals from the LD pathway (mediated by CO) and from the autonomous and vernalization pathways, which promote flowering by relieving the repression of FT and SOC1 that is mediated by FLC (Michaels and Amasino, 2001; Hepworth et al., 2002).
Besides these pathways that promote flowering, Arabidopsis mutants that exhibit early flowering have revealed the existence of genes involved in the repression of flowering. Some of these repressors act independently of environmental factors and are required to inhibit flowering during the initial stages of development. For example, EMBRYONIC FLOWER1 (EMF1) and EMF2 act as strong repressors of flowering, as suggested by the extremely early flowering of emf mutants (Chen et al., 1997). EMF2 encodes a homolog of the Drosophila Polycomb Group (PcG) protein Su(z)12, whereas EMF1 encodes a putative transcription factor (Aubert et al., 2001; Yoshida et al., 2001). CURLY LEAF (CLF), which also encodes a PcG protein, acts by preventing the expression of the floral meristem identity gene AGAMOUS (AG) during vegetative growth, so that clf mutations result in the ectopic expression of AG and premature flowering (Goodrich et al., 1997).
We have shown previously that early bolting in short days (ebs) mutations result in an acceleration of flowering, especially under noninductive SD conditions. In addition to early flowering, ebs mutants show a reduction in seed dormancy and an increase in the level of expression of the floral organ identity genes APETALA3 (AP3), AG, and PISTILATA that can partially rescue the floral phenotype of leafy-6 (lfy-6) mutant plants. These observations suggested that EBS participates as a repressor in several developmental processes, such as germination, induction of flowering, and expression of floral homeotic genes. Genetic analyses demonstrated that both the precocious germination and the early flowering of ebs mutants require GA biosynthesis and that FT function also is required for the premature flowering of ebs mutants (Gómez-Mena et al., 2001).
We have identified the EBS gene and shown that it encodes a nuclear protein with a bromoadjacent homology (BAH) domain and a plant homeodomain (PHD) Zn finger. These motifs are thought to mediate protein–protein interactions and are found in transcriptional regulators and chromatin-remodeling factors in other organisms, suggesting that EBS is part of a transcriptional repressor complex involved in the modulation of chromatin structure. Analysis of the expression of key genes in the regulation of flowering time in ebs mutants indicates that EBS participates in the regulation of floral induction specifically repressing the expression of FT, a crucial gene in the integration of signals from different pathways that control flowering time in Arabidopsis.
RESULTS
Identification of the EBS Gene
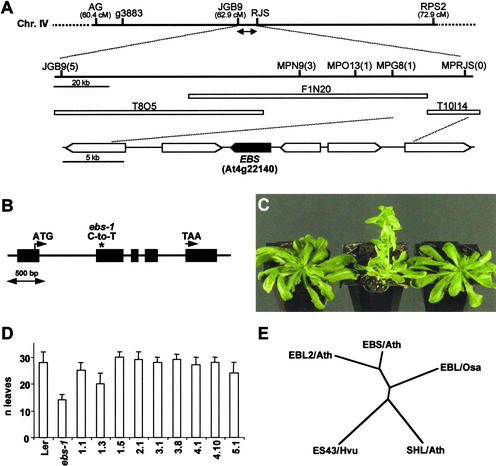
Mutations at the EBS locus cause early flowering in Arabidopsis, especially under SD conditions, indicating that its product is involved in the repression of flowering. Previously, we described the isolation of two mutant alleles of the EBS locus. The mutation present in the ethyl methanesulfonate–induced allele, ebs-1, was mapped initially to the bottom arm of chromosome IV, between markers g3883 and RPS2. Preliminary analysis of the transposon-induced allele, ebs-2, suggested that it contained a chromosomal rearrangement (Gómez-Mena et al., 2001). Detailed molecular analysis of the mutation present in ebs-2 confirmed that a deletion is present in this mutant (see Methods). This deletion spans the region between JGB9 and RJS (∼150 kb), which is included completely in the g3883-RPS2 interval. Subsequently, to identify the EBS locus, the mapping population generated with the ebs-1 allele (904 mutant plants derived from the F2 population of the cross ebs-1 × Columbia) was used to locate the gene within the deletion present in ebs-2. The gene was mapped to a region of ∼25 kb that contained four predicted open reading frames (ORFs), At4g22130 to At4g22160 (Figure 1A). Sequencing of these predicted ORFs in ebs-1 and Landsberg erecta (Ler) allowed the identification of the mutation present in this mutant allele. At4g22140 carried a point mutation in ebs-1 (C to T), which caused an amino acid substitution (Pro-41 to Leu) in the predicted protein (Figure 1B).
Figure 1.
Identification of the EBS Gene and Phylogenic Relationships among EBS-Like Proteins.
(A) Map-based cloning of EBS. The genetic interval, molecular markers, and BAC clones in the EBS region are shown. The number of recombinant events between molecular markers within the JGB9-RJS region, deleted in ebs-2 (double-headed arrow), and EBS are given in parentheses. cM, centimorgan.
(B) Scheme of the EBS gene showing the position of the mutation present in ebs-1 (asterisk). Exons are shown as black boxes. The positions of the start and stop codons are indicated by arrows.
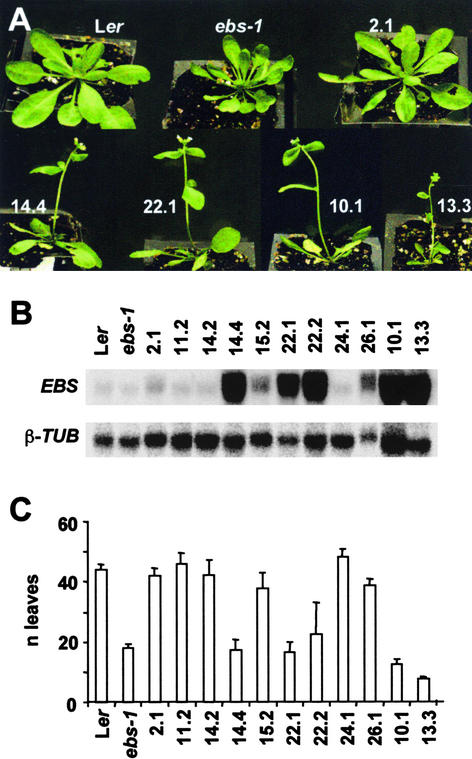
(C) Complementation of the ebs mutant. From left to right: Ler, ebs-1, and TnORF2 transgenic line 1.1 containing the complementing genomic fragment. Plants were grown for 10 weeks under SD conditions. Both Ler and TnORF2 are shown at the time of bolting initiation.
(D) Flowering time, measured as the number of leaves produced before flowering, of Ler, ebs-1, and several TnORF2 transformant lines containing the complementing genomic fragment. Plants were grown under SD conditions. Error bars indicate standard errors.
(E) Phylogenic diagram showing the relatedness of full-length EBS and EBS-like proteins from different species. EBS/Ath is At4g22140, EBL2/Ath is At4g04260, and SHL/Ath is At4g39100 from Arabidopsis (Müssig et al., 2000); ES43/Hvu (Speulman and Salamini, 1995) and EBL/Osa are barley and rice homologs, respectively. CLUSTAL W (www.ebi.ac.uk/clustalw) and TreeView (http://genetics.stanford.edu/~alok/treeview) were used to generate the dendrogram.
To confirm that this ORF was EBS, a genomic fragment containing only At4g22140 was introduced into ebs-1 mutant plants, generating 30 independent transformant lines (TnORF2); in the progeny of these transformant plants, all individuals carrying the T-DNA (1008 of 1292 plants analyzed) showed a wild-type phenotype in LD conditions. Because the flowering-time phenotype of ebs mutants is more conspicuous in noninductive photoperiods, we also scored the number of leaves at flowering of several T2 lines under SD conditions. All of these lines displayed a flowering time close or identical to that of wild-type plants (Figures 1C and 1D); therefore, on the basis of these complementation experiments, we concluded that At4g22140 is EBS.
EBS Is a Member of a Family of Putative Plant Chromatin-Remodeling Factors
Two ESTs corresponding to EBS were identified (F3C6T7A and 135G1T7). Sequencing of the cDNA clones revealed that the EBS gene contains five exons and encodes a protein of 224 amino acids (Figure 1B). The deduced amino acid sequence for the EBS protein contains a BAH domain, a PHD Zn finger, and a nuclear localization signal. Among plant proteins, EBS is highly similar (54% identity) to the barley ES43 protein (Speulman and Salamini, 1995), to a rice predicted protein (64% identity), and to two Arabidopsis proteins: a predicted protein (At4g04260; 70% identity) and SHORT LIFE (57% identity) (Müssig et al., 2000) (Figure 1E). The regions corresponding to the BAH and PHD domains are highly conserved in all five proteins, whereas the N and C termini are more divergent. In addition, ESTs from other plant species, such as cotton, tomato, wheat, and sorghum, also display a high level of identity to the EBS cDNA, suggesting the existence of a family of this type of putative transcriptional regulator in higher plants. No proteins with the same modular architecture as EBS have been found in the animal genomes sequenced to date.
BAH domains appear to mediate protein–protein interactions in protein complexes involved in the silencing of regions of chromatin (Callebaut et al., 1999; Goodwin and Nicholas, 2001). The BAH motif was identified first in the chicken POLYBROMO1 (PB1) protein and also is found in the yeast Rsc1 and Rsc2 (REMODELING STRUCTURE OF CHROMATIN 1 and 2) proteins, which are orthologs of the vertebrate PB proteins and are involved in gene transcriptional regulation (Goodwin and Nicholas, 2001). BAH domains also occur in eukaryotic DNA methyltransferases, in ORIGIN OF REPLICATION COMPLEX1 (Orc1) proteins of several species, and in the yeast-silencing factor Sir3 as well as other transcriptional regulators (Callebaut et al., 1999). Methyltransferases modify cytosine residues in the DNA, and this DNA methylation is associated with transcriptional repression, perhaps by promoting an inactive chromatin conformation (Li et al., 2002). Yeast Orc1 and Sir3 proteins are required to maintain transcriptional silencing by recruiting other silencing factors and compacting regions of chromatin. These observations led to the proposal that the BAH motif plays a role in transcriptional repression through chromatin remodeling (Callebaut et al., 1999).
As with the BAH motif, PHD domains seem to mediate protein–protein interactions and also are present in transcriptional regulators involved in chromatin remodeling (Aasland et al., 1995). PHD fingers are found in the Drosophila NUCLEOSOME REMODELING FACTOR, in the human KAP1 corepressor, which was shown to act by recruiting chromatin-remodeling activities such as histone deacetylase complexes (Schultz et al., 2001), and in Mi-2 proteins, which are involved in transcriptional repression by interacting with the NuRD type of histone deacetylase complex; one Mi-2 protein, corresponding to the protein PICKLE, was isolated in Arabidopsis and shown to be required for the repression of embryonic differentiation genes during postembryonic development (Li et al., 2002). The Drosophila Polycomb-like protein also contains a PHD domain and is involved in the transcriptional repression of homeotic gene expression during development by altering chromatin structure. The presence of both BAH and PHD motifs suggests that EBS participates in a protein complex involved in the transcriptional regulation of gene expression through the modulation of chromatin structure.
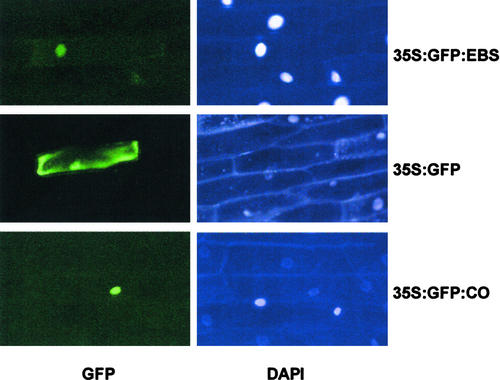
A nuclear localization signal is predicted in the C terminus of the EBS protein. To determine whether EBS is a nuclear protein, a green fluorescent protein (GFP)–EBS fusion protein under the control of the 35S promoter (35S:GFP-EBS) was expressed transiently in epidermal onion cells. In cells that transiently express the fusion protein (GFP-EBS), GFP activity appeared restricted to the nuclei, whereas in cells that express the 35S:GFP construct, GFP activity was detectable in both the nuclei and the cytoplasm (Figure 2). Therefore, the nuclear localization signal present in the EBS protein can drive GFP to the nucleus in onion epidermal cells. The presence of a functional nuclear localization signal in EBS is consistent with its proposed role in the transcriptional regulation of gene expression.
Figure 2.
Nuclear Localization of the EBS Protein.
At left, GFP activity in onion epidermal cells bombarded with 35S:GFP-EBS (top), 35S:GFP (middle), and 35S:GFP-CO (bottom) as a positive control for nuclear localization (Robson et al., 2001). At right, the same groups of cells stained with 4′,6-diamidino-2-phenylindole (DAPI).
EBS Is Expressed Ubiquitously
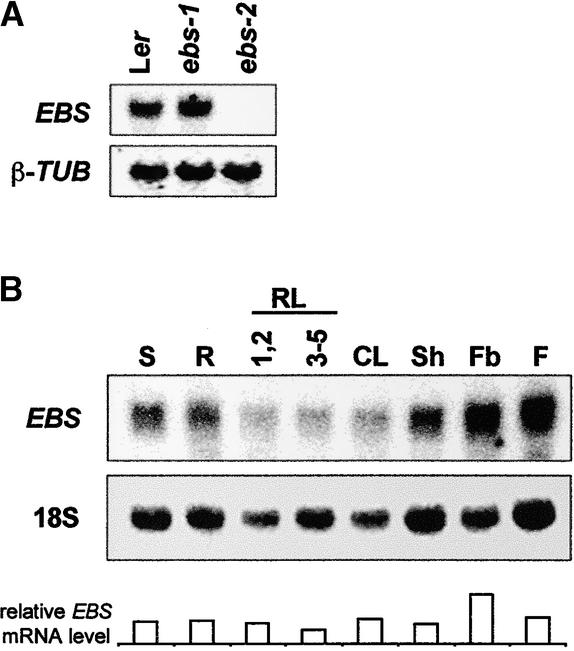
The expression of EBS was examined by both RNA gel blot analysis and in situ hybridization. EBS transcript was as abundant in the ebs-1 mutant as in wild-type plants, whereas, as expected, it was absent in the ebs-2 deletion allele (Figure 3A), which also demonstrates the specificity of the probe. EBS transcript was present at similar levels in all adult organs except for floral buds, in which it appeared to be slightly higher (Figure 3B). The level of EBS mRNA remained constant during the day, indicating that the transcription of EBS is not subject to circadian or light-dark regulation (data not shown). Similarly, EBS mRNA abundance was not sensitive to photoperiod and remained at similar levels throughout development under both LD and SD conditions (data not shown).
Figure 3.
Expression of the EBS Gene.
(A) RNA gel blot showing the expression of EBS in 7-day-old seedlings of Ler, ebs-1, and ebs-2 grown under LD conditions. β-TUB, β-TUBULIN.
(B) Expression of EBS in different organs: S, 7-day-old seedlings; R, roots; RL, rosette leaves (1,2 and 3-5 indicate leaves 1 and 2 and 3 to 5, respectively); CL, cauline leaves; Sh, shoot; Fb, floral buds; F, flowers. The relative abundance of EBS transcript is given for comparison.
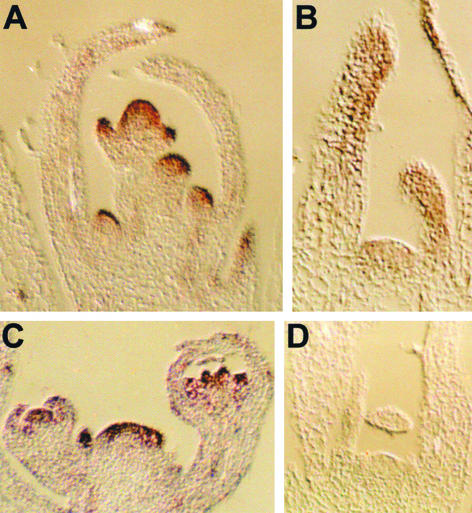
To investigate in detail possible expression patterns of EBS within organs, we performed in situ hybridization experiments (Figure 4). This analysis revealed that EBS transcript was present throughout the shoot apical meristem and young primordia during both vegetative and reproductive development. The relative levels of EBS mRNA seemed to be higher in young inflorescence and floral primordia, particularly in outer cell layers, as well as in floral organ primordia within the developing flower (Figures 4A to 4C). As expected, no hybridization signal was observed on sections of the ebs-2 mutant allele (Figure 4D). The expression pattern of EBS was consistent with the observed pleiotropic effects of ebs mutations and the proposed involvement of the EBS product in the regulation of flowering time and other aspects of plant development (Gómez-Mena et al., 2001).
Figure 4.
Expression Patterns of EBS at Different Developmental Stages.
In situ hybridization was performed on sections prepared from Ler seedlings grown for 14 days under LD conditions (A), Ler seedlings grown for 14 days under SD conditions (B), inflorescences of Ler plants with flower primordia at different developmental stages (C), and ebs-2 seedlings grown for 14 days under SD conditions (D).
Overexpression of EBS Causes Early Flowering
To investigate the effect of increasing the abundance of the EBS transcript, we generated 28 transgenic lines expressing EBS under the control of the 35S promoter (35S:EBS). Approximately 60% of these overexpressor lines showed similar phenotypes to the loss-of-function ebs mutants, namely, early flowering, a dwarf phenotype, and reduced fertility (Figure 5A). Further characterization of some of these lines revealed that those containing higher levels of EBS transcript were earlier flowering and showed a more extreme dwarf phenotype (Figures 5A to 5C). The high abundance of the EBS mRNA in these plants suggested that gene silencing is unlikely to explain the similarity in the phenotype of overexpressor and loss-of-function mutants. As with ebs mutants (Gómez-Mena et al., 2001), the effect of EBS overexpression was more conspicuous under SD conditions. The presence of the BAH and PHD motifs suggests that EBS could be part of a protein complex involved in the modulation of chromatin structure. The accumulation of EBS protein in 35S:EBS lines could result in an impairment of the formation of the putative EBS protein complex, perhaps by sequestering components of the complex into inactive associations, resulting in the deregulation of gene expression similar to that caused by loss-of-function mutations. To further explore this hypothesis, we generated transformant lines overexpressing either the mutant version of the EBS gene present in the ebs-1 mutant (35S:ebs) or a truncated protein containing the PHD domain and the nuclear localization signal (35S:M-PHD). 35S:ebs plants showed a phenotype similar to that of 35S:EBS plants (data not shown), suggesting that the mutant version of the protein retains the ability to impair the proper formation of the EBS complex, perhaps by binding to other proteins normally present in the complex. On the other hand, 35S:M-PHD plants also showed an early-flowering phenotype and dwarfism, although in this case, both phenotypes were less severe than in plants that overexpressed any of the full-length proteins (data not shown). Again, this result suggests that PHD domain overexpression can interfere with the organization or regulation of the complex containing EBS.
Figure 5.
Effect of EBS Overexpression on Flowering Time.
(A) Flowering time phenotypes of several transgenic lines overexpressing EBS (35S:EBS) compared with those of Ler and ebs-1.
(B) Accumulation of EBS transcript in different 35S:EBS lines. β-TUB, β-TUBULIN.
(C) Flowering time of 35S:EBS lines expressed as the number of leaves produced before flowering.
All plants were grown under SD conditions.
EBS Regulates Flowering Time through the Specific Repression of FT Expression
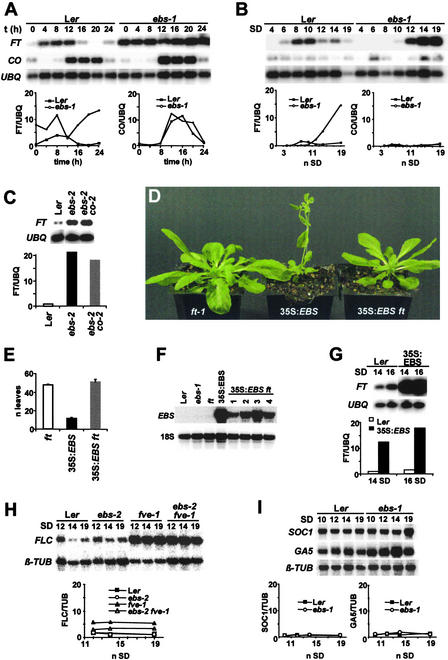
The sequence of the EBS protein suggests that it could be part of a transcriptional repressor complex involved in the modulation of chromatin structure. Because ebs mutations accelerate flowering, especially under SD conditions, EBS could be involved in the repression of genes that promote the transition to flowering in noninductive photoperiods. With respect to flowering time, the ft mutation is epistatic to ebs, indicating that FT is required for the early flowering of ebs mutants (Gómez-Mena et al., 2001). FT expression is promoted by growth under LD conditions through the activity of CO. Under these conditions, FT mRNA cycles with a daily pattern that appears to follow that of CO expression (Suárez-López et al., 2001). Because the overexpression of FT causes early flowering (Kardailsky et al., 1999; Kobayashi et al., 1999), we tested whether the acceleration of flowering observed in ebs mutants is caused by the upregulation or premature induction of FT. The early flowering of ebs mutants is especially clear in SD conditions; therefore, we compared the oscillations in FT expression at different times of a 24-h cycle in Ler and ebs-1 plants grown under SD conditions. As shown in Figure 6A, FT expression in ebs mutants was higher than that in wild-type plants for most of the 24-h period. By contrast, the abundance of CO mRNA was not affected by the ebs mutation (Figure 6A). This increase in FT expression in ebs mutants was further confirmed by collecting samples at dawn after different periods of growth in SD conditions. Again, the level of FT expression in ebs mutants appeared to increase prematurely compared with the level in Ler, and no significant differences in CO expression were detected between Ler and ebs-1 (Figure 6B). These results suggest that EBS represses FT expression under SD conditions independently of CO. Consistent with this conclusion, the abundance of FT mRNA abundance was similar in ebs co double mutant and ebs mutant plants, and in both cases, it was higher than that in wild-type plants (Figure 6C).
Figure 6.
Effect of ebs Mutations and EBS Overexpression on the Expression of Genes Involved in the Control of Flowering Time in Arabidopsis.
(A) FT and CO expression in Ler and ebs-1 mutant plants during a 24-h cycle. Seedlings were grown for 14 short days, and RNA was isolated from tissue collected at the times indicated after dawn. UBQ, UBIQUITIN.
(B) FT and CO expression in Ler and ebs-1 plants collected at different days after sowing under SD conditions.
(C) FT expression in Ler, ebs-2, and ebs-2 co-2 plants after 16 days of growth under SD conditions.
(D) Effect of the ft mutation on the flowering time of 35S:EBS plants.
(E) Flowering time of ft, 35S:EBS, and 35S:EBS ft lines expressed as the number of leaves produced before flowering.
(F) Level of accumulation of EBS transcript in different 35S:EBS ft lines.
(G) FT expression in 35S:EBS lines compared with Ler wild type after 14 and 16 days of growth under SD conditions.
(H) FLC expression in ebs-2 in both Ler and fve-1 mutant background at different days after sowing under SD conditions. β-TUB, β-TUBULIN.
(I) SOC1 and GA5 expression in Ler and ebs-1 mutant plants at different days after sowing under SD conditions.
All values are expressed relative to Ler at 24 h (A), day 19 ([B], [H], and [I]), day 16 (C), or day 14 (G) after normalization using UBIQUITIN ([A] to [C] and [G]) or β-TUBULIN ([H] and [I]). Unless indicated otherwise, samples for RNA isolation were collected immediately after dawn. All experiments were repeated at least twice.
As mentioned above, the overexpression of EBS causes early flowering, and as with ebs mutations, this phenotype is more conspicuous under SD conditions. To determine whether this acceleration of flowering observed in 35S:EBS plants also required FT, we analyzed the effect of ft mutations on the flowering time of 35S:EBS plants. If the accumulation of the EBS product causes a deregulation of FT expression that leads to early flowering in 35S:EBS lines, then in the absence of FT functional product, the flowering time of these transformants should be delayed. The results obtained show that 35S:EBS ft plants grown under SD conditions flowered with a similar number of leaves as ft mutants (Figures 6D and 6E), despite the high levels of EBS mRNA present in 35S:EBS ft plants (Figure 6F), indicating that the early-flowering phenotype of 35S:EBS requires FT. Furthermore, the overexpression of EBS also causes a premature increase of FT expression under SD conditions (Figure 6G), which can explain the observed early-flowering phenotype of 35S:EBS plants. Therefore, the early flowering caused by EBS overexpression appears to be mediated by the effect of EBS on the regulation of FT expression.
In addition to FT, two other genes, SOC1 and FLC, integrate signals from different pathways to regulate flowering time in Arabidopsis (Mouradov et al., 2002; Simpson and Dean, 2002). To determine whether the EBS protein also is involved in their regulation, we monitored the expression of these genes around the time of FT induction in SD-grown ebs mutants. As shown in Figure 6H, the ebs mutation did not significantly alter the expression of FLC in the Ler or fve-1 background, in which the level of FLC transcript is higher. Similarly, no significant differences in the expression of SOC1 were detected between Ler and ebs plants (Figure 6I), indicating that the EBS product is not involved in the transcriptional regulation of either FLC or SOC1. Phenotypic analysis of the double mutant ebs-1 soc1 provided further indication that EBS and SOC1 act in independent pathways. The flowering time of the double mutant, measured as the number of leaves produced before flowering, was intermediate between both parental lines (soc1, 57.4 ± 1.6; ebs-1, 12.8 ± 2.3; ebs-1 soc1, 22.4 ± 2.3), demonstrating the lack of interaction between EBS and SOC1.
The flowering of Arabidopsis under SD conditions is strongly dependent on GA (Wilson et al., 1992). GA biosynthesis also is required for the early flowering of ebs mutants, because mutations in GA biosynthetic enzymes eliminate the acceleration of flowering observed in ebs mutants (Gómez-Mena et al., 2001). GA5 encodes a key enzyme in the regulation of the GA biosynthetic pathway (20-oxidase), and changes in the level of GAs feed back on the level of expression of GA5 (Hedden and Phillips, 2000). Therefore, we examined the effect of the ebs mutation on GA5 expression under SD conditions. As shown in Figure 6I, there was no significant difference in the expression of GA5 between Ler and ebs. These results are consistent with previous genetic data (Gómez-Mena et al., 2001), which suggested that EBS is not involved in the regulation of GA biosynthesis but regulates developmental processes also regulated by GAs, such as flowering under SD conditions. Together, the results obtained from the analysis of the expression of key genes in the regulation of flowering time suggest that EBS acts to delay flowering under SD conditions by repressing specifically the expression of FT, and this repression appears to be independent of the activity of other genes, such as CO or FLC, that are known to participate in the regulation of FT expression.
Mutations in EBS can partially rescue the floral phenotype of lfy mutants, and both petals and stamens, which normally are absent in lfy-6 mutants, differentiate in the double mutant ebs lfy-6 (Gómez-Mena et al., 2001) (Figure 7A). FT appears to be required, together with LFY, in the determination of floral meristem identity (Ruiz-García et al., 1997) (Figure 7B). Because the effect of EBS on flowering time is mediated specifically by FT, it is possible that the effect of EBS on the determination of floral organ identity also requires FT. To test this hypothesis, we generated the triple mutant ebs-1 ft lfy-6. As shown in Figures 7C and 7D, the development of petals and stamens was suppressed completely in the triple mutant plants, suggesting that FT is required for the specification of these floral organs in the ebs lfy-6 background. This observation suggests that FT mediates the effect of EBS on different aspects of reproductive development, such as the transition to flowering and the specification of floral organ identity.
Figure 7.
Inflorescence Phenotype of the ebs-1 ft lfy-6 Triple Mutant.
The partial rescue of the floral phenotype of lfy-6 observed in the ebs-1 lfy-6 double mutant is suppressed in the presence of ft mutations.
(A) The floral phenotype of the double mutant ebs-1 lfy-6 displaying the development of petals and stamens (described previously by Gómez-Mena et al. [2001]) is shown for comparison.
(B) Inflorescence of the lfy-6 ft double mutant.
(C) Inflorescence of the triple mutant ebs-1 lfy-6 ft.
(D) Scanning electron micrograph showing the absence of petals and stamens in the triple mutant. Bar = 500 μm.
DISCUSSION
EBS Belongs to a Family of Plant Proteins with Features Characteristic of Chromatin-Interacting Factors
EBS, a gene involved in the repression of the floral transition and other developmental processes, such as germination and floral organ specification (Gómez-Mena et al., 2001), encodes a nuclear protein that contains a BAH and a PHD domain. Both motifs are thought to be involved in the establishment of protein–protein interactions and are present in several transcriptional regulators that act through chromatin remodeling (Aasland et al., 1995; Callebaut et al., 1999). Therefore, the presence of both BAH and PHD motifs suggests that EBS is part of a nuclear protein complex involved in the chromatin-mediated regulation of gene expression.
A small family of plant proteins share the same modular architecture as EBS—that is, they are made up of a BAH and a PHD domain together with a nuclear localization signal. The first protein of this type to be identified was ES43 from barley (Speulman and Salamini, 1995). Two EBS homologs have been identified in the Arabidopsis genome, SHL (SHORT LIFE) and EBL2 (EBS-LIKE2). Like EBS, SHL is a nuclear protein, and based on the phenotype of transgenic lines with altered levels of expression, it was proposed to be required for proper plant growth and development (Müssig et al., 2000). Observations made in our laboratory indicate that loss-of-function insertion alleles of the SHL gene show no visible effect on flowering time; however, when this mutation was combined with ebs-1, the double mutant flowered much earlier than ebs-1 alone (J. Jarillo, personal communication), suggesting that SHL is partially redundant with EBS.
A number of chromatin-remodeling factors are conserved functionally in animals and plants. However, some of the plant proteins contain different combinations of motifs than their animal counterparts (Wagner, 2003) or belong to novel classes of chromatin-modifying proteins not found in animals (Bartee et al., 2001). Proteins with the same modular architecture as EBS are present only in plants, although the BAH and PHD domains are closely associated in a number of other animal and yeast proteins that also contain other motifs often involved in epigenetic mechanisms of gene transcriptional regulation (such as SET domains) or DNA binding motifs (such as AT hooks) (Callebaut et al., 1999). The functional significance of this association remains unknown but raises the intriguing question of whether those modules present in BAH/PHD-containing proteins from other organisms are provided by different proteins in plants.
EBS Regulates Flowering by Repressing FT Expression
FT plays a crucial role in integrating the information from different pathways that regulate flowering time in Arabidopsis (Mouradov et al., 2002; Simpson and Dean, 2002). FT promotes the initiation of flowering, and its overexpression is sufficient to accelerate flowering under noninductive SD conditions (Kardailsky, et al., 1999; Kobayashi et al., 1999). We have shown previously that the early-flowering phenotype of ebs mutants requires FT (Gómez-Mena et al., 2001). Here, we present evidence demonstrating that FT expression is upregulated in ebs mutants grown under SD conditions and therefore that EBS is required to directly or indirectly repress FT expression in noninductive photoperiods. This regulation is specific for FT, because the expression of other genes, such as CO and FLC, that are known to regulate FT (Samach et al., 2000; Hepworth et al., 2002) is not affected in ebs mutants. This is consistent with previous conclusions from the analysis of double mutants that combine ebs with late-flowering mutants, which indicated that EBS acts independently of CO and the autonomous pathway to regulate flowering time (Gómez-Mena et al., 2001). Our data also indicate that EBS and SOC1 do not interact within the same pathway to regulate the floral transition.
In addition to the requirement for FT, the early-flowering phenotype of ebs mutants under SD conditions also showed an absolute requirement for GA biosynthesis. However, the levels of expression of GA5 under SD conditions were not affected by ebs mutations, confirming previous genetic analyses that indicated that the phenotype of ebs mutants is not likely to be caused by alterations in GA levels (Gómez-Mena et al., 2001). Because EBS is required to repress FT expression but not GA biosynthesis under SD conditions, the requirement of both FT and GAs for the early flowering of ebs mutants suggests that GAs play a regulatory role in FT expression or downstream of it to promote the floral transition under SD conditions. GA-deficient mutants are expected to suppress the flowering of 35S:FT transgenic plants if GAs are required downstream of FT but not if GAs regulate FT expression. Constitutive overexpression of FT can promote flowering under SD conditions in the ga1-3 mutant background (Blázquez et al., 2002), supporting a model in which GAs are involved in the regulation of FT expression under SD conditions. This and our observation that the ga1 mutation suppresses the early-flowering phenotype of ebs under SD conditions are consistent with the proposal that GA is required for the increased expression of FT observed in the ebs mutant. This could be tested by directly analyzing FT expression in ebs ga1 double mutants, but to date, poor germination of these double mutants has prevented us from performing this experiment.
Other phenotypic alterations caused by ebs mutations during reproductive development, such as the partial recovery of petal and stamen development in the lfy background (Gómez-Mena et al., 2001), also appear to be mediated by FT, because ft mutations completely eliminate the rescue of the lfy floral phenotype observed in ebs lfy double mutants. This may be attributable to a reduction in AP1 expression, because ft mutations were shown previously to reduce AP1 mRNA levels when combined with lfy (Ruiz-García et al., 1997). In conclusion, both genetic and molecular data support a role for EBS in the control of reproductive development by specifically repressing FT expression.
The Molecular Mechanism of EBS-Mediated Repression
EBS is required to repress FT expression during vegetative growth in noninductive photoperiods; later in development, this repression must be relieved to allow the onset of reproductive development. The CO protein is required to promote FT expression under LD conditions, and under these conditions, CO may overcome the repression mediated by EBS. Under SD conditions, however, FT activation probably occurs independently of CO, and consistent with this possibility, the peak in FT expression under SD conditions occurred before that in CO. Transcriptional repression of EBS is unlikely to be responsible for the relief of repression of FT, because EBS transcript was present at similar levels in all organs analyzed and in different photoperiod regimes; therefore, other regulatory mechanisms, such as protein modifications or changes in protein–protein interactions, could be invoked to explain this repression release.
The nature of the EBS protein suggests that it is part of a protein complex involved in the control of gene expression through changes in chromatin structure. A number of recent studies have shown that proteins involved in chromatin remodeling, such as plant PcG protein homologs, play an essential role in repressing the expression of key regulatory genes required for the onset of different developmental processes in Arabidopsis, including the reproductive program (Wagner, 2003). PcG proteins such as CLF, EMF2, and FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) appear to repress the expression of floral homeotic genes during vegetative growth to prevent precocious flowering (Goodrich et al., 1997; Kinoshita et al., 2001; Yoshida et al., 2001); VERNALIZATION2, another PcG protein, is required to maintain the repression of the key floral repressor FLC in response to vernalization (Gendall et al., 2001). Other types of chromatin-remodeling factors also are involved in the regulation of different aspects of reproductive development; the SWI/SNF ATPase homolog SPLAYED is a repressor of the floral transition proposed to regulate LFY activity (Wagner and Meyerowitz, 2002). Mutations in the LIKE HETEROCHROMATIN PROTEIN1 (LHP1) gene, a homolog of Drosophila HP1, cause early flowering, which correlates with precocious CO expression (Gaudin et al., 2001) and is dependent on FT. As in ebs, FT is upregulated in lhp1/tfl2 mutants, suggesting that LHP1/TFL2 may be required to repress FT during vegetative development (K. Goto, personal communication) (Wagner, 2003).
The mechanism of repression of PcG proteins and other chromatin-remodeling factors remains largely unknown in plants, although, as in animals, they are likely to form large protein complexes responsible for altering the accessibility of DNA to the transcriptional machinery (Simon and Tamkun, 2002). The domains present in EBS suggest that it could be part of a multiprotein repressor complex. The observed early-flowering phenotype of 35S:EBS plants is consistent with this view. Despite the fact that EBS is required for the repression of flowering, 35S:EBS plants do not flower late, suggesting that EBS is necessary but not sufficient for the repression of floral initiation and that other components probably are required for the repressor activity of EBS. Similar results were obtained in the case of the PcG protein EMF2, because both emf2 mutants and 35S:EMF2 transgenic plants displayed early flowering (Yoshida et al., 2001). The precocious flowering of 35S:EBS plants could be explained by the disruption caused in the formation or function of the EBS complex by the hyperaccumulation of one of the members of the protein complex.
Together, our data suggest that EBS likely is part of a protein-repressor complex involved in the control of gene expression. The ubiquitous pattern of expression of EBS and the pleiotropic phenotype observed in ebs mutants suggest that the EBS protein complex participates in the regulation of different stages of plant development, including flowering time. Further research will be required to increase our understanding of the composition, organization, and regulation of the EBS complex and its role in plant development as well as to identify the target genes of the complex.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana mutant lines used in this work are in the Ler background and were obtained from the ABRC (Ohio State University, Columbus) and the Nottingham Arabidopsis Centre (UK). The late-flowering mutants ft-1, fve-1, and co-2 were described previously (Koornneef et al., 1991). The ebs-1 and ebs-2 mutant alleles were isolated as described previously (Gómez-Mena et al., 2001). Seeds were stratified for 4 to 7 days before sowing on soil containing a mixture of substrate and vermiculite (3:1). Plants were grown under controlled environmental conditions (18°C and 80% RH) and illuminated with cool-white fluorescent lights for either 16 h followed by 8 h of darkness (long days) or 8 h followed by 16 h of darkness (short days).
Molecular Characterization of the ebs-2 Allele and Map-Based Cloning
A chromosomal rearrangement was present in the Dissociation (Ds)-induced ebs-2 allele (Gómez-Mena et al., 2001). To determine the nature of the mutation present in ebs-2, we generated a cosmid library of this mutant using the Gigapack II XL kit (Stratagene). The isolation of several clones containing Ds and the DNA flanking the mobile element on both ends confirmed that a deletion was present in ebs-2. This deletion spans the genomic region between RJS (the inverse PCR fragment isolated from the 3′ end of Ds) and JGB9 (the launching site of Ds) (Long et al., 1997).
To finely map the ebs-1 mutation within the interval deleted in ebs-2, we generated cleaved amplified polymorphic sequence (Bell and Ecker, 1994) and simple sequence length polymorphism (Konieczny and Ausubel, 1993) markers between JGB9 and RJS: MPRJS, a DNA fragment generated by primers RTMP11 (5′-TGGACTGCTCTTTCACGGTTTTC-3′) and RTMP10 (5′-GGAGCCAGCTTCCGTAGG-3′), cleaved with BsaAI; MPO13, a fragment generated by primers MP502.9 (5′-CAAAAGCAATGAAATAGTTGCC-3′) and MP502.10 (5′-AGCGCCGATCCCACC-3′), cleaved with MunI; MPG8, a fragment generated by primers MP502.11 (5′-GCATTAGAATCTCTCCTGC-3′) and MP502.12 (5′-GCTCCGCGAGTGCAC-3′), cleaved with AvaII; and MPN9, a fragment generated by primers MP502.3 (5′-CAGCATTCTCCATTCCCCG-3′) and MP502.4 (5′-GTGTTGAAGCTATTGCCTGTC-3′).
Plasmid Construction
Standard molecular biology procedures were used for plasmid construction. To make the fusion protein GFP-EBS for nuclear localization experiments, we used vector pAVA393 derived from pAVA321 (von Arnim et al., 1998). To insert EBS cDNA in frame with GFP, we used primers 5′-AAAACCAGATCTTCCATGGCG-3′ (which creates a BglII site upstream of the EBS ATG), and 5′-AGAACTCTAGATTACCTTTTTCTGG-3′ (which creates an XbaI site after the stop codon).
To make the 35S:EBS construct, EBS cDNA was inserted into pJIT60 vector, carrying two copies of the 35S promoter. Subsequently, a SacI-XhoI fragment containing the promoter and the EBS cDNA was moved into the binary vector pGREEN(0229) (Hellens et al., 2000). p35S:ebs was constructed to generate a PCR fragment with primer OXF (5′-AAACCCTGTCGACCATGGC-3′), which creates a SalI site upstream of the EBS ATG, and OXR (5′-CCCAAGAATTCGCGATTAC-3′), generating an EcoRI site after the stop. To construct p35S:M-PHD, a PCR fragment was generated with primers OXPHDF (5′-CCTGGTCGACTTGCTATGTACTG-3′) and OXR. OXPHDF creates a Met codon by replacing the first base of the Val-147 codon (GTG) and introduces a SalI site upstream of it. Both PCR fragments were cloned into pJIT60 and moved into pGREEN(0229) as described above for the p35S:EBS construct.
Plant Transformation and Transient Expression Assays
Arabidopsis plants were transformed using the floral-dip method (Clough and Bent, 1998). The Agrobacterium tumefaciens strain used was C58C1 pGV3101 pMP90. Transformant plants were selected on soil by spraying seedlings with BASTA. Transient expression assays in onion epidermal cells have been described (Varagona et al., 1992). DNA-coated particles were bombarded onto onion epidermal peels using a Biolistic PDS-1000/He System (Bio-Rad). After bombardment, onion cell layers were incubated overnight to allow for GFP expression. To visualize nuclei, the onion peels were immersed in 0.1% (v/v) 4′,6-diamidino-2-phenylindole for 5 min. Subsequently, samples were examined by epifluorescence microscopy (Leica DMR, Wetzlar, Germany).
Expression Analyses
RNA was extracted from seedlings at the times indicated in the figures according to procedures described previously (Logemann et al., 1987). For RNA gel blot analysis, 10 μg of RNA was loaded onto 1.2% agarose denaturing formaldehyde gels and transferred onto Hybond NX nylon membranes (Amersham). The EBS probe was a 270-bp HindIII-DdeI fragment from EBS cDNA. The β-TUBULIN and the 18S rRNA probes have been described (Onouchi et al., 2000; Samach et al., 2000). The SOC1 probe was from pAGL20-3 (Samach et al., 2000), and the FLC probe was a cDNA fragment lacking the MADS-box domain (Michaels and Amasino, 1999). The GA5 probe was a PCR fragment (Peng et al., 1997). For low-abundance mRNAs, such as FT and CO, we performed reverse transcriptase–mediated PCR according to described procedures (Blázquez and Weigel, 1999). In situ hybridization experiments were performed as described by Gómez-Mena et al. (2001). EBS probes (+ and −) were generated using T7 polymerase and plasmid pCD124 containing the coding region of EBS cDNA cloned into pBS with inverted orientations.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for the sequences mentioned are as follows: EST F3C6T7A, N96619; EST 135G1T7, T46081; ES43 barley protein, X77575; and rice EBS-like protein, BAC24958.
Acknowledgments
We thank D. Long for her contribution to the isolation of the ebs-2 allele. This work was supported by Grant BIO2001-3891 from the Ministerio de Ciencia y Tecnología (MCYT). Support for research activity at the Centro Nacional de Biotecnología is provided by a specific agreement with Consejo Superior de Investigaciones Científicas–Instituto Nacional de Investigacion y Tecnologia Agraria y Alimentaria (INIA). M.P. was supported by a European Molecular Biology Organization long-term fellowship and Biotechnology and Biological Science Research Council and INIA-MCYT postdoctoral contracts. C.G.-M. was supported by an INIA predoctoral fellowship.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012153.
References
- Aasland, R., Gibson, T.J., and Stewart, A.F. (1995). The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Aubert, D., Chen, L., Moon, Y.H., Martin, D., Castle, L.A., Yang, C.H., and Sung, Z.R. (2001). EMF1, a novel protein involved in the control of shoot architecture and flowering in Arabidopsis. Plant Cell 13, 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee, L., Malagnac, F., and Bender, J. (2001). Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15, 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., Trénor, M., and Weigel, D. (2002). Independent control of gibberellin biosynthesis and flowering time by the circadian clock in Arabidopsis. Plant Physiol. 130, 1770–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez, M.A., and Weigel, D. (1999). Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol. 120, 1025–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut, I., Courvalin, J.-C., and Mornon, J.-P. (1999). The BAH (bromoadjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446, 189–193. [DOI] [PubMed] [Google Scholar]
- Chen, L., Cheng, J.C., Castle, L., and Sung, Z.R. (1997). EMF genes regulate Arabidopsis inflorescence development. Plant Cell 9, 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Gaudin, V., Libault, M., Pouteau, S., Juul, T., Zhao, G., Lefebvre, D., and Grandjean, O. (2001). Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128, 4847–4858. [DOI] [PubMed] [Google Scholar]
- Gendall, A.R., Levy, Y.Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. [DOI] [PubMed] [Google Scholar]
- Gómez-Mena, C., Piñeiro, M., Franco-Zorrilla, J.M., Salinas, J., Coupland, G., and Martínez-Zapater, J.M. (2001). early bolting in short days: An Arabidopsis mutation that causes early flowering and partially suppresses the floral phenotype of leafy. Plant Cell 13, 1011–1024. [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. [DOI] [PubMed] [Google Scholar]
- Goodwin, G.H., and Nicholas, R.H. (2001). The BAH domain, polybromo and the RSC chromatin remodeling complex. Gene 268, 1–7. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Manipulation of hormone biosynthetic genes in transgenic plants. Curr. Opin. Biotechnol. 11, 130–136. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S.R., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Hepworth, S.R., Valverde, F., Ravenscroft, D., Mouradov, A., and Coupland, G. (2002). Antagonistic regulation of flowering time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21, 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2001). Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. USA 98, 14156–14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using codominant ecotype specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Lee, H., Suh, S.S., Park, E., Cho, E., Ahn, J.H., Kim, S.G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Hall, T.C., and Holmes-Davies, R. (2002). Plant chromatin: Development and gene control. Bioessays 24, 234–243. [DOI] [PubMed] [Google Scholar]
- Logemann, J., Schell, J., and Willmitzer, L. (1987). Improved method for isolation of RNA from plant tissue. Anal. Biochem. 163, 16–20. [DOI] [PubMed] [Google Scholar]
- Long, D., Goodrich, J., Wilson, K., Sundberg, E., Martin, M., Puangsomlee, P., and Coupland, G. (1997). Ds elements on all five Arabidopsis chromosomes and assessment of their utility for transposon tagging. Plant J. 11, 145–148. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS-domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous-pathway mutations, but not responsiveness to vernalization. Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 (suppl.), S111.–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig, C., Kauschmann, S.D., Clouse, S.D., and Altmann, T. (2000). The Arabidopsis PHD-finger protein SHL is required for proper development and fertility. Mol. Gen. Genet. 264, 363–370. [DOI] [PubMed] [Google Scholar]
- Onouchi, H., Igeño, M.I., Perilleux, C., Graves, K., and Coupland, G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12, 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson, F., Costa, M.M., Hepworth, S.R., Vizir, I., Piñeiro, M., Reeves, P.H., Puterill, J., and Coupland, G. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631. [DOI] [PubMed] [Google Scholar]
- Ruiz-García, L., Madueño, F., Wilkinson, M., Haughn, G., Salinas, J., and Martínez-Zapater, J.M. (1997). Different roles of flowering time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9, 1921–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Schultz, D.C., Friedman, J.R., and Rauscher, F.J., III (2001). Targeting histone deacetylase complexes via KRAB-zinc finger proteins: The PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15, 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, J.A., and Tamkun, W.T. (2002). Programming on and off states of chromatin: Mechanisms of Polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 12, 210–218. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Speulman, E., and Salamini, F. (1995). A barley cDNA clone with homology to the DNA-binding domain of the steroid hormone receptors. Plant Sci. 106, 91–98. [Google Scholar]
- Suárez-López, P., Wheatly, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Varagona, M.J., Schmidt, R.J., and Raikhel, N.V. (1992). Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein opaque-2. Plant Cell 4, 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim, A.G., Deng, X.-W., and Stacey, M.G. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- Wagner, D. (2003). Chromatin regulation of plant development. Curr. Opin. Plant Biol. 6, 20–28. [DOI] [PubMed] [Google Scholar]
- Wagner, D., and Meyerowitz, E.M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Wilson, R.N., Heckman, J.W., and Somerville, C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100, 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, N., Yanai, Y., Chen, L., Kato, Y., Hiratsuka, J., Miwa, T., Sung, Z.R., and Takahashi, S. (2001). EMBRYONIC FLOWER2, a novel Polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13, 2471–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]