Abstract
Heterotrimeric G proteins are implicated in diverse signaling processes in plants, but the molecular mechanisms of their function are largely unknown. Finding G protein effectors and regulatory proteins can help in understanding the roles of these signal transduction proteins in plants. A yeast two-hybrid screen was performed to search for proteins that interact with Arabidopsis G protein α-subunit (GPA1). One of the identified GPA1-interacting proteins is the cupin-domain protein AtPirin1. Pirin is a recently defined protein found because of its ability to interact with a CCAAT box binding transcription factor. The GPA1–AtPirin1 interaction was confirmed in an in vitro binding assay. We characterized two atpirin1 T-DNA insertional mutants and established that they display a set of phenotypes similar to those of gpa1 mutants, including reduced germination levels in the absence of stratification and an abscisic acid–imposed delay in germination and early seedling development. These data indicate that AtPirin1 likely functions immediately downstream of GPA1 in regulating seed germination and early seedling development.
INTRODUCTION
Heterotrimeric G protein–mediated signal transduction is one of the most conserved eukaryotic signaling mechanisms and is responsible for the transmission of extracellular signals to intracellular effectors. Heterotrimeric guanine nucleotide binding proteins (G proteins) belong to the superfamily of regulatory GTP hydrolases (Kaziro et al., 1991). G proteins consist of three subunits: α (Gα), β (Gβ), and γ (Gγ). In its inactive state, the Gα-subunit is bound to a GDP molecule and is associated with a Gβγ-subunit. Upon activation by a G protein–coupled receptor, the Gα-subunit exchanges GDP for GTP and dissociates from the Gβγ-subunit. As a result, the Gα- and Gβγ-subunits are free to transduce the signal to intracellular effectors (reviewed by Sprang, 1997; Hamm, 1998). In animal systems, effector molecules include adenylyl and guanylyl cyclases, phosphodiesterases, phospholipases, and phosphoinositide 3-kinases (reviewed by Spiegel et al., 1994; Marinissen and Gutkind, 2001). Little is known about plant G protein effectors. Recently, a small GTPase Rac1 was implicated in the downstream functioning of the rice Gα-subunit during the plant's pathogen response (Suharsono et al., 2002).
In contrast to animals, in which multiple α-, β-, and γ-subunits have been found, plant G protein subunits are encoded by single- or double-copy genes. Two Gα-subunit–encoding genes were found in pea (Marsh and Kaufman, 1999), tobacco (Saalbach et al., 1999; Ando et al., 2000), and soybean (Kim et al., 1995; Gotor et al., 1996). The Arabidopsis genome appears to contain only one gene that encodes a canonical Gα-subunit (Ma et al., 1990), one gene that encodes a Gβ-subunit (Weiss et al., 1994), and two genes that encode Gγ-subunits (Mason and Botella, 2000, 2001). Nevertheless, a number of largely pharmacological studies have suggested plant G protein involvement in a variety of processes, including blue light–mediated responses (Warpeha et al., 1991), red light–mediated responses (Romero and Lam, 1993), abscisic acid (ABA) and gibberellin signaling, regulation of ion channels, and pathogen resistance (reviewed by Assmann, 2002).
Recently, two independent T-DNA insertions that are likely to be null mutations were found in the Arabidopsis Gα-subunit–encoding gene GPA1 (Ullah et al., 2001). Analysis of these gpa1 mutants revealed that they have reduced cell division during hypocotyl and leaf formation (Ullah et al., 2001). Additionally, gpa1 mutants are impaired in the inhibition of stomatal opening by ABA (Wang et al., 2001). The gpa1 mutant seeds appear to be hypersensitive to sugars, hyposensitive to gibberellic acid, and insensitive to brassinosteroids. Furthermore, gpa1 mutant seeds exhibit less than wild-type germination levels in the presence of exogenous ABA (Ullah et al., 2002). Multiple phenotypes of gpa1 mutants suggest that GPA1 is an integration point of several signaling pathways. It is likely that GPA1 executes its diverse functions by interacting with a variety of regulatory and effector proteins; however, no GPA1-interacting proteins have been reported to date.
To further investigate the mechanism of Gα-subunit function in Arabidopsis, we performed a yeast two-hybrid screen to identify its interacting partners. One of the GPA1-interacting proteins found in the screen was AtPirin1, an ortholog of the human pirin protein. Pirin is a recently defined protein found as a result of its ability to interact with the CCAAT box binding transcription factor NFI/CTF1 (Wendler et al., 1997). We have established that AtPirin1 interacts specifically with GPA1 in yeast as well as in vitro. AtPirin1 transcript levels were found to be upregulated by ABA and low-fluence red light but not by low-fluence blue light. Two Arabidopsis mutants harboring T-DNA insertions in the AtPirin1 gene were characterized. It was observed that insertions in the AtPirin1 gene cause reduced levels of seed germination in the absence of stratification and that atpirin1 mutant seed germination is hypersensitive to exogenous ABA in a manner similar to that of gpa1 mutants. Additionally, we observed that atpirin1 mutant plants initiate primary shoots and flower earlier than wild-type plants.
RESULTS
The Arabidopsis Gα-Subunit Interacts with AtPirin1 in the Yeast Two-Hybrid Screen
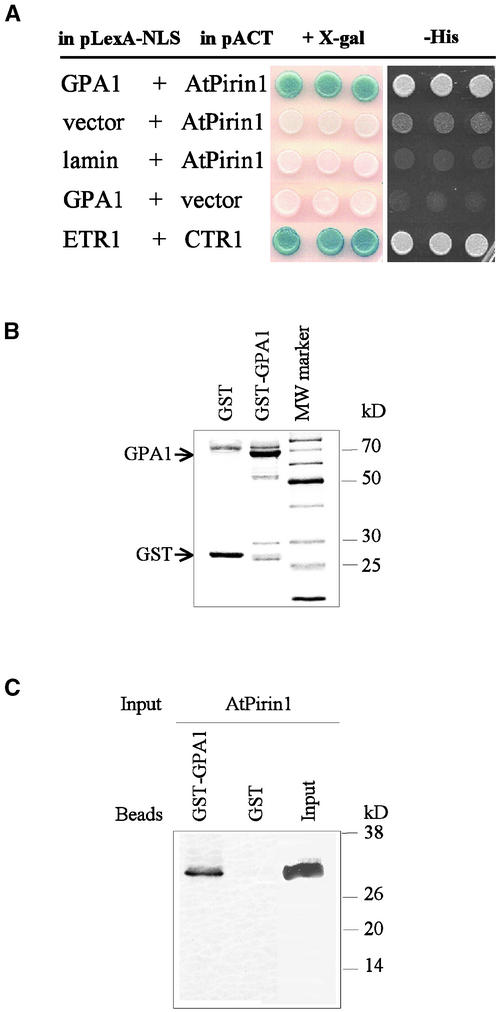
To identify potential components of G protein–mediated signaling pathways in plants, we performed a yeast two-hybrid screen of the Arabidopsis seedling library using a full-length Arabidopsis Gα-subunit (GPA1) as “bait.” Screening of ∼1.9 × 106 transformants yielded 16 positive clones that specifically activate His3 and LacZ reporter genes. Restriction pattern comparison indicated that four of these positive clones contained partial cDNAs of the same gene. Sequence analysis revealed a protein homologous with human pirin (Wendler et al., 1997). Therefore, we named this GPA1-interacting protein AtPirin1. The remaining GPA1-interacting clones will be described elsewhere. AtPirin1 clones caused specific activation of His3 and LacZ reporter genes and did not interact with the lamin protein used as a control bait (Figure 1A). The interaction between the Arabidopsis proteins ETR1 and CTR1 (Clark et al., 1998) served as a positive control for the assay (Figure 1A). The two shortest GPA1-interacting AtPirin1 cDNA clones corresponded to the 170 C-terminal amino acids of the AtPirin1 protein, indicating that the first 117 N-terminal amino acid residues are not critical for the interaction with GPA1 (Figure 2A). The two longest interacting AtPirin1 cDNA clones lack the 64 N-terminal amino acid residues (Figure 2A).
Figure 1.
Interaction between AtPirin1 and GPA1.
(A) Reconstitution of the yeast two-hybrid interaction between AtPirin1 and GPA1. The indicated combinations of the bait (pLexA-NLS) and prey (pACT) constructs were transformed into the yeast reporter strain. Transformants were examined for β-galactosidase activity in the presence of 5-bromo-4-chloro-β-d-galactoside (+X-gal) and for growth in the absence of His (−His).
(B) Coomassie blue staining of the GST-GPA1 fusion protein and GST (positions of the GST-GPA1 and GST proteins are indicated by arrows). MW, molecular mass.
(C) In vitro protein interaction between AtPirin1 and GPA1. GST-GPA1 was overexpressed in E. coli, immobilized on glutathione-Sepharose 4B beads, and incubated with 35S-labeled AtPirin1 protein obtained by coupled in vitro transcription/translation. AtPirin1 binds to GST-GPA1 but not to GST alone.
Figure 2.
AtPirin1 Amino Acid Sequence Analysis, AtPirin1 Gene Structure, and Locations of the T-DNA Insertions.
(A) Alignment of the deduced amino acid sequences of AtPirin1, AtPirin2, AtPirin3, AtPirin4, tomato pirin (TOMpirin), and human pirin (HUMpirin). The alignment was generated using the CLUSTAL X program with default parameters. Identical amino acids are highlighted with a black background, and similar residues are highlighted with a gray background. Regions corresponding to motifs 1 and 2 of the cupin domain are indicated. The single and double asterisks mark residues corresponding to the beginning of the longest and shortest GPA1-interacting AtPirin1 clones, respectively, identified in the two-hybrid screen. Arrows indicate the positions of T-DNA insertions in the atpirin1-1 and atpirin1-2 mutants.
(B) Scheme of the AtPirin1 gene. Relative positions of potential cis elements mentioned in the text are shown. The positions of the atpirin1-1 and atpirin1-2 T-DNA insertions are indicated. Boxes represent exons, and motifs 1 and 2 of the cupin domain are marked by vertical and horizontal hatching, respectively.
AtPirin1 Interacts with GPA1 in Vitro
To confirm that AtPirin1 binds directly to GPA1, we studied their interactions using an in vitro binding assay. For these experiments, the GPA1 coding region was overexpressed as a glutathione S-transferase (GST) fusion in Escherichia coli and immobilized on glutathione-Sepharose beads. Beads coated with GST alone served as a control (Figure 1B). In vitro binding assays were performed with AtPirin1 obtained by coupled in vitro transcription/translation. After multiple washes, bound proteins were separated on SDS-containing polyacrylamide gels. Autoradiographic detection revealed that AtPirin1 interacts with GPA1 and not with GST alone (Figure 1C).
Additionally, we tested whether AtPirin1 interaction with GPA1 depends on the GPA1 activation state. For this analysis, we examined the in vitro activation of GPA1 by incubation with GTPγS (a nonhydrolyzable GTP analog) before the binding assay. In this experiment, AtPirin1 displayed no significant preference for either the active or the inactive form of GPA1 (data not shown).
AtPirin1 Is a Conserved Protein Belonging to the Cupin Superfamily
Conceptual translation of AtPirin1 cDNA revealed that it encodes a 287–amino acid protein. Database searches identified several more pirin genes in the Arabidopsis genome. AtPirin1 is closely related to two of its family members, AtPirin2 and AtPirin3, with 66% identity and 75% amino acid similarity (Figure 2A). Sequence alignments revealed that pirin is present in mammals, plants, and a variety of prokaryotic species, whereas there are no putative pirin orthologs in three model eukaryotic species with fully sequenced genomes: Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster.
Pirin does not appear to have any known functional domains; however, it contains a two-motif sequence characteristic of the cupin domain (Figure 2A). Based on the three-dimensional structure of several cupin proteins, it was established that the two-motif cupin domain contains residues implicated in forming a β-barrel fold and in metal binding (Gane et al., 1998; Dunwell et al., 2000). The β-barrel fold also is associated with the high levels of thermal stability of cupin superfamily members (Dunwell et al., 2001). The superfamily of cupin proteins is the most functionally diverse of any family described to date and includes enzymes, transcription factors, seed storage proteins, auxin binding proteins, stress-related proteins (spherulins and germin-like proteins), and other proteins (Dunwell et al., 2000, 2001).
The fact that the two shortest GPA1-interacting Atpirin1 cDNA clones found in the two-hybrid screen do not contain the cupin domain (Figure 2A) indicates that the cupin domain is not required for interaction with the Gα-subunit.
AtPirin1 mRNA Levels Are Regulated by Abscisic Acid and Red Light
Sequence analysis of the 5′ regulatory region of AtPirin1 revealed the presence of several potential cis-acting regulatory elements, including two ACGT elements and one TT motif at 308, 430, and 512 bp upstream of the AtPirin1 ATG codon (Figure 2B). ACGT elements usually are defined as 8- to 10-bp sequences with the core sequence ACGT (Busk and Pages, 1998; Leung and Giraudat, 1998); they include the ABRE-like elements (abscisic acid–responsive elements) and the G-box elements. ABREs are found in numerous promoters of ABA- and stress-inducible genes (Leung and Giraudat, 1998). The strong ABRE (CACGTG), found 430 bp upstream of the AtPirin1 gene ATG codon, is identical to the core sequence of the G-box element. G boxes are found in promoters of several light-regulated genes, such as CAB, RBCS, and CHS (Busk and Pages, 1998).
In contrast to the ACGT elements, little is known about the TT motif. The TT motif, also called the TCGT motif (TTTCGTGT), was found originally in the distal promoter region of the desiccation- and ABA-inducible Dc3 gene of carrot and appears to be involved in ABA-induced gene expression in vegetative tissues (Busk and Pages, 1998).
The finding of potential cis-regulatory elements in the promoter of the AtPirin1 gene prompted us to test the effects of ABA and different light conditions on AtPirin1 transcript levels. RNA gel blot analysis of poly(A)+ RNA did not reveal any transcript; therefore, we performed relative quantitative reverse transcription (RT)–PCR.
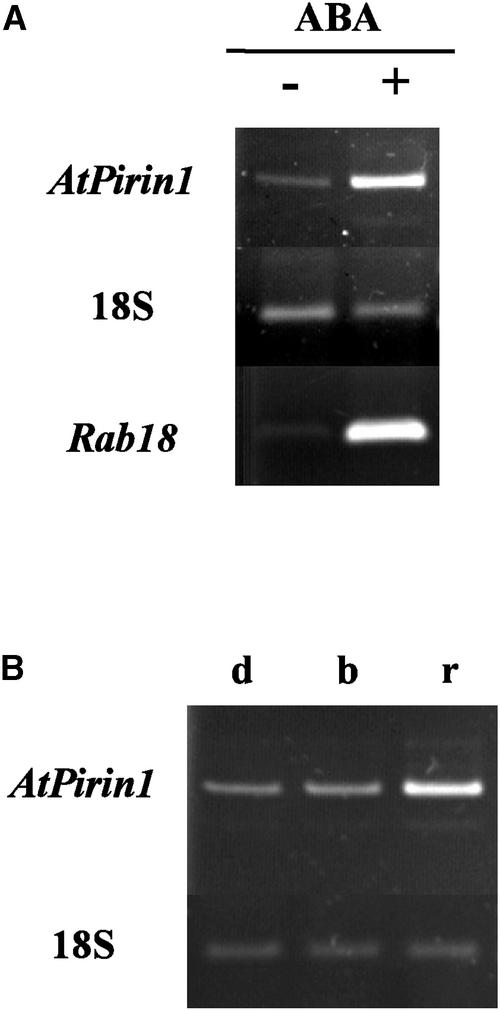
RT-PCR revealed that AtPirin1 is present at similar levels in Arabidopsis seedlings and mature plants (data not shown). The effect of ABA on AtPirin1 transcript levels was tested by spraying 9-day-old plants with 100 μM ABA and harvesting tissue 4 h later. This ABA treatment increased AtPirin1 transcript levels (Figure 3A). The increase was approximately fivefold, as determined by RT-PCR product densitometric analysis of AtPirin1 and 18S relative quantitative amplification standards. This result suggests that ABA-inducible cis-acting regulatory elements in the AtPirin1 promoter region are functional.
Figure 3.
AtPirin1 Transcript Levels Are Regulated by ABA and Low-Fluence Red Light.
(A) Transcript levels of the AtPirin1 gene are induced by ABA. AtPirin1 transcript levels were examined by relative quantitative RT-PCR in 9-day-old light-grown Arabidopsis plants sprayed with 100 μM ABA (+) or distilled water (−). The ABA-responsive Rab18 transcript (Lang and Palva, 1992) served as a positive control for the assay.
(B) Transcript levels of the AtPirin1 gene are induced by red light. AtPirin1 transcript levels were examined by relative quantitative RT-PCR in seedlings grown for 6 days in the dark and treated with a mock pulsed light (d) or irradiated with a single pulse of low-fluence blue light (b) or low-fluence red light (r).
18S rRNA was used as an endogenous RT-PCR standard. Representative experimental replicates are shown.
When we tested AtPirin1 transcript levels under different light conditions, we found that a single pulse of low-fluence red light (104 μmol·m−2·s−1) causes an increase in AtPirin1 transcript levels compared with those in dark-grown seedlings (Figure 3B). A single pulse of low-fluence blue light did not elicit a noticeable response (Figure 3B). Densitometric analysis of AtPirin1 and 18S amplification levels indicated that the pulse of low-fluence red light leads to an approximately threefold increase in AtPirin1 transcript levels in dark-grown Arabidopsis seedlings.
Identification of atpirin1 T-DNA Insertional Mutants
To examine the role of AtPirin1 in plants, we obtained two different mutant lines, atpirin1-1 and atpirin1-2, that carry a T-DNA insertion in the AtPirin1 gene. The atpirin1-1 mutant harboring a T-DNA insertion in intron 5 of the AtPirin1 gene (Figure 2B) was found in a PCR screen of a collection of 60,480 T-DNA–inserted Arabidopsis lines using an AtPirin1-specific primer and a T-DNA left border–specific primer (Krysan et al., 1999). The genetic background for this mutant line is ecotype Wassilewskija (Ws). The atpirin1-2 mutant harboring a T-DNA insertion in exon 4 of the AtPirin1 gene (Figure 2B) was obtained from the Salk Institute Genomic Analysis Laboratory. The genetic background for this mutant line is ecotype Columbia (Col). DNA gel blot and PCR analyses of atpirin1 mutants and their progeny revealed that there is only one T-DNA insertion in each of these mutant lines (data not shown).
No homozygous atpirin1-1 insertional mutants were found because of the developmental arrest of embryos at the globular stage of development, whereas the development of heterozygous atpirin1-1 mutant seeds was indistinguishable from that of the wild type (data not shown). Introduction of the AtPirin1 transgene under a moderate protochlorophyllide oxidoreductase A (PorA) promoter or a constitutive 35S promoter of Cauliflower mosaic virus did not lead to complementation of the embryo-lethal phenotype in atpirin1-1 mutants. atpirin1-2 homozygous mutants do not display the embryo-lethal phenotype. Together, these data suggest that the atpirin1-1 homozygous lethality probably is caused by a mutation in a linked gene.
For phenotypic analysis, comparisons were made between heterozygous atpirin1-1 and heterozygous and homozygous atpirin1-2 mutant plants as well as between two null alleles of GPA1, gpa1-1 and gpa1-2. Note that atpirin1-1 and atpirin1-2 seeds, referred to herein as “heterozygous,” are in fact the progeny of heterozygous plants and, as determined by PCR analysis, represent an ∼2:1 mixture of heterozygous and wild-type seeds in the case of atpirin1-1 and an ∼1:2:1 mixture of homozygous, heterozygous, and wild-type seeds in the case of atpirin1-2.
Germination of atpirin1 Mutant Seeds Is Delayed in the Absence of Stratification
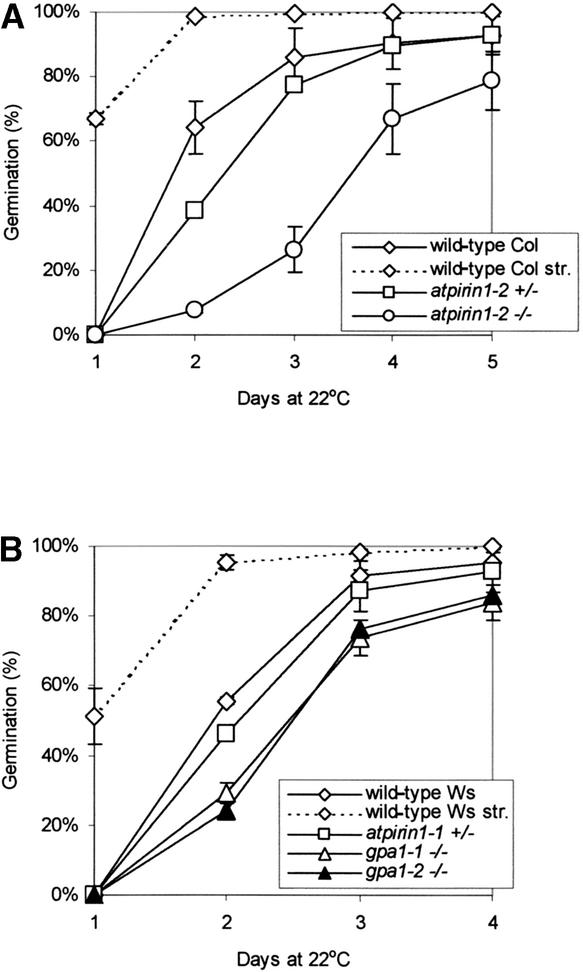
In the absence of stratification treatment (4°C for 48 h), germination of atpirin1-2 homozygous mutant seeds was delayed compared with that of wild-type seeds (Figure 4A). There was a slight delay in the germination of atpirin1-1 (Figure 4B) and atpirin1-2 (Figure 4A) heterozygous seeds as well. Stratification brought atpirin1-1 and atpirin1-2 germination to wild-type levels (Figure 4). Importantly, germination of the GPA1 homozygous mutant lines gpa1-1 and gpa1-2 also was delayed somewhat under the conditions tested (Figure 4B). As with atpirin1, stratification increased gpa1-1 and gpa1-2 germination to wild-type levels (Figure 4B). Germination profiles of the atpirin1 and gpa1 mutants suggest an increased state of dormancy that is alleviated by stratification, which is known to be a dormancy-breaking factor.
Figure 4.
Seed Germination of atpirin1 and gpa1 Mutants.
(A) Germination of wild-type Col and atpirin1-2 mutant seeds.
(B) Germination of wild-type Ws, atpirin1-1, gpa1-1, and gpa1-2 mutant seeds.
Matched seed lots on premoistened filter paper were placed directly at 22°C under continuous white light for germination. Values presented are mean percentages of germination from three independent experimental replicates (each replicate included 70 to 150 seeds per line) with standard errors. Heterozygous and homozygous mutants are indicated by +/− and −/−, respectively, next to the corresponding mutant genotype. Germination data for stratified wild-type Col seeds (wild type Col str.) and stratified wild-type Ws seeds (wild type Ws str.) are included in (A) and (B), respectively. Upon stratification, there was no statistically significant difference in germination rates between wild-type and corresponding mutant seeds.
Germination of atpirin1 Mutant Seeds Is Hypersensitive to ABA
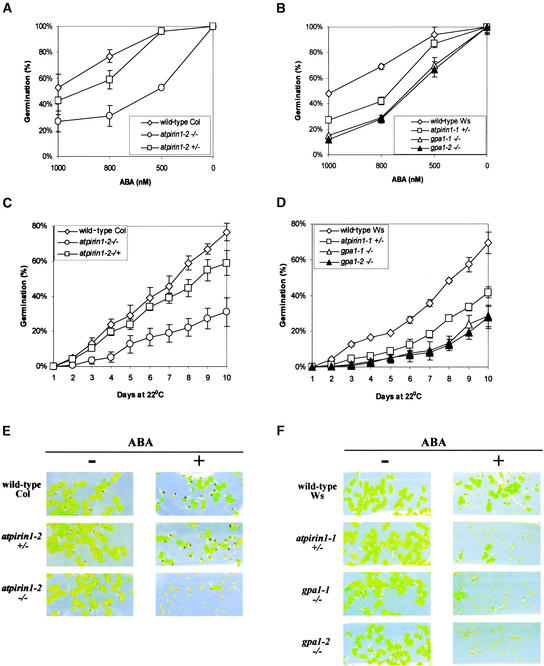
In the presence of 500 to 1000 nM ABA, germination of atpirin1 mutant seeds was delayed (Figures 5A to 5D). An ABA-imposed inhibition of gpa1 seed germination was observed recently, although under somewhat different growth conditions (Ullah et al., 2002). To fully compare the atpirin1 and gpa1 mutant phenotypes, all seeds were harvested, stored, and tested under identical conditions. Matched seed lots were planted on half-strength Murashige and Skoog (1962) medium lacking sucrose and Gamborg vitamins, stratified, and germinated at room temperature under continuous white light. On day 10, almost 80% of all wild-type Col seeds germinated, although only 31% of atpirin1-2 homozygous mutant seeds germinated, in the presence of 800 nM ABA (Figure 5C). Under identical conditions, >50% of the atpirin1-1 heterozygous seeds and >70% of gpa1-1 and gpa1-2 homozygous mutant seeds failed to germinate by day 10; however, only 31% of wild-type Ws seeds did not germinate under the same conditions (Figure 5D).
Figure 5.
Seed Germination and Early Seedling Development of atpirin1 and gpa1 Mutants Are Hypersensitive to ABA.
Matched seed lots on half-strength Murashige and Skoog (1962) agarose medium supplemented with ABA were stratified for 48 h at 4°C before being placed at 22°C under continuous white light for germination. Heterozygous and homozygous mutants are indicated by +/− and −/−, respectively, next to the corresponding mutant genotype.
(A) Germination of wild-type Col seeds and atpirin1-2 mutant seeds in the presence of the indicated concentrations of ABA at day 10 after stratification.
(B) Germination of wild-type Ws seeds and atpirin1-1, gpa1-1, and gpa1-2 mutant seeds in the presence of the indicated concentrations of ABA at day 10 after stratification.
(C) Germination of wild-type Col seeds and atpirin1-2 mutant seeds in the presence of 800 nM ABA over time (days after stratification).
(D) Germination of wild-type Ws seeds and atpirin1-1, gpa1-1, and gpa1-2 mutant seeds in the presence of 800 nM ABA over time (days after stratification).
Values shown in (A) to (D) are mean percentages of germination from three independent experimental replicates (each replicate included 70 to 150 seeds per line) with standard errors.
(E) Early seedling development of atpirin1-2 mutants is inhibited by ABA. No ABA (−) or 500 nM ABA (+) was added to the growth medium in phytatrays. Note that >50% of the mutant seeds shown are germinated as judged by radicle emergence (cf. with [A]).
(F) Early seedling development of atpirin1-1, gpa1-1, and gpa1-2 mutants is inhibited by ABA. No ABA (−) or 800 nM ABA (+) was added to the growth medium in phytatrays. Note that ∼30 to 40% of the mutant seeds shown are germinated as judged by radicle emergence (cf. with [D]).
Photographs for (E) and (F) were taken on day 10.
In addition to a reduced rate of seed germination, we found that ABA also inhibited early seedling development: it significantly delayed the rate of cotyledon expansion and greening in atpirin1 and gpa1 mutant seedlings. In the presence of exogenous ABA, even when the radicles of many atpirin1 and gpa1 mutant seedlings emerged, the majority of cotyledons failed to expand and to green (Figures 5E and 5F). Note that under the growth conditions tested, exogenous ABA inhibited the early seedling development of atpirin1 and gpa1 mutants to a much greater extent than it inhibited their germination rate (compare Figure 5C with 5E and Figure 5D with 5F); therefore, the seedling developmental delay cannot be explained simply by a delay in germination.
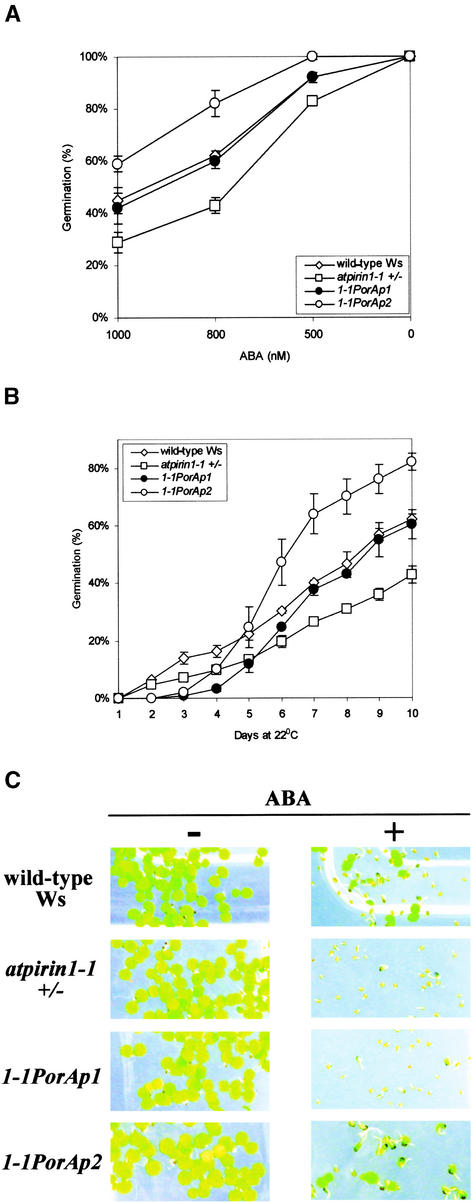
We performed a complementation test with atpirin1-1 heterozygous mutants to confirm that the observed ABA-sensitive phenotype is attributable to the genetic lesion in the AtPirin1 gene. Heterozygous atpirin1-1 plants were transformed with the AtPirin1 cDNA under the control of the PorA gene promoter. The PorA promoter is active predominantly in etiolated seedlings, and PorA mRNA disappears within the first 4 h of greening (Armstrong et al., 1995; Runge et al., 1996). Figures 6A and 6B show that the PorA-AtPirin1 transgene was able to complement the ABA-imposed reduction in germination levels of the atpirin1-1 mutant. Moreover, in one of the complementation lines, 1-1PorAp2, both ABA-related phenotypes of atpirin1-1 plants (reduced germination and inhibition of early seedling development) were complemented (Figure 6C). This complementation test confirms that the observed atpirin1 mutant phenotypes result from genetic lesions in the AtPirin1 gene.
Figure 6.
Complementation of the atpirin1-1 ABA-Hypersensitive Phenotype in Plants Carrying the AtPirin1 Transgene.
Matched seed lots on half-strength Murashige and Skoog (1962) agarose medium supplemented with ABA were stratified for 48 h at 4°C before being placed at 22°C under continuous white light for germination. The atpirin1-1 heterozygous mutant genotype is indicated by +/−.
(A) Germination of two independent complementation lines, 1-1PorAp1 and 1-1PorAp2, carrying AtPirin1 cDNA under the control of the PorA promoter in the atpirin1-1 mutant background. Germination was tested in the presence of the indicated concentrations of ABA at day 10 after stratification. Wild-type Ws seeds and atpirin1-1 mutant seeds were included for comparison.
(B) Germination of two independent complementation lines, 1-1PorAp1 and 1-1PorAp2, wild-type Ws seeds, and atpirin1-1 mutant seeds in the presence of 800 nM ABA over time (days after stratification).
Values presented in (A) and (B) are mean percentages of germination from three independent experimental replicates with standard errors.
(C) Early seedling development of the 1-1PorAp1 and 1-1PorAp2 complementation lines. Wild-type (Ws) and atpirin1-1 mutant seeds were included for comparison. No ABA (−) or 800 nM ABA (+) was added to the growth medium in phytatrays.
In addition to being necessary for the regulation of several events during seed development, ABA is involved in responses to environmental stresses, including desiccation and high salt (Busk and Pages, 1998; Leung and Giraudat, 1998). No difference was noted in atpirin1 seed germination, seedling growth, or seedling development in the presence of 400 mM mannitol, 200 mM NaCl, or 10% polyethylene glycol. Similarly, mutant plants displayed no difference in wilting compared with wild-type plants (data not shown). Together, these data suggest that the role of AtPirin1 with regard to ABA-related processes is limited to seed germination and early seedling development.
Additionally, we tested atpirin1 sensitivity to gibberellic acid (GA), because GA is known to function as an antagonist of ABA during seed germination and gpa1 mutant seed germination appears to be less sensitive to GA (Ullah et al., 2002). However, under the conditions tested, we observed no statistically significant difference from the wild type in atpirin1 germination and early seedling development (data not shown).
The Transition from Vegetative to Reproductive Growth Is Accelerated in atpirin1 Mutants
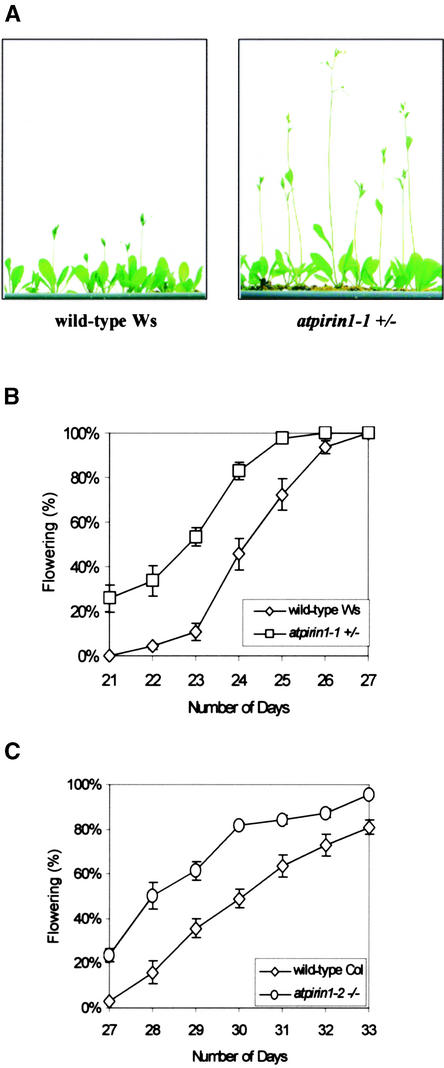
atpirin1-1 mutant plants initiate primary shoots and start flowering earlier than wild-type plants, such that a few days before the onset of flowering, atpirin1 mutants produced primary shoots of significant length, whereas wild-type plants had very short shoots (Figure 7A). To analyze this phenotype further, we measured the time needed for the transition from vegetative to reproductive growth in atpirin1 mutants and wild-type plants. Time to flowering was determined by counting the number of days after stratification until the day the first flower opened. Under long-day growth conditions, which are known to induce flowering in Arabidopsis plants, 50% of atpirin1-1 plants were flowering in <23 days; however, only 10% of wild-type Ws plants were flowering at the same time (Figure 7B). atpirin1-2 homozygous plants also displayed a similar flowering phenotype (Figure 7C). No statistically significant difference in flowering time was noted when atpirin1 plants were grown under short-day conditions (data not shown).
Figure 7.
Bolting and Flowering Time of atpirin1-1 and atpirin1-2 Plants in Long-Day Growth Conditions.
(A) At day 17, atpirin1-1 plants have longer bolts and start flowering, whereas the majority of wild-type Ws plants are just initiating bolting.
(B) Results are plotted as a percentage of plants in the wild-type Ws and atpirin1-1 heterozygous populations that flower on a particular day.
(C) Results are plotted as a percentage of plants in the wild-type Col and atpirin1-2 homozygous populations that flower on a particular day.
Flowering was scored when the first flower opened. Values presented in (B) and (C) are mean percentages of germination from three independent experimental replicates (each replicate included 25 to 40 plants per line) with standard errors. Number of days was counted from the day when planted seeds were shifted to long-day conditions after stratification. Heterozygous and homozygous mutants are indicated by +/− and −/−, respectively, next to the corresponding mutant genotype.
These results support the hypothesis that AtPirin1 functions as a moderate negative regulator of flowering under long-day conditions but is not involved in primary shoot and flowering initiation. gpa1 mutants do not display any alterations in flowering time under the conditions tested (data not shown), suggesting that AtPirin1 is involved in regulating flowering time in a GPA1-independent manner.
DISCUSSION
It is likely that the Arabidopsis Gα-subunit, GPA1, integrates multiple signaling pathways (Assmann, 2002) and therefore interacts with numerous effector and regulatory proteins. To date, other than the G protein β-subunit, no GPA1-interacting proteins have been reported. We have identified the 31-kD AtPirin1 protein as a GPA1-interacting partner that, along with GPA1, plays a defined role in seed germination and early seedling development.
Pirin was isolated initially from mammals through its physical interaction with the CAAT-box binding transcription factor NFI/CTF1 (Wendler et al., 1997). Subsequently, pirin was found to interact with the ankyrin-repeat domain of the proto-oncoprotein Bcl-3, a nucleus-localized member of the IκB family of transcriptional regulators (Dechend et al., 1999). It is well established that G proteins regulate the activity of a number of transcription factors through various effectors and/or other intermediates (Neves et al., 2002). GPA1 interaction with AtPirin1 may represent an example of a direct connection between G proteins and transcriptional regulation.
A perfect CAAT-box sequence along with several of the surrounding bases defines the core element of the low-fluence blue light response element in the pea Lhcb1*4 promoter (Folta and Kaufman, 1999). Plant pirin's potential to interact with a Gα-subunit and a CAAT-box factor, both of which are implicated in the low-fluence blue light system activation of Lhcb1*4 (Warpeha et al., 1991; Folta and Kaufman, 1999), suggests a role in blue light signal transduction in addition to the ABA pathways identified here.
Two cis-acting elements, an ACGT core and a TT motif, implicated in ABA- and light-mediated transcription regulation are found in the 5′ regulatory region of the AtPirin1 gene. The ACGT core element is thought to be the target of basic domain/Leu zipper transcription factors (Menkens et al., 1995; Leung and Giraudat, 1998). It has been shown that a phytochrome-interacting factor, PIF3, a member of the basic helix-loop-helix family, also binds an ACGT element in a sequence-specific manner and regulates the expression of photoresponsive genes (Martinez-Garcia et al., 2000). AtPirin1 transcript levels increase in response to a single pulse of low-fluence red light and exogenous ABA, suggesting that at least some of the potential upstream cis elements are active. These data, along with the recent finding that overexpression of GPA1 enhances phytochrome-mediated responses during seedling development (Okamoto et al., 2001), suggest that both AtPirin1 and GPA1 may be involved in the light regulation of seedling development.
Two atpirin1 mutant lines, atpirin1-1 and atpirin1-2, that harbor distinct T-DNA insertions were used in this study. Both mutant lines display a set of very similar phenotypes with regard to seed germination, ABA sensitivity, and flowering time, indicating that these pleiotropic effects are the result of T-DNA insertions in the AtPirin1 gene. Also, the PorA-Pirin transgene complements the ABA-imposed delay in the germination and early seedling development of atpirin1-1.
The atpirin1 and gpa1 mutants display a significant reduction in germination rates in the absence of stratification. When germination rates were tested in the presence of exogenous ABA, both atpirin1 and gpa1 mutants displayed less than wild-type germination rates. Furthermore, exogenous ABA inhibited the expansion and greening of cotyledons in atpirin1 and gpa1 mutant seedlings. A subset of atpirin1 mutant phenotypes correspond to gpa1 mutant phenotypes, confirming that these interacting proteins function in the same pathway.
The reduced germination rates of the atpirin1 and gpa1 mutants likely are the result of the enhanced dormancy of these mutant seeds, which is further facilitated by the application of exogenous ABA, a plant hormone known to establish seed dormancy during embryo maturation (Bonetta and McCourt, 1998; Koornneef et al., 2002). The recent observation that overexpression of the Arabidopsis G protein–coupled receptor GCR1 abolishes seed dormancy (Colucci et al., 2002) further implicates the plant G protein pathway in the regulation of seed dormancy.
The two alleles have similar genetic characteristics with respect to flowering and germination in the absence of cold or ABA treatment. By contrast, atpirin1-1 and atpirin1-2 display apparent dominant and recessive natures, respectively, with regard to ABA effects on seed germination and early seedling development. The differences between atpirin1-1 and atpirin1-2 may reflect varietal differences with respect to ABA sensitivity and/or the maternally based ABA-mediated expression of the Atpirin1 gene.
atpirin1 and gpa1 are similar to a small group of mutants, including era1, fiery1, abh1, and sad1, that display increased seed dormancy and ABA hypersensitivity (Cutler et al., 1996; Hugouvieux et al., 2001; Xiong et al., 2001a, 2001b). These mutations are recessive, indicating that the proteins involved might be negative regulators of ABA action. Like atpirin1 and gpa1, these mutations have multiple phenotypes, some without evident connection to ABA signaling, suggesting that these proteins may be involved in the regulation of a number of different processes in plants.
It was suggested recently that gpa1 mutant seeds display lower germination rates in the presence of ABA as a result of the reduced sensitivity of these mutants to GA (Ullah et al., 2002). Under the germination conditions used in the present study (half-strength Murashige and Skoog [1962] medium lacking sucrose and Gamborg vitamins, cold pretreatment, and germination in continuous white light), mutants of the potential GPA1 effector, AtPirin1, had increased ABA sensitivity but did not display a reduction in GA sensitivity, suggesting that ABA hypersensitivity and GA hyposensitivity during gpa1 seed germination may result from lesions in two separate pathways.
Besides delaying the germination rates of atpirin1 and gpa1 mutants, ABA delays their overall seedling development: the majority of cotyledons failed to green and expand. Therefore, even though a significant fraction of mutant seeds germinated in the presence of low ABA concentrations, as judged by the full penetration of the seed coat by the root tip (Figures 5A to 5D), the seedlings still were growth arrested (Figures 5E to 5F). A similar phenotype has been reported for two other ABA-hypersensitive mutants, fiery1 and sad1 (Xiong et al., 2001a, 2001b). Previously, it was suggested that ABA inhibits cotyledon greening and cell expansion more strongly than the breakdown of the seed coat (Steber et al., 1998).
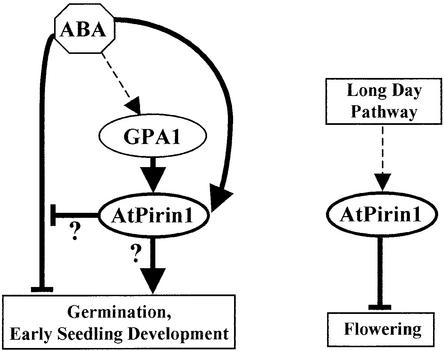
In addition to germination-related phenotypes, atpirin1 mutants exhibit a moderate early-flowering phenotype when grown under long-day conditions. Early-flowering mutant phenotypes are thought to result from the inactivation of genes required to repress flowering (Coupland, 1995). Overexpression of the putative Arabidopsis G protein–coupled receptor GCR1 accelerates flowering as well as the expression of the flowering-inducing gene LFY (Colucci et al., 2002). However, we observed no difference in flowering time in gpa1 mutants, suggesting that AtPirin1-mediated flowering regulation is a G protein–independent process. A model illustrating AtPirin1's involvement in flowering time regulation is presented in Figure 8 (at right).
Figure 8.
Schemes of AtPirin1 Modes of Action.
Together with GPA1, AtPirin1 positively regulates seed germination and early seedling development by overcoming the negative effects of ABA and/or activating germination-promoting pathway(s). AtPirin1 mRNA levels are upregulated by ABA, suggesting the existence of a negative feedback regulatory loop of the ABA signaling pathway. Independent of GPA1, AtPirin1 is involved in the regulation of flowering time. Thick arrows indicate either known pathways or pathways established in this study.
Based on the AtPirin1 gene characterization and atpirin1 mutant phenotypes, there are two conceivable models for AtPirin1's mode of action. The first model postulates that, with regard to germination and early seedling development, AtPirin1 functions in a negative feedback regulatory loop of ABA signaling and is involved in resetting the ABA signal transduction pathway (Figure 8). According to the second model, AtPirin1 also relieves the ABA-imposed inhibition of germination, although indirectly, through the activation of germination-promoting processes (Figure 8). These models are not mutually exclusive and, importantly, GPA1 can be envisioned to function together with AtPirin1 in both of them.
The isolation and characterization of the AtPirin1 protein and the characterization of its mutant phenotypes provide biochemical and genetic evidence that AtPirin1 is a multifunctional regulatory protein that likely operates immediately downstream of its interacting partner, GPA1, in regulating seed germination and early seedling development.
METHODS
Plant Materials and Growth Conditions
Both Columbia (Col) and Wassilewskija (Ws) ecotypes of Arabidopsis thaliana were used as wild-type controls for phenotype comparison. For germination assays, seeds were placed in 15-cm Petri dishes on filter paper saturated with distilled water and, where specified, stratified (4°C for 48 h) before being placed at room temperature in continuous white light (20 μmol·m−2·s−1) for germination. For hormonal assays, stress tolerance assays, and RNA isolation from abscisic acid (ABA)–treated plants, seeds were planted in phytatrays (Sigma) on half-strength Murashige and Skoog (1962) 0.8% (w/v) agarose medium lacking sucrose and Gamborg vitamins, stratified, and germinated as described above. For flowering tests, plant transformation, and bulking up of seeds, plants were grown on Metro-Mix 200 growing soil medium (Scotts, Maysville, OH) under white light supplied by cool-white fluorescent bulbs (90 μmol·m−2·s−1). The atpirin1-1 T-DNA insertion line was isolated by screening the α population of the Arabidopsis T-DNA insertion lines generated in the Ws ecotype at the Arabidopsis Knockout Facility at the University of Wisconsin (Madison). The atpirin1-2 T-DNA insertion line in the Col ecotype background was obtained from the Salk Institute Genomic Analysis Laboratory through the ABRC (Columbus, OH).
Yeast and Bacterial Strains
For the yeast two-hybrid experiments, the Saccharomyces cerevisiae L40 strain was used. The partial genotype of L40 is MATa his3-200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-LacZ GAL4 (Vojtek et al., 1993). Escherichia coli strain KC8 (Clontech Laboratories, Palo Alto, CA), which is auxotrophic for Leu and Trp, was used for plasmid recovery from yeast. E. coli protease-deficient strain BL21 was used for glutathione S-transferase (GST) fusion protein expression.
Plasmid Construction
For the two-hybrid assay, protein fusions to the DNA binding domain of the bacterial repressor LexA were made using the plasmid pLexA-NLS, a modified version of the pBTM116 plasmid (Vojtek et al., 1993) containing nuclear localization signal. The GPA1 cDNA was obtained by PCR from pCIT1828 plasmid using primers designed to incorporate partial EcoRI and XhoI restriction sites. The GPA1 bait plasmid was created by cloning the GPA1 cDNA into the pLexA-NLS plasmid (EcoRI-SalI) after exoIII (Promega) modification of PCR product ends (Kaluz et al., 1992). For in vitro protein association assays, the restriction fragment of GPA1 cDNA (EcoRI-XhoI) was subcloned into the GST fusion expression vector pGEX-4T-1 (Amersham Pharmacia Biotech). For the complementation test of the atpirin1-1 mutant, the AtPirin1 cDNA was obtained by reverse transcription (RT)–PCR using primers designed to incorporate partial NcoI restriction sites. After exoIII modification of the PCR product ends, the AtPirin1 cDNA was cloned into the vector pBSK101.4PorA (NcoI), a modified version of pBSK101.3 (Folta and Kaufman, 1999) in which the β-glucuronidase (GUS; uidA) coding region was replaced with a multiple cloning site (EcoRI-HindIII-NcoI) and the PorA gene promoter was released from the pAG3/6 plasmid and inserted into HindIII-NcoI sites. The PorA-AtPirin1 fusion was moved into the plant transformation vector CLS9600, a derivative of pPZP100 (Hajdukiewicz et al., 1994). CLS9600 contains an additional cassette consisting of the minimal blue light promoter (the −95 to +2 fragment of the PsLhcb1*4 promoter), the GUS gene, and the nopaline synthase terminator (Folta and Kaufman, 1999).
Yeast Two-Hybrid Screening
The GPA1 bait construct was used to screen the Kim and Theologis λACT two-hybrid cDNA library (ABRC). Yeast transformations were performed using the lithium acetate/polyethylene glycol–based method (Gietz and Sugino, 1988). Standard yeast growth medium was prepared as described in the Yeast Protocol Handbook from Clontech. The S. cerevisiae reporter strain L40 was transformed first with the GPA1 bait vector and subsequently with the Arabidopsis two-hybrid cDNA library. Transformants were plated on 15-cm synthetic dextrose plates (Trp−, Leu−, Lys−, His−) to select for His prototrophy. The His+ colonies were restreaked on 5-bromo-4-chloro-β-d-galactoside–containing selective medium, and LacZ reporter gene activity was monitored visually. Library plasmids from His+LacZ+ yeast colonies were rescued by direct retransformation from LacZ+ cells into the E. coli strain KC8 according to a standard protocol. E. coli colonies grown on medium lacking Leu were selected. Positive clones were sequenced at the Cancer Research Center DNA Sequencing Facility at the University of Chicago.
Expression of the GST-GPA1 Fusion Protein
The GST-GPA1 fusion protein was expressed in E. coli strain BL21. Protein expression was induced with 100 μM isopropyl-β-d-thiogalactopyranoside, and extracts were prepared by treating cells for 3 min at 0°C with lysozyme solution (1 mg/mL lysozyme and 2.5 mM Tris, pH 8.0) followed by a brief sonication. Recombinant proteins were affinity-purified on glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech). Samples were visualized on SDS-PAGE gels stained with Coomassie Brilliant Blue G 250.
In Vitro Binding Assay
Expression of the GST-GPA1 fusion proteins was induced as described above. The radiolabeled AtPirin1 protein was produced by coupled in vitro transcription/translation using the TNT T7 Coupled Wheat Germ Extract System (Promega). Translation-grade 35S-Met (Amersham International Redivue) was obtained from Amersham Pharmacia Biotech. A typical binding reaction was prepared by mixing 20 μL of 50% GST-fused protein immobilized on glutathione-Sepharose 4B beads, 300 μL of binding assay buffer (10 mM Tris, pH 7.2, 50 mM potassium acetate, pH 7.0, 0.01% Triton X-100, and 4 mM magnesium acetate), and 20 μL of AtPirin1 in vitro translation reaction. Binding reactions were agitated on an orbital shaker for 2 h at room temperature. The beads were pelleted and washed four times with the binding assay buffer. Samples were resolved on a 10% SDS-PAGE gel, and the resulting gel was fixed and fluorographically enhanced with EN3HANCE Autoradiography Enhancer (DuPont–New England Nuclear Life Science Products) according to the manufacturer's instructions and visualized by autoradiography.
In specified cases, the GST-GPA1 fusion was loaded with GTPγS (Sigma) before addition to a binding reaction. GST-GPA1 loading with GTPγS was achieved by incubating 15 μL of washed GST-GPA1–Sepharose beads with 10 μL of 10 mM GTPγS in 90 μL of wash buffer (50 mM Tris, 100 mM NaCl, 0.1% Triton X-100, and 10 mM magnesium acetate) and agitating the reaction overnight on an orbital shaker at 4°C.
Expression Studies by Relative Quantitative RT-PCR
To study the effects of ABA, 9-day-old Arabidopsis seedlings were sprayed with 100 μM ABA or sterile distilled water and plant tissue was harvested 4 h later. To study the effects of light, planting was performed as described (Folta and Kaufman, 1999). Six-day-old dark-grown Arabidopsis seedlings were irradiated with a single pulse of low-fluence blue or red light (104 μmol·m−2·s−1). Plant tissue was harvested 2 h later. Total RNA was isolated with TRI Reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer's instructions. RNA was used as a template for first-strand DNA synthesis using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen Life Technologies, Carlsbad, CA). PCR amplification was performed using Elongase Enzyme Mix (Invitrogen Life Technologies); the optimal concentration of Mg2+ for these amplification reactions was 1.3 mM. PCR amplifications were performed in a final volume of 23 μL with RtpirinD (5′-TGACGTATGAGAACAATAGTGTAC-3′) and pirinTrev (5′-GTAACCTCTAACACTTGGCCATTTCAAC-3′), AtPirin1-specific primers that distinguish between the AtPirin1 transcript and genomic DNA contaminants. Conditions were chosen in the linear range of amplification for the products and were as follows: 30 cycles of 94°C for 20 s, 94°C for 15 s, 60°C for 30 s, and 72°C for 2 min, followed by 72°C for 2 min. QuantumRNA 18S Internal Standards (Ambion, Austin, TX) were used as a relative quantitative amplification control. To achieve levels of amplification comparable to those of the AtPirin1 PCR products, 18S-specific primers were equilibrated with 18S PCR Competimers in a ratio of 2:8, which corresponds to the abundance levels of rare transcripts. The PCR products were resolved on a 1% agarose gel, and amplification levels were determined using the one-dimensional multidensitometry tool on an AlphaImager 2000 (Alpha Innotech, San Leandro, CA).
Identification of the atpirin1-1 T-DNA Insertion Mutant
The α population of ∼64,000 Arabidopsis T-DNA insertion lines at the University of Wisconsin Arabidopsis Knockout Facility was screened using pirinTdir primer (5′-ATACATGCTACAGGGAGGTATCATTCACA-3′) according to the facility's protocol (www.biotech.wisc.edu/Arabidopsis/). The progeny of a plant heterozygous for a T-DNA insertion in the AtPirin1 gene were genotyped by PCR using pirinTdir primer and JL202 T-DNA left border–specific primer (www.biotech.wisc.edu/Arabidopsis/). As a result of the embryonic lethality of plants homozygous for a T-DNA insertion in the AtPirin1 gene, heterozygous plants were distinguished by histochemical GUS staining of flowers and selected for propagation. Flowers were chosen for GUS staining because the T-DNA insert vector contains the GUS reporter gene driven by the flower-specific apetala3 promoter. GUS histochemical staining was conducted essentially as described (Jefferson et al., 1987). In brief, freshly excised flowers were immersed in GUS staining solution containing 8 mM 5-bromo-4-chloro-β-d-glucuronide cyclohexylammonium salt (Gold BioTechnology, St. Louis, MO) and 2% Triton X-100 in 50 mM sodium phosphate buffer, pH 7.2, and then incubated at 37°C overnight.
Germination Assay
Germination rates were compared between seed lots that were produced, harvested, and stored under identical conditions. Seventy to 150 seeds from wild-type (Col and Ws), atpirin1-1, gpa1-1, gpa1-2, and atpirin1-2 plants were planted in triplicate. Seeds were considered germinated when the radicles completely penetrated the seed coat. Germination was scored daily for 10 days after seeds were placed at room temperature.
Hormonal and Stress Tolerance Assays
The effects of ABA on seed germination were studied by determining the germination rates of 70 to 150 seeds planted in triplicate on growth medium containing ABA (mixed isomers; Sigma). For direct comparisons of germination rates at a particular ABA concentration, each phytatray was subdivided and all seed lines were planted on the same phytatray.
The effects of GA on seed germination were determined as described for ABA except that 1 or 10 μM GA (Sigma) was added to the medium. The effects of mannitol, polyethylene glycol, and NaCl were studied in a similar manner.
Complementation Analysis
Complementation constructs carrying the AtPirin1 cDNA under the control of the PorA promoter are described above. atpirin1-1 heterozygous Arabidopsis plants were transformed by vacuum infiltration (Bechtold et al., 1993). Transgenic plants were selected by resistance to the herbicide Finale (Roussel Uclaf, Montvale, NJ) and by histochemical GUS staining of leaves. atpirin1-1 heterozygous plants carrying the PorA-AtPirin1 transgene were selected by PCR.
Flowering Time Analysis
Wild-type and mutant plants for corresponding experimental replicates were grown in the same tray, which was rotated 180° every 24 h. For each experimental replicate, 25 to 40 plants were analyzed per line. Long-day conditions consisted of 16 h of light followed by 8 h of darkness. The onset of flowering was defined as the day when the first flower opened. atpirin1-1 heterozygous plants were selected by histochemical GUS staining of flowers as described above.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
Accession numbers for the sequences shown in Figure 2A are as follows: AtPirin1 (NP_191481), AtPirin2 (NP_191485), AtPirin3 (NP_030851), AtPirin4 (NP_175474), tomato pirin (TOMpirin; AAF22236), and human pirin (HUMpirin; CAA69195).
Acknowledgments
We thank Alan Jones for providing the gpa1-1 and gpa1-2 mutant seed lines, Stanley Hollenberg for the pLexA-NLS plasmid, Hong Ma for the pCIT1828 plasmid, Gregory Armstrong for the pAG3/6 plasmid, Caren Chang for the ETR1 bait construct, and Joseph Kieber for the CTR1 prey construct. We also thank Mary Beth Anderson, Kevin Folta, and John Marsh for their assistance in the cultivation and transformation of Arabidopsis and for their useful discussions, Dmitri Pestov and Sanghamitra Saha for their useful suggestions, and Sam Hawkins for technical assistance. This work was supported by grants from the U.S. Department of Agriculture and the National Science Foundation to L.S.K.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011890.
References
- Ando, S., Takumi, S., Ueda, Y., Ueda, T., Mori, N., and Nakamura, C. (2000). Nicotiana tabacum cDNAs encoding alpha and beta subunits of a heterotrimeric GTP-binding protein isolated from hairy root tissues. Genes Genet. Syst. 75, 211–221. [DOI] [PubMed] [Google Scholar]
- Armstrong, G.A., Runge, S., Frick, G., Sperling, U., and Apel, K. (1995). Identification of NADPH:protochlorophyllide oxidoreductases A and B: A branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol. 108, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann, S.M. (2002). Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 14 (suppl.), S355.–S373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Bonetta, D., and McCourt, P. (1998). Genetic analysis of ABA signal transduction pathways. Trends Plant Sci. 3, 231–235. [Google Scholar]
- Busk, P.K., and Pages, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37, 425–435. [DOI] [PubMed] [Google Scholar]
- Clark, K.L., Larsen, P.B., Wang, X., and Chang, C. (1998). Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. USA 95, 5401–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colucci, G., Apone, F., Alyeshmerni, N., Chalmers, D., and Chrispeels, M.J. (2002). GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc. Natl. Acad. Sci. USA 99, 4736–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland, G. (1995). Genetic and environmental control of flowering time in Arabidopsis. Trends Genet. 11, 393–397. [DOI] [PubMed] [Google Scholar]
- Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S., and McCourt, P. (1996). A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273, 1239–1241. [DOI] [PubMed] [Google Scholar]
- Dechend, R., Hirano, F., Lehmann, K., Heissmeyer, V., Ansieau, S., Wulczyn, F.G., Scheidereit, C., and Leutz, A. (1999). The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene 18, 3316–3323. [DOI] [PubMed] [Google Scholar]
- Dunwell, J.M., Culham, A., Carter, C.E., Sosa-Aguirre, C.R., and Goodenough, P.W. (2001). Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26, 740–746. [DOI] [PubMed] [Google Scholar]
- Dunwell, J.M., Khuri, S., and Gane, P.J. (2000). Microbial relatives of the seed storage proteins of higher plants: Conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta, K.M., and Kaufman, L.S. (1999). Regions of the pea Lhcb1*4 promoter necessary for blue-light regulation in transgenic Arabidopsis. Plant Physiol. 120, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gane, P.J., Dunwell, J.M., and Warwicker, J. (1998). Modeling based on the structure of vicilins predicts a histidine cluster in the active site of oxalate oxidase. J. Mol. Evol. 46, 488–493. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gotor, C., Lam, E., Cejudo, F.J., and Romero, L.C. (1996). Isolation and analysis of the soybean SGA2 gene (cDNA), encoding a new member of the plant G-protein family of signal transducers. Plant Mol. Biol. 32, 1227–1234. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hamm, H.E. (1998). The many faces of G protein signaling. J. Biol. Chem. 273, 669–672. [DOI] [PubMed] [Google Scholar]
- Hugouvieux, V., Kwak, J.M., and Schroeder, J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477–487. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluz, S., Kolble, K., and Reid, K.B. (1992). Directional cloning of PCR products using exonuclease III. Nucleic Acids Res. 20, 4369–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaziro, Y., Itoh, H., Kozasa, T., Nakafuku, M., and Satoh, T. (1991). Structure and function of signal-transducing GTP-binding proteins. Annu. Rev. Biochem. 60, 349–400. [DOI] [PubMed] [Google Scholar]
- Kim, W.Y., Cheong, N.E., Lee, D.C., Je, D.Y., Bahk, J.D., Cho, M.J., and Lee, S.Y. (1995). Cloning and sequencing analysis of a full-length cDNA encoding a G protein α subunit, SGA1, from soybean. Plant Physiol. 108, 1315–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, V., and Palva, E.T. (1992). The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20, 951–962. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Ma, H., Yanofsky, M.F., and Meyerowitz, E.M. (1990). Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 87, 3821–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinissen, M.J., and Gutkind, J.S. (2001). G-protein-coupled receptors and signaling networks: Emerging paradigms. Trends Pharmacol. Sci. 22, 368–376. [DOI] [PubMed] [Google Scholar]
- Marsh, J.F., and Kaufman, L.S. (1999). Cloning and characterisation of PGA1 and PGA2: Two G protein alpha-subunits from pea that promote growth in the yeast Saccharomyces cerevisiae. Plant J. 19, 237–247. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Mason, M.G., and Botella, J.R. (2000). Completing the heterotrimer: Isolation and characterization of an Arabidopsis thaliana G protein gamma-subunit cDNA. Proc. Natl. Acad. Sci. USA 97, 14784–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, M.G., and Botella, J.R. (2001). Isolation of a novel G-protein γ-subunit from Arabidopsis thaliana and its interaction with Gβ. Biochim. Biophys. Acta 1520, 147–153. [DOI] [PubMed] [Google Scholar]
- Menkens, A.E., Schindler, U., and Cashmore, A.R. (1995). The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Neves, S.R., Ram, P.T., and Iyengar, R. (2002). G protein pathways. Science 296, 1636–1639. [DOI] [PubMed] [Google Scholar]
- Okamoto, H., Matsui, M., and Deng, X.W. (2001). Overexpression of the heterotrimeric G-protein α-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell 13, 1639–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, L.C., and Lam, E. (1993). Guanine nucleotide binding protein involvement in early steps of phytochrome-regulated gene expression. Proc. Natl. Acad. Sci. USA 90, 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge, S., Sperling, U., Frick, G., Apel, K., and Armstrong, G.A. (1996). Distinct roles for light-dependent NADPH:protochlorophyllide oxidoreductases (POR) A and B during greening in higher plants. Plant J. 9, 513–523. [DOI] [PubMed] [Google Scholar]
- Saalbach, G., Natura, B., Lein, W., Buschmann, P., Dahse, I., Rohrbeck, M., and Nagny, F. (1999). The α-subunit of a heterotrimeric G-protein from tobacco, NtGPα1, functions in K+ channel regulation in mesophyll cells. J. Exp. Bot. 50, 53–61. [Google Scholar]
- Spiegel, A.M., Jones, T.L.Z., Simmonds, W.F., and Weinstein, L.S. (1994). G Proteins. (Austin, TX: R.G. Landes).
- Sprang, S.R. (1997). G protein mechanisms: Insights from structural analysis. Annu. Rev. Biochem. 66, 639–678. [DOI] [PubMed] [Google Scholar]
- Steber, C.M., Cooney, S.E., and McCourt, P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suharsono, U., Fujisawa, Y., Kawasaki, T., Iwasaki, Y., Satoh, H., and Shimamoto, K. (2002). The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 99, 13307–13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Wang, S., and Jones, A.M. (2002). Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 129, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Young, J.C., Im, K.H., Sussman, M.R., and Jones, A.M. (2001). Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292, 2066–2069. [DOI] [PubMed] [Google Scholar]
- Vojtek, A.B., Hollenberg, S.M., and Cooper, J.A. (1993). Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74, 205–214. [DOI] [PubMed] [Google Scholar]
- Wang, X.Q., Ullah, H., Jones, A.M., and Assmann, S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292, 2070–2072. [DOI] [PubMed] [Google Scholar]
- Warpeha, K.M., Hamm, H.E., Rasenick, M.M., and Kaufman, L.S. (1991). A blue-light-activated GTP-binding protein in the plasma membranes of etiolated peas. Proc. Natl. Acad. Sci. USA 88, 8925–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, C.A., Garnaat, C.W., Mukai, K., Hu, Y., and Ma, H. (1994). Isolation of cDNAs encoding guanine nucleotide-binding protein β-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc. Natl. Acad. Sci. USA 91, 9554–9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler, W.M., Kremmer, E., Forster, R., and Winnacker, E.L. (1997). Identification of pirin, a novel highly conserved nuclear protein. J. Biol. Chem. 272, 8482–8489. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Gong, Z., Rock, C.D., Subramanian, S., Guo, Y., Xu, W., Galbraith, D., and Zhu, J.K. (2001. a). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1, 771–781. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Lee, B., Ishitani, M., Lee, H., Zhang, C., and Zhu, J.K. (2001. b). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]