Abstract
From an enhancer trap screen for genes expressed in Arabidopsis embryos, we identified a gene expressed from the octant stage onward in the boundary between the two presumptive cotyledons and in a variety of postembryonic organ and meristem boundaries. This gene, CUP-SHAPED COTYLEDON3 (CUC3), encodes a putative NAC-domain transcription factor that is homologous with CUC1 and CUC2. Analysis of a CUC3 hypomorph and a putative cuc3 null mutant indicates that CUC3 function is partially redundant with that of CUC1 and CUC2 in the establishment of the cotyledon boundary and the shoot meristem, thus revealing an even higher degree of redundancy in this class of genes than was thought previously. The CUC3 expression pattern, the cuc3 phenotypes, and CUC3 expression in a series of shoot meristem mutants and transgenes suggest a primary role for CUC3 in the establishment of boundaries that contain cells with low proliferation and/or differentiation rates. The CUC-mediated establishment of such boundaries may be essential for the initiation of shoot meristems.
INTRODUCTION
The basic body architecture of higher plants is established during embryogenesis; however, unlike the situation in animals, almost the entire adult plant is produced postembryonically from two meristems situated at opposite ends of the embryo (Jürgens et al., 1991). The establishment of the shoot and root meristems during embryogenesis is crucial for proper plant development and thus should be tightly regulated. The developmental process through which cell groups in the embryo acquire distinct developmental fates according to their relative positions has been termed pattern formation (Jürgens et al., 1991). The final shape of the embryo is a result of the elaboration of the pattern, through the regional regulation of cellular proliferation and differentiation. For example, in dicot embryos, two cotyledonary primordia arise through increased cellular proliferation in two apical regions of the globular embryo, flanking a region of restricted proliferation. The latter region forms the boundary between the developing cotyledons and harbors the cells that contain the shoot apical meristem (SAM) (Barton and Poethig, 1993).
The elaboration of pattern through the regional regulation of cellular proliferation rates occurs not only during embryogenesis but is a recurrent mechanism during plant development. For instance, floral organs arise in concentric whorls from the floral meristem as bulges of cells separated by boundary regions with a low rate of cellular proliferation (Furner, 1996). Proliferating leaf primordia are separated from the shoot meristem by slower proliferating boundary regions (Callos and Medford, 1994), and trichomes arise through highly increased proliferation of individual leaf epidermal cells compared with the surrounding cells (Hulskamp et al., 1994). Several genes have been identified that are expressed specifically in organ or meristem boundaries. The Arabidopsis CUP-SHAPED COTYLEDON1 (CUC1) (Takada et al., 2001) and CUC2 (Aida et al., 1997) and the petunia NO APICAL MERISTEM (NAM) (Souer et al., 1996) genes are expressed in boundaries between floral organ primordia and in the boundary between the cotyledons. Mutations in these genes cause defects in the establishment of several boundaries, resulting in organ fusions. This fact suggests that boundaries are actively established and maintained. The CUC1, CUC2, and NAM genes all encode putative transcription factors of the NAC-domain class (Aida et al., 1997). NAC genes constitute a family of ∼100 putative members in Arabidopsis (Arabidopsis Genome Initiative, 2000; Duval et al., 2002) and are not found in organisms other than plants.
Besides defects in the establishment of boundaries, mutations in CUC1, CUC2, and NAM also affect the initiation of the SAM (Souer et al., 1996; Aida et al., 1997; Takada et al., 2001), which becomes histologically and functionally distinct in the boundary between the embryonic cotyledons during normal development. cuc1 and cuc2 single mutants occasionally show cotyledon fusion on one side and display weak fusions of sepals and stamens in the flower. cuc1 cuc2 double mutant seedlings have cotyledons fused on both sides and completely lack a SAM (Aida et al., 1997). Thus, CUC1 and CUC2 are required for boundary and SAM formation and are partially functionally redundant (Takada et al., 2001). CUC1 and CUC2 are required for the expression of another gene involved in SAM formation, SHOOT MERISTEMLESS (STM), which encodes a putative homeodomain transcription factor (Long et al., 1996; Aida et al., 1999). stm mutants also lack a SAM and display weak cotyledon fusions. STM is expressed in the SAM and initially also in the boundary between the cotyledons. Thus, the CUC/STM regulatory pathway is critical for the establishment of the boundary between the cotyledons and for the initiation of the SAM (Long and Barton, 1998; Aida et al., 1999).
Once established, the SAM harbors a population of stem cells over almost the entire life cycle of the plant (Weigel and Jürgens, 2002). Shoot meristem homeostasis is maintained by a tightly controlled balance between slowly dividing stem cells in the meristem center and cells that are displaced to the periphery and undergo differentiation (reviewed by Haecker and Laux, 2001). Molecular genetic studies have identified WUSCHEL (WUS) and CLAVATA as components of a negative feedback loop that controls this balance (Schoof et al., 2000). WUS encodes a transcription factor that functions in the specification of stem cells, and wus mutants fail to initiate a SAM (Laux et al., 1996; Mayer et al., 1998).
In this study, we identified the CUC3 gene, which encodes a NAC-domain protein highly similar to CUC1 and CUC2. CUC3 was identified in an enhancer trap screen for genes involved in pattern formation during embryogenesis (Vroemen et al., 1998). CUC3 is expressed during embryogenesis from the octant stage onward in the boundary between the presumptive cotyledons and later during development in a wide variety of plant organ and meristem boundaries. Analysis of a cuc3 hypomorph and a putative cuc3 null mutant shows that CUC3 functions in the establishment of the boundary between the cotyledons and in the formation of the SAM. Moreover, CUC3 function appears to be partially redundant with that of CUC1 and CUC2. Thus, the identification of CUC3 suggests an even greater degree of redundancy among CUC genes than was thought previously on the basis of CUC1 and CUC2. Based on the results of CUC3 expression analyses in shoot meristem mutants and on a transgenic background displaying ectopic shoot meristems, we propose a model in which the establishment of boundaries is a prerequisite for SAM formation.
RESULTS
Identification of Enhancer Trap Line WET368 and Molecular Cloning of CUC3
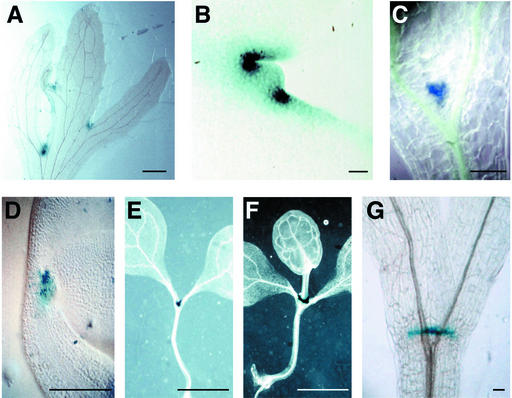
Line Wageningen Enhancer Trap 368 (WET368) was identified in a screen for genes expressed during Arabidopsis embryogenesis that encompassed 431 enhancer trap lines (Vroemen et al., 1998) and was selected for in-depth analysis based on its restricted expression in a region containing the embryonic SAM (Figures 1A to 1I). DNA gel blot analysis demonstrated the presence of a single DsE element insertion (Vroemen et al., 1998). The name WET368 refers to both the plant line carrying this enhancer trap insertion and to the actual DsE element inserted in this line.
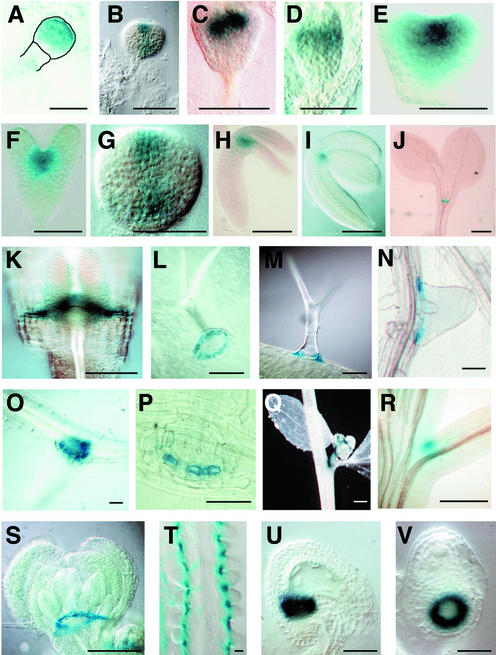
Figure 1.
GUS Expression in WET368 Plants.
(A) Early globular embryo (the embryo and the upper part of the suspensor are outlined for clarity).
(B) Triangular-stage embryo.
(C) Triangular-stage embryo (oblique view).
(D) Triangular-stage embryo (side view).
(E) Heart-stage embryo.
(F) Torpedo-stage embryo.
(G) Bending cotyledon-stage embryo (viewed from above).
(H) Bending cotyledon-stage embryo.
(I) Mature embryo.
(J) Seedling at 5 days after germination.
(K) Seedling at 7 days after germination (side view). The frontal cotyledon has been removed to expose the SAM and the boundaries of the cotyledons.
(L) Base of a leaf trichome viewed from below.
(M) Leaf trichome viewed from the side.
(N) Lateral root.
(O) and (P) Lateral root viewed through its base.
(Q) Axillary bud in the axil of a cauline leaf.
(R) Axil of inflorescence stem and pedicel.
(S) Flower bud.
(T) Placenta with ovule primordia.
(U) Mature ovule at approximately the time of fertilization.
(V) The same ovule as in (U) viewed through its chalazal end.
Bars = 40 μm in (A) to (P) and (T) to (V) and 2 mm in (Q) to (S).
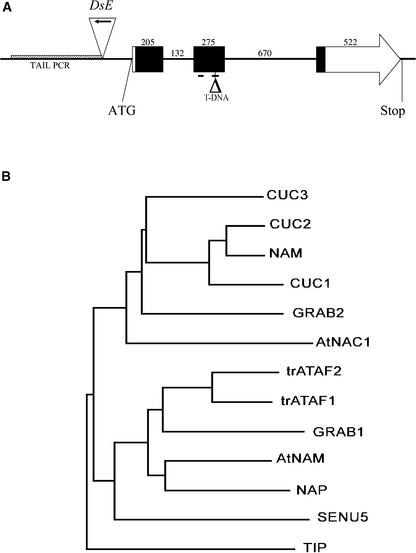
Genomic DNA flanking DsE was amplified by thermal asymmetric interlaced PCR to isolate the gene identified by the WET368 enhancer trap and used to screen an Arabidopsis genomic library. The insert of one of the positive phages was sequenced, and BLAST (Basic Local Alignment Search Tool) searches identified two overlapping BACs, F14G6 and F15M4, spanning the phage insert. A predicted open reading frame encoding a novel NAC-domain protein of 334 amino acids was located on the overlapping region of the two BACs, 220 bp downstream of the 3′ end of the WET368 DsE element (Figure 2A). We named this predicted gene NAC368, but based on sequence similarity with the previously identified CUC1 and CUC2 genes (Aida et al., 1997; Takada et al., 2001) and functional data presented here, it was renamed CUC3. CUC3 is located on the lower arm of chromosome 1, and the coding sequence consists of three exons of 205, 275, and 522 bp separated by two introns of 132 and 670 bp (Figure 2A). The NAC domain, which spans amino acids 22 to 171, is encoded by most of the first, the second, and the beginning of the third exons. The second exon contains a putative bipartite nuclear localization signal sequence (Kikuchi et al., 2000; Xie et al., 2000) that spans amino acids 84 to 96 and 124 to 141, suggesting that CUC3 is able to enter the nucleus (Figure 2A).
Figure 2.
The CUC3 Gene and Predicted Protein.
(A) Structure of CUC3. Boxes represent the open reading frame, the arrow at the end of the box at right corresponds to the C terminus of the predicted CUC3 protein, and lines indicate introns or untranslated regions. Black boxes represent nucleotides that encode the NAC domain, and numbers represent the lengths of exons and introns (in nucleotides). Lines below the second exon indicate putative bipartite nuclear localization signals. ATG and Stop indicate the start and stop codons; the triangle at left (with arrow) represents the DsE element in the WET368 (cuc3-1) line, and the arrow indicates the GUS gene; the shaded box represents the thermal asymmetric interlaced (TAIL) PCR product used to isolate CUC3; and the triangle at right indicates the Versailles T-DNA insertion in cuc3-2.
(B) Phylogenetic tree of the NAC domains of Arabidopsis CUC3, CUC2, CUC1, AtNAC1, ATAF1, ATAF2, AtNAM, NAP, and TIP, petunia NAM, wheat GRAB1 and GRAB2, and tomato SENU5. ATAF1 and ATAF2 are preceded by “tr” to indicate that the protein sequences were obtained by translation of mRNA sequences, leading to predicted proteins that are 60 amino acids longer than the protein sequences stored in GenBank.
A phylogenetic tree of the NAC domains of CUC3 and previously described NAC-domain proteins revealed that CUC3 is most similar to the Arabidopsis CUC1 and CUC2 and the petunia NAM proteins (Figure 2B). The NAC domain of CUC3 shares 66, 71, and 69% amino acid sequence identity with CUC1, CUC2, and NAM, respectively (Souer et al., 1996; Aida et al., 1997; Takada et al., 2001), whereas the CUC1 and CUC2 NAC domains are 82% identical.
WET368 β-Glucuronidase Is Expressed in Organ and Meristem Boundaries
We examined the WET368 β-glucuronidase (GUS) expression pattern by means of which the CUC3 gene was identified in detail. In embryos, GUS staining was detected first at the early globular stage, during which it was strongest in the apical part of the embryo (Figure 1A). We were unable to reliably detect GUS staining at earlier stages of embryogenesis. During the triangular (Figures 1B to 1D), heart (Figure 1E), and torpedo (Figure 1F) stages, GUS expression resolved to the center of the apical part of the embryo. From oblique (Figure 1C) and side (Figure 1D) views of triangular embryos, it appeared that WET368 GUS was expressed in a central stripe across the apical half of the embryo, representing the boundary between the emerging cotyledons. Between the torpedo and bending cotyledon stages, GUS staining became more prominent in the boundaries of the cotyledon margins than in the SAM region (Figure 1G). WET368 GUS expression remained associated with the embryo and seedling apex during further embryo and seedling development (Figures 1H to 1J). In the seedling apex, GUS staining was absent from the SAM itself and restricted to the boundaries of the cotyledon margins and the boundaries between the SAM and the cotyledons (Figure 1K).
Because WET368 GUS expression seemed restricted to (presumptive) boundary regions during embryo and seedling development, we next examined whether WET368 GUS also was expressed in other boundaries during plant development. WET368 GUS expression was detected in a one-cell-wide ring at the boundary between trichomes and leaf epidermis cells (Figures 1L and 1M), whereas no expression was detected in trichomes themselves (Figure 1M). This “support cell”–specific expression was not found in any other enhancer trap line in our collection (data not shown). Lateral roots were delimited at their bases by a one-cell-wide ring of cells expressing WET368 GUS (Figures 1N to 1P). This expression ring appeared only after the lateral root primordium emerged from the main root. In aerial plant parts, WET368 GUS expression was found in the adaxial axils of secondary inflorescences (Figure 1Q) and pedicels (Figure 1R). Furthermore, GUS expression was observed in axillary buds (Figure 1Q). WET368 GUS staining in flower buds was detected in a ring at the bases of sepals and petals (Figure 1S). In the carpel, WET368 GUS expression marked the boundaries between ovule primordia (Figure 1T). Analysis of serial optical sections (data not shown) revealed that ovule primordia were encircled at their bases and separated from each other by WET368 GUS–expressing cells. In ovules, WET368 GUS expression appeared only at the approximate time of fertilization, in a ring at the boundary between the nucellus and the chalaza (Figures 1U and 1V).
In conclusion, a variety of boundaries during Arabidopsis development are marked by WET368 GUS expression. In general, these boundaries separate two proliferating organs or a proliferating organ or cell from its surrounding cells.
Expression of CUC3 during Embryo Sac, Embryo, and Seedling Development
We used in situ mRNA hybridization to validate whether WET368 GUS expression faithfully mimicked CUC3 expression. In the mature embryo sac, CUC3 mRNA was detected in the two polar nuclei of the central cell (Figure 3A). The signal in the seed coat in Figures 3A to 3H also was seen in sense controls (data not shown), indicating that it does not relate to the presence of CUC3 mRNA. No clear signal was detected in the endosperm during seed development (Figures 3B to 3L). CUC3 mRNA was not detected in the zygote (data not shown) or in two-celled (Figure 3B) or four-celled (data not shown) embryos. During embryogenesis, CUC3 mRNA was detected first at the octant stage, at which it was restricted to the apical half of the embryo, above the O line (Figure 3C). In early globular embryos, CUC3 mRNA was most abundant in cells of the upper tier, although a weak signal also was detected in cells of the lower tier (Figure 3D).
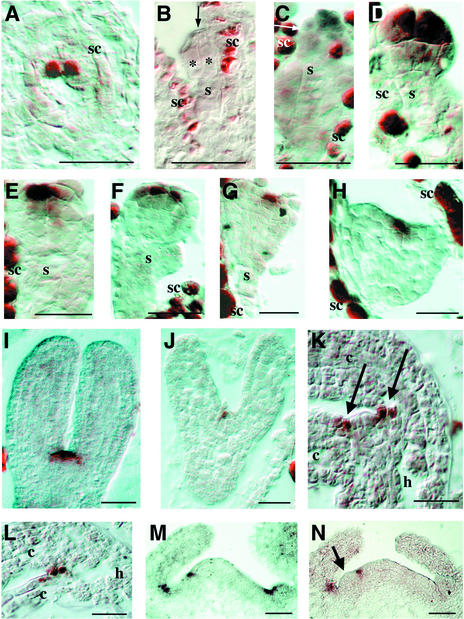
Figure 3.
In Situ Localization of CUC3 mRNA.
Sections are frontal and longitudinal unless stated otherwise.
(A) Mature embryo sac, transverse section through the polar nuclei of the central cell.
(B) Two-celled embryo (asterisks indicate the two embryo cells, and the arrow indicates the cell wall separating them).
(C) Octant-stage embryo (the arrow indicates the O line separating the upper and lower tiers).
(D) Early globular embryo.
(E) Globular embryo.
(F) Globular embryo (sagittal section).
(G) Triangular-stage embryo.
(H) Heart-stage embryo.
(I) Torpedo-stage embryo (central section through the SAM).
(J) Torpedo-stage embryo (superficial section through the boundary of the cotyledons).
(K) Mature embryo (central section through the SAM; arrows indicate the boundaries between the SAM and the cotyledons).
(L) Mature embryo (section through the boundary of the cotyledons).
(M) pt-1 seedling apex at 5 days after germination.
(N) pt-1 seedling apex at 5 days after germination with emerging leaf primordium (arrow).
c, cotyledon; h, hypocotyl; s, suspensor; sc, seed coat. Bars = 20 μm.
From the globular stage onward, CUC3 expression became restricted to the center of the apical part of the embryo (Figure 3E). Expression was strongest in epidermal cells, and from sagittal sections it appeared that CUC3 expression localized to a central stripe across the apical half of the embryo (Figure 3F). This stripe represents an early manifestation of bilateral symmetry, which becomes morphologically visible only at the triangular stage, along with the appearance of the cotyledon primordia (Figure 3G). Expression in the central apical stripe continued through the triangular (Figure 3G), heart (Figure 3H), and torpedo (Figure 3I) stages of embryogenesis, remaining strongest in epidermal cells, and corresponded to the WET368 GUS–expressing stripe during the same stages of embryogenesis (Figures 1B to 1F). During these stages, cotyledons developed on both sides of the CUC3-expressing cells.
Central sections through torpedo-stage embryos showed CUC3 expression in the presumptive SAM, extending into the boundaries between the SAM and the cotyledons (Figure 3I). Sections that were more superficial indicated that outside of the presumptive SAM, CUC3 was expressed in a one-cell-wide boundary separating the cotyledon margins (Figure 3J). In mature embryos, CUC3 mRNA was excluded form the SAM upon its first morphologically apparent proliferation (Figure 3K). CUC3 expression became restricted to the boundaries between the SAM and the cotyledons (Figure 3K, arrows) and the boundaries of the cotyledon margins (Figure 3L). In both boundaries, the expression domain was one cell wide and included the epidermal and maximally one hypodermal cell layer.
Seedlings of primordia timing (pt-1) were used to examine CUC3 expression in the seedling apex. These seedlings have an enlarged SAM (Mordhorst et al., 1998), and therefore were anticipated to give a higher spatial resolution of CUC3 expression compared with the wild type. CUC3 expression in pt-1 was identical to that in the wild type, as determined by WET368 GUS expression (data not shown). As in mature embryos, CUC3 expression was detected in the boundary between the SAM and the cotyledons in pt-1 seedlings (Figure 3M). In addition, CUC3 expression on the flank of the SAM marked the boundary between the vegetative meristem and an incipient leaf primordium (Figure 3M). Slightly later in development, proliferation of such a leaf primordium from the region enclosed by this boundary became morphologically visible (Figure 3N, arrow).
The combined WET368 GUS expression analysis and CUC3 in situ hybridization data indicate that WET368 GUS expression (Figures 1A to 1K) faithfully mimics the CUC3 expression pattern, although the GUS expression analysis has a somewhat lower cellular resolution and the in situ hybridization detects expression at a slightly earlier stage of embryogenesis. In the embryo, CUC3 expression precedes the appearance of the morphologically visible boundary between the cotyledons. The SAM is initiated from cells within this boundary, and once initiated, its surface is isolated from the neighboring cotyledons by a CUC3-expressing boundary. In the seedling apex, CUC3-expressing cells mark the boundary between the SAM and the emerging leaf primordia. Based on this CUC3 expression pattern and the extensive range of boundaries in adult plants marked by WET368 GUS expression (Figure 1), we classified CUC3 as a boundary gene.
WET368 GUS Expression in cuc Mutants
Based on the observation that CUC3 was expressed in boundaries, we next examined whether CUC3 expression was affected in boundary control mutants. cuc mutants display variable defects in cotyledon separation and SAM formation (Aida et al., 1997). The majority of cuc1 and cuc2 single mutants are phenotypically normal. However, in ∼0.5% of them, cotyledons are fused to a variable extent along one side, affecting the boundary of the cotyledon margins that normally expresses CUC3. These seedlings are called “heart-shaped seedlings” (Figure 4A). In heart-shaped cuc2 seedlings, WET368 GUS expression was detected in the boundary of the SAM and in the boundary of the cotyledon margins on the nonfused side (Figures 4B and 4C), essentially as in wild-type seedlings. On the fused side, WET368 GUS expression was restricted to a few cells at the top of the cotyledon fusion, marking the depression that partially separates the two fused cotyledons (Figures 4B and 4D). The same WET368 GUS expression pattern was observed in heart-shaped cuc1 seedlings (data not shown).
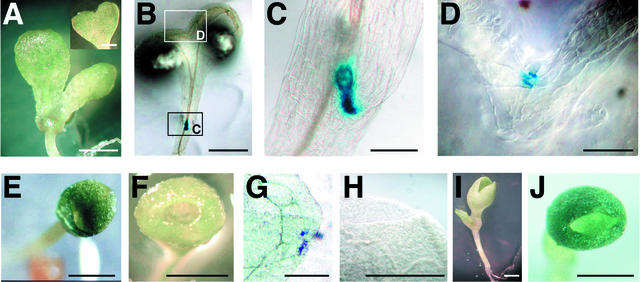
Figure 4.
WET368 GUS Expression in WET368 cuc Mutant Backgrounds.
(A) Heart-shaped cuc2 seedling. The inset shows a heart-shaped cotyledon with more extensive cotyledon fusion.
(B) Heart-shaped WET368 cuc2 seedling after 24 h of GUS staining.
(C) Magnification of section C in (B) containing the SAM and the nonfused boundary of the cotyledons.
(D) Magnification of section D in (B) containing the top of the one-sided fusion of the cotyledons.
(E) s-cuc seedling.
(F) f-cuc seedling.
(G) Split of a cuc1/cuc1 cuc2/cuc2 WET368/+ s-cuc seedling after 6 days of GUS staining.
(H) Flat rim of a cuc1/cuc1 cuc2/cuc2 WET368/WET368 f-cuc seedling after 6 days of GUS staining.
(I) cuc3-2 cup-shaped cotyledon seedling. The leaf at left originated from the SAM.
(J) Cup-shaped cotyledon seedling from the progeny of a cuc1/+ cuc3-2/+ plant.
Bars = 1 mm in (A), (B), (E), (F), (I), and (J) and 250 μm in (C), (D), (G), and (H).
In cuc1 cuc2 double mutant embryos and seedlings, the cotyledons are fused along both sides, resulting in the cup-shaped cotyledon phenotype from which the mutant's name was derived (Aida et al., 1997). The upper region of almost all cuc1 cuc2 cup-shaped cotyledons splits in two, because the cotyledons are not fused over their entire length (Figure 4E). In contrast to the single mutants, cuc1 cuc2 double mutants always lack a SAM. Interestingly, two classes of the cup-shaped cotyledon phenotype could be distinguished among cuc1 cuc2 seedlings carrying the WET368 Ds insertion (see below and Figure 5). One was the previously described cup-shaped cotyledon phenotype, in which the upper region of the cup-shaped cotyledon is split in two, as observed predominantly in cuc1 cuc2 mutant seedlings. We called this phenotype the split cup–shaped cotyledon (s-cuc) phenotype (Figure 4E). A second, novel phenotypic class was represented by seedlings in which the upper region of the cup-shaped cotyledon was not split but entirely flat, without any sign of cotyledon separation. We called this phenotype the fully cup–shaped cotyledon (f-cuc) phenotype, which can be considered a stronger cup-shaped cotyledon phenotype than the s-cuc phenotype (Figure 4F). A similar phenotype also was reported recently in pin-formed1 (pin1) cuc1 cuc2, pin1 cuc1, and pin1 stm mutant seedlings (Aida et al., 2002). In s-cuc seedlings, WET368 GUS was expressed very weakly in a few cells in the depression that partially separates the cotyledons (Figure 4G). By contrast, WET368 GUS expression was never observed in the upper region of fully cup–shaped cotyledons (Figure 4H). Together, these results indicate that CUC3 expression coincides with the boundary between partially or completely separated cotyledons in cuc mutant seedlings. A full loss of cotyledon separation was correlated with a complete loss of WET368 GUS expression along the cup-shaped cotyledon rim. The observation that s-cuc seedlings displayed only weak GUS expression suggests that WET368 enhancer trap activation is reduced in cuc1 cuc2 seedlings. This finding suggests that CUC1 and CUC2 normally stimulate the activation of the CUC3 promoter.
Figure 5.
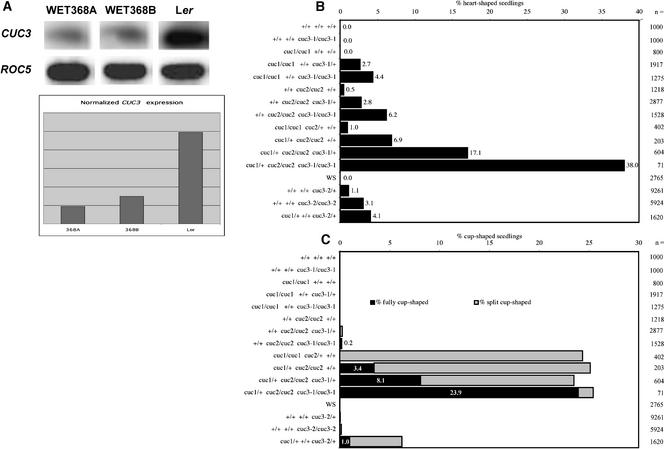
Phenotypic Effects of Reduced CUC3 Expression.
(A) Semiquantitative RT-PCR of CUC3 expression in homozygous WET368A and WET368B plants and in wild-type Ler. ROC5 expression was used to normalize CUC3 expression levels.
(B) and (C) Relative abundance of seedling phenotypes among the progeny of plants with the indicated genotypes of CUC1, CUC2, and CUC3. + indicates wild-type alleles, cuc1 and cuc2 indicate mutant alleles, cuc3-1 indicates the CUC3 hypomorph allele caused by the WET368 Ds insertion, and cuc3-2 indicates the CUC3 mutant allele caused by the T-DNA insertion in exon 2. Numerals beside the bars indicate the total number of seedlings analyzed. WS, wild-type Wassilewskija.
(B) Percentages of heart-shaped seedlings.
(C) Percentages of cup-shaped cotyledon seedlings. f-cuc seedlings are shown in black, and s-cuc seedlings are shown in gray. Data labels indicate the percentages of f-cuc seedlings among the entire progeny.
Phenotypic Effects of Reduced CUC3 Expression
We determined whether CUC3 expression was affected by the DsE insertion in WET368 plants. CUC3 mRNA levels in two individual homozygous WET368 plants, WET368A and WET368B, were compared with that of wild-type Landsberg erecta (Ler) plants using reverse transcriptase–mediated (RT) PCR. Quantitative analysis showed that the CUC3 mRNA level in homozygous WET368 plants was reduced to ∼20% of the wild-type CUC3 mRNA level (Figure 5A). As a result, CUC3 function could be reduced in homozygous WET368 plants, and the WET368 line can be considered a CUC3 hypomorph. We named the CUC3 hypomorph allele cuc3-1. No obvious mutant phenotypes were observed in hemizygous or homozygous cuc3-1 plants. In view of the redundancy between CUC1 and CUC2 (Aida et al., 1997) and the similarities in sequence and expression pattern among CUC3, CUC1, and CUC2, we speculated that the failure to detect obvious phenotypes in cuc3-1 plants could be attributable to redundancy between CUC3 and the CUC1 and CUC2 genes. Therefore, we investigated whether the WET368 Ds insertion had phenotypic effects in cuc1 and cuc2 mutant backgrounds.
We examined the effect of hemizygous or homozygous WET368 Ds insertions (indicated as cuc3-1/+ and cuc3-1/cuc3-1) in cuc1, cuc2, and cuc1/+ cuc2/cuc2 plants on the frequencies of heart-shaped, s-cuc, and f-cuc seedlings among their progeny. Progeny of wild-type Ler, homozygous cuc3-1, cuc1, and cuc2 plants, and cuc1/cuc1 cuc2/+ and cuc1/+ cuc2/cuc2 plants were included in the analysis as references. cuc1 plants homozygous for cuc3-1 produced heart-shaped seedlings at a frequency of 4.4%, whereas we detected no heart-shaped seedlings among the progeny of cuc1 plants (Figure 5B). cuc1 plants hemizygous for cuc3-1 produced an intermediate frequency of heart-shaped seedlings (Figure 5B). Such an intermediate frequency is expected because the progeny of cuc1/cuc1 cuc3-1/+ plants segregates for cuc3-1 in a 1:2:1 ratio (cuc3-1/cuc3-1:cuc3-1/+:+/+). Likewise, cuc2 plants homozygous for cuc3-1 showed a 12-fold increase (0.5 to 6.2%) in the frequency of heart-shaped seedlings compared with cuc2 plants (Figure 5B). cuc2 plants hemizygous for cuc3-1 exhibited an intermediate frequency of heart-shaped seedlings (Figure 5B). A similar effect of homozygous and hemizygous cuc3-1 was seen in the cuc1/+ cuc2/cuc2 mutant background. Here, the frequency of heart-shaped seedlings increased from 6.9 to 17.1 to 38% in the progeny of cuc1/+ cuc2/cuc2 +/+, cuc1/+ cuc2/cuc2 cuc3-1/+, and cuc1/+ cuc2/cuc2 cuc3-1/cuc3-1 plants, respectively (Figure 5B). These data suggest that reduced CUC3 mRNA levels caused an increase in the percentages of heart-shaped seedlings in cuc mutant backgrounds.
To ensure that the observed phenotypic effects were caused by reduced CUC3 expression, we studied the phenotypes of a recently identified Arabidopsis line with a T-DNA insertion in CUC3. This line from the Institut National de la Recherche Agronomique Versailles collection has a single T-DNA insertion (data not shown) in the second exon of CUC3, just before nucleotide 543 of the genomic sequence (Figure 2A). We named this cuc3 insertion allele cuc3-2. No CUC3 mRNA was detected in homozygous cuc3-2 plants by RT-PCR (data not shown), suggesting that cuc3-2 represents a null allele. Among the progeny of homozygous cuc3-2/cuc3-2 plants, 3.1% of the seedlings were heart shaped (Figure 5B). cuc3-2/+ plants produced 1.1% heart-shaped seedlings, whereas wild-type Wassilewskija (Ws) plants produced no heart-shaped seedlings (Figure 5B). The fact that the independent cuc3-2 insertion in CUC3 resulted in phenotypes similar to those found for cuc3-1 corroborates our conclusion that these phenotypes are caused by a reduction in CUC3 expression. Notably, the cuc3-2 mutant displayed a higher frequency of heart-shaped seedlings than the cuc1 and cuc2 single mutants. In addition, in contrast to the cuc1 and cuc2 single mutant plants, cuc3-2/cuc3-2 and cuc3-2/+ plants produced cup-shaped cotyledon seedlings at low frequencies (0.15 and 0.04%, respectively; Figure 5C). These cup-shaped cotyledon seedlings were similar to those found in the cuc1 cuc2 double mutants described above with respect to the cotyledon fusion on both sides, but in contrast to these double mutants, the cuc3-2 cup-shaped cotyledon seedlings always formed a shoot meristem (Figure 4I).
Thus, the cuc3-2 cup-shaped cotyledon phenotype can be considered stronger than the heart-shaped phenotype but weaker than the s-cuc and f-cuc phenotypes. The higher percentage of heart-shaped seedlings and the occurrence of cup-shaped cotyledon seedlings in cuc3-2 single mutants indicate that the cuc3-2 mutant has a somewhat stronger phenotype than the cuc1 and cuc2 mutants. Thus, the contribution of CUC3 to the establishment of the cotyledon boundary appears greater than that of CUC1 and CUC2. Alternatively, because cuc3-2 is in the Ws background and cuc1 and cuc2 are in Ler, there may be greater penetrance of cotyledon boundary phenotypes in the Ws background than in Ler.
cuc1/cuc1 cuc2/+ and cuc1/+ cuc2/cuc2 plants produced 25% cuc1/cuc1 cuc2/cuc2 progeny, recognizable by the cup-shaped cotyledon phenotype. The cuc3-1 mutation did not influence the percentage of cup-shaped cotyledon seedlings among the progeny of cuc1/+ cuc2/cuc2 plants, but the frequency of f-cuc seedlings increased from 3.4 to 23.9% in the presence of a homozygous cuc3-1 mutation (Figure 5C). In the progeny of cuc1/+ cuc2/cuc2 cuc3-1/+ plants, an intermediate frequency of 8.1% f-cuc seedlings was observed (Figure 5C). The observed WET368 GUS expression in the “rim depression” of split cup–shaped cotyledons (Figure 4G) and the increase in the relative frequency of f-cuc seedlings induced by the WET368 Ds insertion suggest that CUC3 function accounts for the partial cotyledon separation in cuc1 cuc2 split cup–shaped cotyledons.
Cup-shaped cotyledon seedlings were observed not only in cuc1 cuc2 double mutant and cuc1 cuc2 cuc3-1 triple mutant backgrounds but also at low frequency among the progeny of cuc2 plants homozygous or hemizygous for cuc3-1 (Figure 5C). These cuc2 cuc3-1 cup-shaped cotyledon seedlings always lacked a SAM, as judged by the seedling morphology and the absence of leaves and inflorescence stems later in development. The low penetrance of cup-shaped cotyledon seedlings in the cuc2 cuc3-1 background may be the result of the 20% residual CUC3 expression even in homozygous cuc3-1 plants. In contrast to cuc2 cuc3-1, cup-shaped cotyledon seedlings were not observed in the cuc1 cuc3-1 background (Figure 5C). To determine whether this was the result of residual CUC3 expression in the cuc3-1 allele, we determined whether cup-shaped cotyledon seedlings occurred in the cuc1 cuc3-2 background. Among the progeny of double heterozygous cuc1/+ cuc3-2/+ plants, 6.2% of the seedlings were cup shaped (Figures 4J and 5C). This percentage represents 1/16th of the seedlings analyzed and corresponds to the expected percentage of the cuc1/cuc1 cuc3-2/cuc3-2 genotype among the seedlings. In contrast to the cup-shaped cotyledon seedlings in the cuc1 cuc2, cuc2 cuc3-1, and cuc1 cuc2 cuc3-1 backgrounds, cuc1 cuc3-2 cup-shaped cotyledon seedlings formed a functional SAM. The majority of cuc1 cuc3-2 cup-shaped cotyledons were split cup shaped (Figure 5C), in agreement with our observation that f-cuc seedlings occur mainly when the function of all three CUC genes is absent or strongly reduced. Besides cup-shaped cotyledon seedlings, cuc1/+ cuc3-2/+ plants produced heart-shaped seedlings at a frequency of 4.1% (Figure 5B).
Therefore, cup-shaped cotyledon seedlings occur in the cuc3-2 single mutant, the cuc1 cuc2, cuc1 cuc3-2, and cuc2 cuc3-1 double mutants, and the cuc1 cuc2 cuc3-1 triple mutant. However, only in double and triple mutant combinations containing the cuc2 mutation (i.e., cuc1 cuc2, cuc2 cuc3-1, and cuc1 cuc2 cuc3-1) did these cup-shaped cotyledon seedlings lack a functional SAM. This observation suggests that among the three cuc mutations, only cuc2 leads to the absence of the SAM, although only combined with mutations in CUC1 and/or CUC3.
In summary, the results of our phenotypic analysis, which included two independent insertional alleles of cuc3, suggest a function of the CUC3 gene in the establishment of the cotyledon boundaries and in shoot meristem formation. Its function appears to be (partially) redundant with the functions of CUC1 and CUC2.
WET368 GUS Expression in Shoot Meristem Mutant and 35S:KNAT1 Plants
Transgenic plants ectopically expressing the Arabidopsis homeobox gene KNAT1 form lobed leaves, and ectopic shoot meristems occasionally develop within the boundaries, or “sinuses,” between the lobes (Chuck et al., 1996). Leaf lobes in 35S:KNAT1 plants have been proposed to result from the formation of ectopic meristem boundaries in the sinus regions (Ori et al., 2000). Because CUC3 is expressed in meristem boundaries, we determined whether the boundaries in 35S:KNAT1 leaves also expressed CUC3. Progeny plants of 35S:KNAT1 crossed to WET368 reliably displayed the leaf phenotype seen in 35S:KNAT1 plants. Cauline leaves of these plants displayed ectopic WET368 GUS expression in all sinuses. This expression preceded occasional ectopic meristem proliferation (Figure 6A). Upon meristem proliferation in a sinus, WET368 GUS expression resolved to the meristem boundary (Figure 6B), analogous to the situation in the proliferating embryonic SAM (Figure 3K). Thus, the appearance of morphologically visible shoot meristems in 35S:KNAT1 leaves was preceded by the establishment of ectopic boundaries expressing CUC3.
Figure 6.
WET368 GUS Expression Patterns in Transgenic and Mutant Backgrounds.
(A) WET368 35S:KNAT1 cauline leaf before ectopic SAM proliferation from its sinuses.
(B) Ectopic SAM in the sinus of a WET368 35S:KNAT1 cauline leaf.
(C) WET368 stm-1 seedling apex at 5 days after germination.
(D) Mature WET368 zll-3 embryo.
(E) WET368 zll-3 seedling at 5 days after germination.
(F) WET368 zll-3 seedling at 10 days after germination with terminal leaf.
(G) WET368 wus-1 seedling at 10 days after germination viewed from the side.
Staining times were 24 h in (A), (B), and (D) to (F) and 3 h in (C) and (G). Bars = 1 mm in (A), (E), and (F) and 50 μm in (B) to (D) and (G).
We tested whether CUC3 expression is influenced by the establishment of a functional shoot meristem by studying WET368 GUS expression in shoot meristem mutants. shoot meristemless-1 (stm-1) seedlings are characterized by the lack of a shoot meristem (Barton and Poethig, 1993). WET368 GUS expression in stm-1 seedlings localized to a narrow region that forms the boundary between the cotyledons, just above the point at which the vascular bundle splits in two (Figure 6C). Expression in the center of the boundary between the cotyledons correlates with the absence of a proliferating SAM in stm-1. Most embryos and seedlings of zwille-3/pinhead (zll-3) and wuschel-1 (wus-1) have a dysfunctional SAM that terminates precociously with or without the formation of one or two terminal leaves in the place normally occupied by the SAM (Laux et al., 1996; Moussian et al., 1998). WET368 GUS expression was not affected in zwille/pinhead embryos (Figure 6D) or seedlings (Figure 6E), except for the persistence of CUC3 expression in the center in the absence of meristem proliferation. Proliferation of a terminal leaf from the dysfunctional SAM in zll-3 seedlings resulted in the exclusion of GUS expression from the central domain, here occupied by the terminal leaf, analogous to the situation in the wild-type seedling apex, where the central domain is occupied by the SAM (Figure 6F). The same pattern of WET368 GUS expression was observed in wus seedlings with a terminating apex (Figure 6G) or a terminal leaf (data not shown). Thus, STM, ZLL, and WUS functions are not required for CUC3 expression, although the absence of a proliferating SAM in stm, zll, and wus correlates with persistent central CUC3 expression. These observations indicate that CUC3 expression is not dependent on the formation of a functional SAM.
DISCUSSION
CUC3 Encodes a Putative NAC-Domain Transcription Factor That Is Involved in Organ and Meristem Boundary Formation
The predicted CUC3 protein contains a putative bipartite nuclear localization signal within the highly conserved N-terminal NAC-domain that is characteristic of the NAC family of transcription factors (Aida et al., 1997; Duval et al., 2002) and a less conserved C-terminal domain. Phylogenetic analysis indicates that CUC3 is most similar to the Arabidopsis CUC1 and CUC2 and the petunia NAM proteins. These proteins also are most similar to CUC3 in expression pattern and function (Souer et al., 1996; Aida et al., 1999; Takada et al., 2001; this study).
During embryogenesis, the CUC3 expression pattern largely overlaps with that of CUC1 and CUC2 (Aida et al., 1999; Takada et al., 2001). However, there are two main differences. First, CUC3 is expressed earlier than both CUC1 and CUC2: CUC3 expression starts at the octant stage, whereas expression of the CUC1 and CUC2 genes was detected only from the globular stage onward. Second, CUC3 expression is strongest in the epidermal cell layer, whereas CUC1 is expressed only weakly in the epidermis and CUC2 is not expressed in the epidermis at all. cuc3, cuc1, and cuc2 mutations have additive phenotypic effects. Together, these results show that, although CUC3, CUC1, and CUC2 function in the same developmental processes, they have unique expression patterns and their functions are only partially redundant.
Besides expression in embryonic boundaries, CUC3 is expressed in a wide variety of boundaries in the seedling and adult plant, generally separating two proliferating organs or a proliferating organ or cell from its surrounding cells. None of the Arabidopsis CUC1 (Takada et al., 2001), Arabidopsis CUC2 (Ishida et al., 2000), and petunia NAM (Souer et al., 1996) genes has been reported to be expressed at the base of lateral roots and around trichomes. Thus, analysis of CUC3 expression reveals an even broader spectrum of boundaries in plants than was thought previously. Trichomes not only differentiate differently but also grow much faster than the surrounding epidermal cells. Thus, also around trichomes, CUC3-expressing cells may represent a boundary that separates a highly proliferative structure from its surroundings. Interestingly, the recently identified LATERAL ORGAN BOUNDARIES (LOB) gene is expressed in a pattern similar to that of CUC3 in the boundaries of lateral organs formed from the SAM and also in a ring of cells at the bases of lateral roots (Shuai et al., 2002). However, LOB expression commences later and is less localized during embryogenesis, because it was first detected in all cells of the torpedo-stage embryo.
The Identification of CUC3 Reveals a Higher Degree of Redundancy among CUC Genes in Boundary and SAM Formation
cuc3-1 plants did not show any obvious phenotypes, whereas cuc3-2 plants produced heart-shaped seedlings at low frequency. This finding indicates that the residual CUC3 activity in cuc3-1 plants was sufficient for normal development. The phenotypic analysis of cuc3-2 corroborates our conclusion that the enhancement of cuc1 and cuc2 phenotypes by cuc3-1 was caused by a reduction in CUC3 function. The greater frequency of heart- and cup-shaped cotyledon seedlings in the cuc3-2 single mutant, compared with cuc1 and cuc2, suggests a greater contribution of CUC3 to the establishment of the cotyledon boundary compared with CUC1 and CUC2. With respect to SAM formation, only cup-shaped cotyledon seedlings in double and triple mutant backgrounds containing the cuc2 mutation lacked a functional shoot meristem. This finding indicates a greater contribution of CUC2 to SAM formation compared with CUC1 and CUC3. Because all cuc2 single mutant seedlings form a functional SAM, a mutation in CUC2 alone is not sufficient to abolish SAM formation. Only in combination with cuc1 and/or cuc3 does a mutation in CUC2 lead to the absence of a functional SAM.
Our results show that CUC3 function is partially redundant with that of CUC1 and CUC2. Thus, the identification of CUC3 suggests an even greater degree of redundancy among CUC genes than was thought previously on the basis of CUC1 and CUC2. Bearing in mind the variety of boundaries that are marked by CUC3 expression, CUC3 also most likely is involved in boundary establishment and/or maintenance in adult plants, whether or not in conjunction with CUC1 and CUC2. Indeed, preliminary phenotypic observations suggest a role for the CUC3 gene in the establishment of boundaries in adult plants as well (data not shown). Our and previous studies (Aida et al., 1997, 1999; Takada et al., 2001) suggest that the extent to which the normal developmental program of boundary and SAM formation is executed is determined by the collective activities of the CUC1, CUC2, and CUC3 gene products. These gene products could act independently to provide threshold levels sufficient for normal development. Alternatively, in cells in which all three CUC proteins are present, the CUC1, CUC2, and CUC3 gene products may be part of a multivalent transcription factor complex that is fully active only if all components are present or in which the reduction or absence of individual components can be partially complemented by the remaining components. This scenario is similar to the situation in the control of Antirrhinum floral architecture. Ternary complexes of the MADS-box transcription factors SQUAMOSA (SQUA), DEFICIENS (DEF), and GLOBOSA (GLO) have a higher target DNA binding affinity than the DEF/GLO heterodimers or SQUA/SQUA homodimers, whereas target gene activation is lost completely in the absence of all three transcription factors (Egea-Cortines et al., 1999).
Relation of CUC3 to Other Genes Involved in Shoot Meristem Formation
Our observation that WET368 GUS expression does not require the activities of the STM, WUS, CUC1, and CUC2 genes is in agreement with the fact that the expression of all of these genes is detected later than that of CUC3 (Long and Barton, 1998; Mayer et al., 1998; Aida et al., 1999; Takada et al., 2001). Only ZLL expression precedes CUC3 expression, but according to our results, it is not required for CUC3 expression. Although STM is not required for CUC3 expression per se, it does influence its expression level and pattern. At early stages of embryogenesis, STM expression overlaps with CUC3 expression, although STM expression is initiated later (Long and Barton, 1998; Aida et al., 1999). By contrast, late in embryogenesis, STM expression appears to be complementary to that of CUC3, because STM expression resolves to the SAM while it disappears from the boundary of the cotyledon margins. In the absence of STM function, CUC3 expression is maintained in the center of the cotyledon boundary, from which it is excluded in wild-type embryos upon the proliferation of the SAM. Thus, during normal development, either STM is directly involved in the downregulation of CUC3 expression in the presumptive SAM or the downregulation of CUC3 is an indirect consequence of STM-mediated SAM proliferation.
Boundaries and the Establishment of Shoot Meristems
CUC3 is expressed in a wide variety of plant organs, yet only in a restricted number of cells. What do cells that express CUC3 have in common? CUC3-expressing cells are located in a boundary that separates two proliferating organs or a proliferating organ or cell from its surrounding cells. CUC3-expressing cells in the embryo also share this common characteristic, in that they form the boundary between the (presumptive) cotyledons. Reduction of CUC1, CUC2, and/or CUC3 function results in the incomplete establishment of this boundary. Based on these observations, we propose that CUC3 (and CUC1 and CUC2) functions primarily in the establishment and/or maintenance of boundaries at various stages of plant development. Because boundaries contain cells with restricted proliferation and/or differentiation rates, the primary function of CUC3 may be the restriction of cellular proliferation and/or differentiation.
CUC3 expression precedes SAM formation in the center of the embryo apex, and reduction of CUC3, CUC1, and CUC2 function affects not only the cotyledon boundary but also the formation of the SAM. How is the establishment of the SAM affected by the CUC3, CUC1, and CUC2 genes? The study of transgenic plants that induce ectopic shoot meristems may provide clues to answer this question. First, plants that ectopically express the homeobox gene KNAT1 occasionally form ectopic shoot meristems on leaf surfaces (Chuck et al., 1996). These meristems are not distributed randomly over the leaf surface but correlate strictly to ectopic boundaries that give the leaves their lobed appearance. Expression of KNAT1 and KNAT2 in these boundaries has given rise to the idea that the lobed-leaf phenotype results from the establishment of ectopic meristem boundaries in leaves (Ori et al., 2000). We found that CUC3 is expressed in the ectopic leaf boundaries as well, and given its function in the establishment of the cotyledon boundary during embryo and seedling development, CUC3 may be instrumental in the establishment of boundaries in 35S:KNAT1 leaves. In those sinuses in which ectopic meristems are formed, CUC3 expression precedes morphologically visible meristem formation, as it does during the establishment of the embryonic shoot meristem. Hence, CUC3 expression and boundary formation appear to precede shoot meristem formation in 35S:KNAT1 leaves.
Second, 35S:CUC1 plants form lobed cotyledons. The boundaries that separate these lobes contain cells that are smaller than normal cotyledon cells (Takada et al., 2001), and ectopic shoot meristems are induced from cells within these boundaries. Thus, ectopic shoot meristems in 35S:KNAT1 and 35S:CUC1 backgrounds are localized to ectopic boundaries. This situation is similar to the developmental context in which the shoot meristem is established during normal embryogenesis. Our results suggest that the establishment of the boundary between the presumptive cotyledons represents one of the first patterning events that lead to shoot meristem development in the embryo. The cells within this boundary are marked by CUC3 (and later CUC1, CUC2, and STM) expression (Long and Barton, 1998; Aida et al., 1999; Takada et al., 2001) and are characterized by low proliferation compared with the surrounding cells, which are destined to develop into cotyledons (Barton and Poethig, 1993). The SAM appears within the boundary, and failure to correctly establish the boundary correlates with failure to establish the SAM in plants with reduced CUC3, CUC1, CUC2, and/or STM function. Interestingly, tobacco seedlings with fused cotyledons resulting from the overexpression of phytochrome A also often lack a SAM (Emmler and Schäfer, 1997). Thus, the establishment of a boundary that contains cells with low proliferation and/or differentiation rates apparently is required for the establishment and/or maintenance of the shoot meristem.
The relatively undifferentiated cells in the boundary between the two presumptive cotyledons may provide a favorable niche for the establishment and/or maintenance of a population of stem cells. In animals, the maintenance of stem cells is dependent on the local environment, resulting in the concept of stem cell niches (Watt and Hogan, 2000; Weigel and Jürgens, 2002). Secondary inflorescence meristems in Arabidopsis arise in the adaxial boundaries between cauline leaves and the primary stem, which also express CUC3. Although the CUC and STM genes, according to this idea, are required for the establishment and/or maintenance of a niche from which the SAM is initiated and maintained, they are not the only genes involved. For example, studies with the ZWILLE/PINHEAD gene (Moussian et al., 1998; Lynn et al., 1999), with members of the YABBY gene family (Siegfried et al., 1999), and with the phabulosa-1d mutant (McConnell and Barton, 1998) revealed the existence of signals from the adaxial sides of cotyledons and leaves that promote SAM initiation and/or maintenance.
How general is the proposed requirement for CUC transcription factor activity in the establishment of meristems in Arabidopsis? Although primary and secondary shoot meristems all arise from cellular regions that express CUC3, the root meristem presents a different case. None of the CUC genes is expressed in cells from which the root meristem is initiated, and reduced activity of these genes does not affect the establishment and/or maintenance of a functional root meristem (Aida et al., 1997, 1999; Takada et al., 2001; this study). A fundamental difference between the shoot and root meristems is that the former is established in the bilaterally symmetric apical part of the embryo, whereas the latter is initiated and maintained in a radially symmetric architecture (Scheres et al., 1996). In view of the possible dependence of embryonic SAM formation on the establishment of the cotyledon boundary, and thus on bilateral symmetry, it is not surprising that root meristem initiation and maintenance are governed by different regulatory networks. None of the key regulators of shoot meristem initiation described to date affects the root meristem, and vice versa (for reviews, see Scheres and Wolkenfelt, 1998; Vroemen and De Vries, 1998; Haecker and Laux, 2001). Thus, differences between meristem regulatory networks in shoots and roots may emanate from or underlie fundamental differences in local plant architecture.
METHODS
Plant Strains and Growth Conditions
The Wageningen Enhancer Trap (WET) 368 line was identified from an enhancer trap transposon-tagging screen that has been described elsewhere (Sundaresan et al., 1995; Vroemen et al., 1998). Line WET368 was obtained from a cross between starter lines Ac2 and DsE3. The wild-type and all mutant and transgenic lines were in the Landsberg erecta (Ler) background, except for the 35S:KNAT1 line, which was in the Nossen background, and the Versailles T-DNA line cuc3-2, which was in the Wassilewskija background. Plants were grown in a growth chamber at 22°C with a 16-h daylength. Homozygous WET368 plants were crossed to plants heterozygous for stm-1 or wus-1 or homozygous for zll-3, pt-1, cuc1, cuc2, or 35S:KNAT1.
β-Glucuronidase Staining
β-Glucuronidase staining was performed as described previously (Vroemen et al., 1996). The duration of staining varied from 3 h to 6 days. Photographs were taken through a Nikon binocular or a Nikon Optiphot-2 microscope with Normarski optics (Tokyo, Japan). Photographs were taken on Ektachrom 16T film (Kodak) or using a Nikon Coolpix 990 digital camera.
Cloning of the CUC3 Gene
Genomic DNA was isolated according to Bouchez et al. (1996). DNA flanking the DsE element in WET368 was amplified by thermal asymmetric interlaced PCR according to Parinov et al. (1999). A set of three nested primers for the 5′ end of Ds, Ds5-1, Ds5-2, and Ds5-3 (Grossniklaus et al., 1998), was used in combination with arbitrary primers AD1 and AD4 (Liu et al., 1995). Ten nanograms of genomic DNA was used as a substrate. After three rounds of nested amplification, reaction products were separated on a 3% (w/v) agarose gel. Secondary and tertiary reaction products were purified using the High Pure PCR Product Purification Kit (Roche, Indianapolis, IN) and cloned into the pGEM-T vector (Promega). DNA sequences were determined using an ABI 373A (Applied Biosystems, Foster City, CA) automatic sequencer using the DYEnamic ET Terminator Cycle Sequencing Kit (Amersham Pharmacia). The product obtained from reactions with AD4 was 204 bp, and from reactions with AD1 two products were obtained, one of 287 bp and one of 604 bp.
The 604-bp product was used to screen 6 × 104 clones (nine genome equivalents) of an Arabidopsis Ler genomic library in λ FIX II (Stratagene) according to Sambrook et al. (1989). The insert of a positive phage was subcloned into pGEM-4Z (Promega) and partially sequenced. Basic Local Alignment Search Tool (BLAST) searches against The Arabidopsis Information Resource (TAIR) database (www.arabidopsis.org) identified two overlapping BAC sequences, F14G6 and F15M4. GENSCAN 1.0 (genes.mit.edu/GENSCAN.html) analysis of the DNA sequence identified a single open reading frame close to the DsE insertion. BLAST searches at TAIR or the National Center for Biotechnology Information identified this open reading frame as a member of the NAC transcription factor gene family; therefore, we named it NAC368. Based on the functional data presented here, the NAC368 gene was renamed CUP-SHAPED COTYLEDON3 (CUC3).
PCR primers were designed to generate nine overlapping fragments covering the entire CUC3 gene. These primers were as follows: MKnac1 (5′-GGGTGATTTGCAGAGTGTT-3′), MKnac2 (5′-CTAGTAGCATGTGAAGAG-3′), MKnac3 (5′-CCTCGAAAACGACCATTC-3′), MKnac4 (5′-CATCACCTTTTGTATCATTGC-3′), MKnac5 (5′-TAACTTTTAAGAAATTTGTTCG-3′), nacf1 (5′-GAAGAAAGGAACGAGAGAGG-3′), nacr2 (5′-ACGTGTGGCGGTGTGAATGG-3′), and nacr3 (5′-GAAGGTTCTATAAGCAAAGG-3′). PCR products were cloned into pGEM-T and sequenced, resulting in an average threefold sequence coverage of the CUC3 gene. CUC3 cDNA was obtained by reverse transcriptase–mediated (RT) PCR using primers nac368cDNA5 (5′-GTTATATTAAGTAAAAGATGATGC-3′) and nac368cDNA3 (5′-CAAGGGCCAAGATTCTACAGC-3′). Sequence analyses and alignments were performed using DNASTAR (Madison, WI). Phylogenetic trees and multiple protein alignments were constructed using CLUSTAL X (1.64b).
RT-PCR Analysis
Total RNA was isolated from mixed flower buds, flowers, and siliques using TRIzol (Invitrogen, Carlsbad, CA). RT-PCR was performed according to Albrecht et al. (1998). Parallel reactions in which the RT step was omitted were used as negative controls. The constitutively expressed cyclophilin gene ROC5 was used as a normalization control (Chou and Gasser, 1997). Linearity of PCR was determined at 30 cycles using undiluted and 4-, 16-, and 64-fold dilutions of the first-strand cDNA as templates. The primers were nacf1 and nacr3 for CUC3 and roc5-5 (5′-TCTCTCTTCCAAATCTCC-3′) and roc5-3 (5′-AAGTCTCTCACTTTCTCACT-3′) for ROC5. Hybridization signals were detected with a Storm 840 PhosphorImager and quantified using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
In Situ Hybridization
In situ hybridization was performed according to Mayer et al. (1998). Two templates were used for CUC3 antisense and sense probes. nacex3 is a 487-bp DNA fragment from the third exon amplified from genomic DNA using MKnac1 and MKnac2 primers, and nacgen13 is a 1992-bp DNA fragment spanning the entire genomic sequence amplified from genomic DNA using MKnac4 and MKnac5 primers. Both probes gave the same qualitative and quantitative results.
Phenotypic Analyses
Homozygous WET368 plants were crossed to homozygous cuc1 or cuc2 plants, and F2 plants homozygous for cuc1 or cuc2 were selected. The cuc1 mutation was detected by cleaved amplified polymorphic sequence analysis, and the cuc2 mutation was detected by PCR genotyping of individual seedlings as described by Takada et al. (2001). The cuc3-1 genotype of F2 plants was determined by plating F3 seeds on Murashige and Skoog (1962)–kanamycin plates. Seeds from cuc1/cuc1 cuc3-1/cuc3-1, cuc1/cuc1 cuc3-1/+, cuc2/cuc2 cuc3-1/cuc3-1, and cuc2/cuc2 cuc3-1/+ parents were plated, and the frequency of heart-shaped, split cup–shaped, and fully cup–shaped cotyledon seedlings was determined. cuc3-1 plants in the cuc1 cuc2 background were obtained by crossing cuc1/cuc1 cuc2/+ plants to cuc2/cuc2 cuc3-1/cuc3-1 plants. F2 seeds were selected on kanamycin, and the genotypes of the cuc1, cuc2, and cuc3-1 loci were determined. F3 seeds were analyzed phenotypically as described above. Seeds from cuc3-2/cuc3-2 and cuc3-2/+ parents were plated and analyzed phenotypically as described above. cuc3-2/cuc3-2 plants were crossed to homozygous cuc1 plants, and the progeny of the resulting double heterozygous F1 plants were analyzed phenotypically as described above.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The sequence of CUC3 has been deposited in GenBank with accession number AF543194. The accession numbers for the two overlapping BACs are AC015450 (F14G6) and AC012394 (F15M4). The accession numbers for the proteins listed in Figure 2B are as follows: Arabidopsis CUC2 (BAA19529), CUC1 (BAB20598), AtNAC1 (AAF21437), ATAF1 (derived from X74755), ATAF2 (derived from X74756), AtNAM (AAD17314), NAP (CAA10955), and TIP (AAF87300); petunia NAM (X92205); wheat GRAB1 (CAA09371) and GRAB2 (CAA09372); and tomato SENU5 (T07182).
Acknowledgments
We thank Masao Tasaka, Mitsuhiro Aida, and Ken-ichiro Hibara for a gift of cuc1 and cuc2 seeds and for helpful discussions; Sarah Hake for 35S:KNAT1 seeds; and Matthieu Simon and Françoise St. Drenant for the Institut National de la Recherche Agronomique Versailles T-DNA line representing cuc3-2. Thanks to Rob Martienssen, Venkatesan Sundaresan, and Patty Springer for sharing the Cold Spring Harbor enhancer trap system before publication; to Achim Haecker and Thomas Laux for their help with in situ hybridization; and to Boudewijn van Veen for assistance with artwork. Kerstin Gühl, Said Hussin, Ahmed A. Ahmed, Peter Uijtdewilligen, Leonie Smeenk, Rikkert Seele, and Karin Boer are thanked for technical assistance; Megan Griffith, Henk Franssen, Ton Bisseling, Vered Raz, and Annemarie Lekkerkerker are thanked for comments on the manuscript; and Maarten Koornneef and Hans de Jong are thanked for advice concerning phenotypic analyses. This work was supported by grants from Wageningen University, the European Union Biotechnology Program (ERBIO4-CT96-0689), and the Dutch Organization for Scientific Research.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012203.
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–1570. [DOI] [PubMed] [Google Scholar]
- Aida, M., Vernoux, T., Furutani, M., Traas, J., and Tasaka, M. (2002). Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129, 3965–3974. [DOI] [PubMed] [Google Scholar]
- Albrecht, C., Geurts, R., Lapeyrie, F., and Bisseling, T. (1998). Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of early nodulin genes PsENOD5 and PsENOD12A. Plant J. 15, 605–614. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome se-quence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119, 823–831. [Google Scholar]
- Bouchez, D., Vittorioso, P., Courtial, B., and Camilleri, C. (1996). Kanamycin rescue: A simple technique for the recovery of T-DNA flanking sequences. Plant Mol. Biol. Rep. 14, 115–123. [Google Scholar]
- Callos, J.D., and Medford, J.I. (1994). Organ positions and pattern formation in the shoot apex. Plant J. 6, 1–7. [Google Scholar]
- Chou, I.T., and Gasser, C.S. (1997). Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol. Biol. 35, 873–892. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval, M., Hsieh, T.-F., Kim, S.Y., and Thomas, T.L. (2002). Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50, 237–248. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmler, K., and Schäfer, E. (1997). Maternal effect on embryogenesis in tobacco overexpressing rice phytochrome A. Bot. Acta 110, 1–8. [Google Scholar]
- Furner, I.J. (1996). Cell fate in the development of the Arabidopsis flower. Plant J. 10, 645–654. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Haecker, A., and Laux, T. (2001). Cell-cell signaling in the shoot meristem. Curr. Opin. Plant Biol. 4, 441–446. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Misera, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Ishida, T., Aida, M., Takada, S., and Tasaka, M. (2000). Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 41, 60–67. [DOI] [PubMed] [Google Scholar]
- Jürgens, G., Mayer, U., Torres-Ruiz, R.A., Berleth, T., and Miséra, S. (1991). Genetic analysis of pattern formation in the Arabidopsis embryo. Development 1 (suppl.), 27.–38. [Google Scholar]
- Kikuchi, K., Ueguchi-Tanaka, M., Yoshida, K.T., Nagato, Y., Matsusoka, M., and Hirano, H.-Y. (2000). Molecular analysis of the NAC gene family in rice. Mol. Gen. Genet. 262, 1047–1051. [DOI] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F.X., Berger, J., and Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Liu, Y.-G., Mitsukawa, N., and Oosumi, T. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125, 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126, 469–481. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F.X., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- Mordhorst, A.P., Voerman, K.J., Hartog, M.V., Meijer, E.A., van Went, J., Koornneef, M., and de Vries, S.C. (1998). Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian, B., Schoof, H., Haecker, A., Jürgens, G., and Laux, T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17, 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Parinov, S., Sevugan, M., Ye, D., Yang, W.-C., Kumaran, M., and Sundaresan, V. (1999). Analysis of flanking sequences from Dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, R. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Scheres, B., Mckhann, H.I., and Vandenberg, C. (1996). Roots redefined: Anatomical and genetic analysis of root development. Plant Physiol. 111, 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres, B., and Wolkenfelt, H. (1998). The Arabidopsis root as a model to study plant development. Plant Physiol. Biochem. 36, 21–32. [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F.X., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Peña, C.G., and Springer, P.S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY family specify abaxial fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The No Apical Meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis thaliana regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Vroemen, C.W., Aarts, N., In der Rieden, P.M.J., Van Kammen, A., and De Vries, S.C. (1998). Identification of genes expressed during Arabidopsis thaliana embryogenesis using enhancer trap and gene trap Ds-transposons. In Cellular Integration of Signalling Pathways in Plant Development, Vol. 104, F. Lo Schiavo, R.L. Last, G. Morelli, and N.V. Raikhel, eds (Berlin: Springer-Verlag), pp. 207–232.
- Vroemen, C.W., and De Vries, S.C. (1998). Flowering plant embryogenesis. In Development: Genetics, Epigenetics and Environmental Regulation, E. Russo, D. Cove, L. Edgar, R. Jaenisch, and F. Salamini, eds (Heidelberg, Germany: Springer-Verlag), pp. 121–132.
- Vroemen, C.W., Langeveld, S., Mayer, U., Ripper, G., Jürgens, G., Van Kammen, A., and De Vries, S.C. (1996). Pattern formation in the Arabidopsis embryo revealed by position-specific lipid transfer protein gene expression. Plant Cell 8, 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt, F.M., and Hogan, B.L.M. (2000). Out of Eden: Stem cells and their niches. Science 287, 1427–1430. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Jürgens, G. (2002). Stem cells that make stems. Nature 415, 751–754. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.-H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]