Abstract
Protein acetylation is important in regulating DNA-templated processes specifically and protein–protein interactions more generally in eukaryotes. The geminivirus movement protein NSP is essential for virus movement, shuttling the viral DNA genome between the nucleus and the cytoplasm. We have identified a novel Arabidopsis protein, AtNSI, that interacts with NSP. AtNSI is highly conserved among widely divergent plants. Biochemical studies show that its interaction with NSP is direct and that AtNSI acetylates histones, but not NSP, in vitro. Rather, AtNSI specifically acetylates the viral coat protein. AtNSI is a nuclear protein but does not act as a transcriptional coactivator in vitro, which distinguishes it from known eukaryotic histone acetyltransferases. Its overexpression enhances the efficiency of infection by Cabbage leaf curl virus. These findings suggest a role for protein acetylation in coordinating replication of the viral DNA genome with its export from the nucleus.
INTRODUCTION
Protein acetylation is important in regulating a variety of DNA-templated processes in eukaryotes. The best characterized of these is the role of histone hyperacetylation in the chromatin remodeling associated with transcriptional activation. Many transcriptional coactivators, including yeast GCN5, p300 and the closely related CBP (often termed p300/CBP), PCAF, and ACTR, contain intrinsic histone acetyltransferase activity. Extensive biochemical and genetic evidence correlates histone hyperacetylation with transcriptionally active regions of chromatin (Cheung et al., 2000; Chen et al., 2001b, and references cited therein), and recent in vivo studies of GCN5 identify distinct events in this process (Reinke et al., 2001). GCN5 transiently hyperacetylates histones at the PHO8 promoter. This acetylation is not required for transcription factor binding, nor does it directly remodel nucleosomes at the PHO8 promoter. Rather, it marks these nucleosomes for remodeling by a subsequent group of ATP-dependent enzymes, and hyperacetylation is lost once remodeling occurs and the promoter is transcriptionally active.
Although histone acetylation has been best characterized in terms of the chromatin remodeling required for eukaryotic transcriptional activation and elongation, disruption of nucleosomes to provide increased access to DNA for trans-acting factors is required more generally for DNA-templated events. Thus, in addition to transcriptional processes, including activation (GCN5, PCAF, ATF-2, p300/CBP), elongation (Elp3), dosage compensation (MOF), and silencing (Sas2 and Sas3), nuclear histone acetyltransferases have been implicated in DNA replication (HBO1) (Iizuka and Stillman, 1999; Burke et al., 2001), V(D)J recombination required to generate the T-cell receptor in T cells and immunoglobulins in B cells (McMurry and Krangel, 2000), and DNA repair and apoptosis (TIP60) (Ikura et al., 2000).
As first suggested by studies of GCN5 and the recruitment of chromatin-remodeling complexes to activate the PHO8 promoter, protein acetylation also can be important more generally in protein–protein interactions. In particular, p300/CBP and PCAF have been reported to acetylate an increasing number of nonhistone nuclear proteins, including other coactivators and transcription factors, to either stimulate or inhibit their activities depending on the domain acetylated within the target protein (Chen et al., 2001a). For example, acetylation of p53 by p300 can stabilize the transcription initiation complex to increase p53 DNA binding activity, whereas p300-catalyzed acetylation of c-Jun appears to have the opposite effect of destabilizing the promoter complex to repress transcription from the collagenase promoter (Barlev et al., 2001; Vries et al., 2001). Similarly, p300/CBP acetylation of the coactivator ACTR disrupts its interaction with the estrogen receptor to downregulate estrogen receptor–mediated transcription, but PCAF acetylation of MyoD stimulates its DNA binding activity to promote myogenesis (Chen et al., 1999; Sartorelli et al., 1999).
Beyond transcription, acetylation of nonhistone targets by p300/CBP and PCAF has been implicated in protein stabilization, nuclear import and retention, and cell cycle progression and differentiation. PCAF acetylation of E2F1, the key regulator of cell cycle progression, can stabilize the “free” form of E2F1 not bound to the retinoblastoma tumor-suppressor protein Rb (Martinez-Balbas et al., 2000). Acetylation of the nuclear localization sequences in the transcription factors HNF-4 and CIITA by CBP or PCAF, respectively, has been reported to increase nuclear retention (Soutoglou et al., 2000; Spilianakis et al., 2000). CBP also has been reported to acetylate the nuclear import factors importin-α and Rch1 in vitro, as well as Rb, although the functional consequences of these activities remain to be defined (Bannister et al., 2000; Chan et al., 2001). Thus, by acetylating target proteins to disrupt or enhance molecular interactions, p300/CBP and PCAF can potentially impact a wide range of biological processes. This potential has led to the suggestion that protein acetylation may rival phosphorylation as a mechanism for transducing cellular regulatory signals (Kouzarides, 2000; Chen et al., 2001b).
Plant viruses encode novel movement proteins that direct the viral genome to the cortical cytoplasm and across the barrier of the cell wall to propagate infection in the host. For single-stranded DNA (ssDNA) bipartite geminiviruses such as Squash leaf curl virus (SqLCV) and Cabbage leaf curl virus (CLCV), this process requires two movement proteins, a nuclear shuttle protein (NSP) and a cell-to-cell movement protein (MP), to cooperatively move the viral genome from its site of replication in the nucleus to the cytoplasm and into adjacent plant cells. NSP is a ssDNA binding protein that shuttles newly replicated viral genomic DNA between the nucleus and the cytoplasm (Pascal et al., 1994; Sanderfoot et al., 1996; Ward and Lazarowitz, 1999). MP traps these NSP-genome complexes in the cytoplasm and redirects them to and across the plant cell wall (Noueiry et al., 1994; Sanderfoot and Lazarowitz, 1995; Ward et al., 1997). In the adjacent cell, NSP-genome complexes are released and NSP targets the viral genome to the nucleus to initiate new rounds of infection. This process has been visualized by confocal microscopy using a transient expression assay in plant protoplasts to establish that the mechanism of nuclear shuttling is highly conserved in plants, as well as in animal cells and yeast (Sanderfoot and Lazarowitz, 1995; Sanderfoot et al., 1996; Ward and Lazarowitz, 1999). These studies showed that SqLCV NSP is a typical rapidly shuttling nuclear protein that contains two classic basic nuclear localization sequences and an essential Leu-rich nuclear export sequence of the type found in shuttle proteins such as HIV Rev and TFIIIa. We have further shown that this process of movement is highly regulated. Post-translational modifications of SqLCV NSP regulate NSP interactions with MP, and the SqLCV coat protein (CP) synergistically aides NSP in the nuclear phase of virus movement by sequestering newly replicated progeny ssDNA genomes away from the replication pool to make them available for export by NSP (Sanderfoot et al., 1996; Qin et al., 1998).
To further characterize NSP, we used the classic yeast two-hybrid screen to identify cellular proteins that interact with NSP. This assay could identify host proteins involved in the nuclear import and export of NSP, its interactions with MP, or its binding to or release of the viral genome. We report here the identification and functional characterization of a novel acetyltransferase from Arabidopsis that interacts directly with NSP encoded by CLCV, for which Arabidopsis is a natural host (Hill et al., 1998). We show that this protein is highly conserved in plants and is a nuclear protein that can acetylate histones H2A and H3 in vitro. It also specifically acetylates the viral CP, but does not act as a transcriptional coactivator in vitro. Our studies suggest that this acetyltransferase regulates the nuclear export of the viral genome and potentially other nontranscriptional nuclear events in plant cells.
RESULTS
Identification of AtNSI, a Protein That Interacts with NSP
CLCV NSP fused in frame to the GAL4 DNA binding domain was used as bait in yeast two-hybrid screens of three independent Arabidopsis cDNA libraries fused to the GAL4 activation domain expressed from a 2-μm plasmid or a CEN-ARS vector (Durfee et al., 1993; Kim et al., 1997; Samach et al., 1999). From 3.5 × 106 transformants screened, 13 clones were identified in 16 separate screens based on His and/or adenine prototrophy and strong β-galactosidase activity. Nine clones were shown to contain the same cDNA, varying only in their length upstream of the polyadenylation sequence. This cDNA insert was designated AtNSI (Arabidopsis thaliana nuclear shuttle protein interactor), and the longest clone, an ∼900-bp XhoI insert, was characterized further.
The interaction between NSP and AtNSI was confirmed using additional plasmid combinations and constructs to test for autonomous activation of the HIS3 and lacZ reporter genes, for possible direct interactions with the GAL4 binding or activation domain, and for potential artifacts caused by high expression of the encoded GAL4 fusion protein (Table 1). Neither pGBT9:NSP (bait) nor pACT:AtNSI (prey) alone activated the HIS3 or lacZ reporter gene. AtNSI (expressed from pACT:AtNSI) did not interact with the GAL4 binding domain itself, nor did NSP (expressed from pGBT9:NSP) interact with the GAL4 activation domain when the latter was expressed either from pACT or from the out-of-frame construct pGAD424:AtNSI(of). AtNSI fused to the GAL4 activation domain (pACT:AtNSI) still interacted with NSP fused to the GAL4 binding domain when the latter was expressed at reduced levels from pGAD424. NSP and AtNSI also interacted with each other when the domains to which they were fused were exchanged—that is, when pGBT9:AtNSI and pGAD424:NSP were coexpressed in Y190 cells. Thus, expression of both AtNSI and NSP fused in frame to the appropriate GAL4 activation or binding domain was required for interaction, irrespective of promoter strength or the GAL4 domain to which each coding sequence was fused.
Table 1.
Interaction of AtNSI and NSP in Yeast Cells
| GAL4 BDa Vector | GAL4 ADb Vector | His | β-Galactosidase |
|---|---|---|---|
| pGBT9:NSP | —c | − | − |
| —c | pACT:AtNSI | − | − |
| pGBT9 | pACT:AtNSI | − | − |
| pGBT9:NSP | pACT | − | − |
| pGBT9:NSP | pACT:AtNSI | + | + |
| pGBT9:NSP | pGAD424:AtNSI(if)d,e | + | + |
| pGBT9:NSP | pGAD424:AtNSI(of)e,f | − | − |
| pGBT9:AtNSI | pGAD424:NSP | + | + |
BD, binding domain.
AD, activation domain.
No coexpressed vector.
if, AtNSI fused in frame to the GAL4 activation domain.
pGAD424, truncated ADH1 promoter to reduce fusion protein expression.
of, AtNSI cloned out of frame with the GAL4 activation domain.
NSP and AtNSI Interact Directly in Vitro
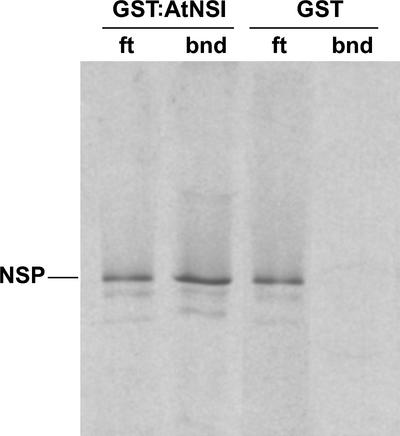
To demonstrate that NSP and AtNSI interacted directly, we tested the ability of NSP to bind to a glutathione S-transferase (GST):AtNSI fusion protein in vitro. 35S-Met–labeled NSP, synthesized by coupled in vitro transcription and translation, was incubated with bacterially expressed GST:AtNSI or unfused GST bound to glutathione-Sepharose beads. After extensive washing, bound protein was eluted and analyzed by SDS-PAGE and autoradiography. As shown in Figure 1, NSP bound specifically to GST:AtNSI. No NSP was detected binding to GST alone. In independent experiments, the amount of NSP bound to GST:AtNSI varied from 5 to 15% of the input, comparable to results reported for other proteins using this type of assay. Thus, NSP interacted directly with AtNSI in vitro in the absence of any additional proteins.
Figure 1.
NSP Interacts with AtNSI in Vitro.
In vitro–synthesized 35S-Met–labeled NSP was incubated with GST:AtNSI or GST bound to glutathione-Sepharose resin. Unbound (flow-through; ft) and bound (bnd) proteins were resolved on 12% SDS-PAGE gels and detected by autoradiography. Ten percent of the unbound protein is shown in the flow-through lane.
Similar results were obtained when a GST:NSP fusion and AtNSI fused to the GAL4 activation domain were cloned into a baculovirus vector and coexpressed in Sf21 insect cells. As expected if NSP and AtNSI interacted in vivo, both proteins, when coexpressed, were coimmunoprecipitated by NSP antiserum, and this antiserum did not immunoprecipitate AtNSI when it was expressed alone in Sf21 cells (data not shown).
AtNSI Is a Single-Copy Gene That Is Expressed Predominantly in Leaves
Sequence analysis of the 900-bp AtNSI cDNA XhoI insert identified an open reading frame of 765 nucleotides in frame with the GAL4 activation domain and showed this cDNA to be an incomplete clone. Using this XhoI fragment to screen a λ-GEM11 Arabidopsis genomic library, we identified three hybridizing plaques, each of which contained a single distinct SstI insert that included the identical genomic sequence corresponding to the AtNSI cDNA. Comparison of the AtNSI genomic and cDNA sequences showed that the latter appeared to lack only the first 12 nucleotides of the coding region. Consistent with this finding, RNA gel blot analyses of extracts from Arabidopsis leaves and stems identified a single AtNSI-hybridizing transcript of ∼1 kb (data not shown).
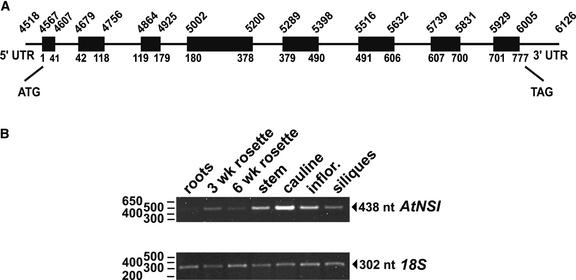
We used 5′ and 3′ rapid amplification of cDNA ends to verify this finding and to clone full-length AtNSI cDNA from Arabidopsis poly(A)-containing RNA. Clones for a single full-length cDNA of 946 nucleotides, predicted to encode a protein of 258 amino acids, were obtained. This cDNA contained the 777-nucleotide complete AtNSI coding sequence, flanked by a 49-nucleotide 5′ untranslated region and a 121-nucleotide 3′ untranslated region, which confirmed that the 5′ proximal ATG in the genomic clones was the start of the AtNSI coding sequence. The AtNSI genomic clone contained seven introns ranging in size from 72 to 118 nucleotides, with all of the inferred splice donor and acceptor sites conforming to the consensus sequences for Arabidopsis (Brown et al., 1996). This gene corresponds to and is colinear with nucleotides 4518 to 6126 of BAC T12O21 on chromosome 1 of the Arabidopsis genome sequence. The annotation of this genomic region based on our data is shown in Figure 2A.
Figure 2.
AtNSI Genomic Locus and Expression Pattern in Arabidopsis.
(A) Annotated AtNSI genomic locus based on genomic library analyses and the full-length cDNA sequence. Numbers indicate positions on Arabidopsis chromosome 1 (BAC T12O21). Boxes mark the positions of exons, with the nucleotides at the splice sites as indicated. Introns are shown as connecting lines. The 5′ and 3′ untranslated regions (UTR) and translational start and stop sites also are shown.
(B) Semiquantitative reverse transcriptase–mediated PCR analysis of AtNSI gene expression in Arabidopsis roots, rosette leaves (3 and 6 weeks after germination), cauline leaves, stems, inflorescences, and siliques. The 18S PCR product, as an internal control, was reverse transcribed in the same reaction as the corresponding 438-nucleotide AtNSI using gene-specific primers. nt, nucleotide.
The pattern of AtNSI expression was examined by semiquantitative reverse transcriptase–mediated PCR (Weigel and Glazebrook, 2002) using gene-specific primers to amplify AtNSI from total RNA isolated from the organs of mature Arabidopsis plants at 40 days after germination. AtNSI transcript levels were highest in cauline leaves (Figure 2B). AtNSI was expressed at lower (∼40%) but comparable levels in stems, siliques, inflorescences, and rosette leaves, and at very low levels in roots. This expression pattern of AtNSI is consistent with the pattern of infection by bipartite geminiviruses such as CLCV, which infect leaf tissue and move through the phloem but do not invade developing embryos or roots.
AtNSI Is a Novel Acetyltransferase That Does Not Act as a Transcriptional Coactivator
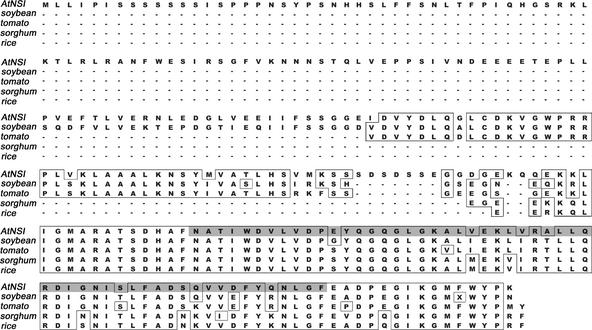
Sequence analysis of AtNSI showed it to be an uncharacterized gene predicted to encode a novel protein. However, we detected striking sequence identity between AtNSI and the largest in-frame open reading frame predicted to be encoded by ESTs from rice, soybean, tomato, and sorghum, as shown in Figure 3. No such striking similarity was found between AtNSI and any proteins predicted to be encoded by nonplant species, including yeast, Drosophila melanogaster, Caenorhabditis elegans, and humans, suggesting that AtNSI is highly conserved among divergent plant species.
Figure 3.
AtNSI Is Highly Conserved among Plants.
The predicted amino acid sequence of AtNSI is aligned with the longest predicted in-frame polypeptides for ESTs from soybean (Glycine maximus), tomato (Lycopersicon esculentum), sorghum (Sorghum propinquum), and rice (Oryza sativa). Identical amino acids (consensus from a minimum of three sequences) are boxed. The predicted GNAT-type acetyltransferase domain in AtNSI is shaded in gray.
A position-specific iterative Basic Local Alignment Search Tool (PSI-BLAST) search predicted residues 175 to 244 of AtNSI to contain an acetyltransferase domain belonging to the GCN5-like N-acetyltransferase (GNAT) family. The primary sequence of AtNSI was only distantly related to that of characterized GNAT family members, including the Saccharomyces cerevisiae histone acetyltransferase GCN5 and its homologs from Drosophila and Arabidopsis. Conserved residues in the active site of GCN5 that are essential for its enzymatic activity were present within the predicted AtNSI acetyltransferase domain (Smith et al., 1998). However, AtNSI lacked a bromodomain, which plays a role in anchoring nuclear histone acetyltransferases and other transcriptional coactivators, including those in the GNAT family, onto active chromatin (Dhalluin et al., 1999; Jacobson et al., 2000; Owen et al., 2000).
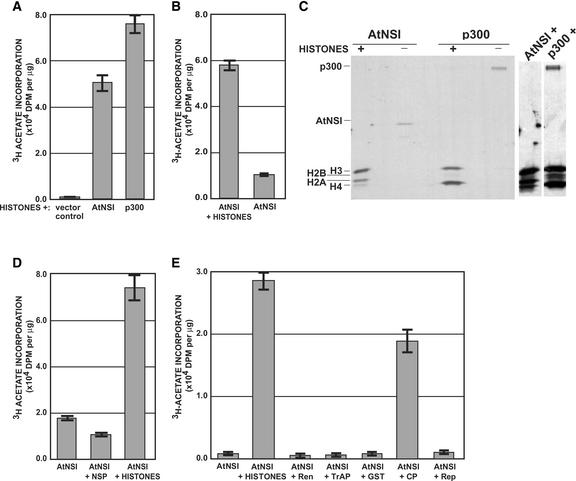
We tested the ability of AtNSI to acetylate histones in vitro. A GST:AtNSI fusion protein with a thrombin cleavage site inserted between the GST and AtNSI sequences was purified from Escherichia coli lysates by affinity chromatography on glutathione-Sepharose beads, and AtNSI was released from the resin-bound GST by thrombin digestion. GST expressed in E. coli and treated identically was used as a negative control, and p300 from Drosophila was included as a positive control. As shown in Figure 4A, AtNSI acetylated histones with a specific activity comparable to that of p300. Similar to p300 and other transcriptional coactivators with acetyltransferase activity, AtNSI also exhibited autoacetylation activity when assayed in the absence of histones: 3H-acetate was incorporated into AtNSI to ∼10 to 15% of the levels observed when histones were present (Figures 4B and 4D). The in vitro acetylated proteins were analyzed by SDS-PAGE and autoradiography, which confirmed the autoacetylation of AtNSI (Figure 4C). This further showed that AtNSI strongly acetylated histone H3 and, to a lesser extent, histone H2A; however, it only weakly acetylated histone H4 (Figure 4C). This was in striking contrast to p300 and other characterized transcriptional coactivators, which predominantly acetylate histones H3 and H4 and weakly acetylate H2A and H2B in vitro, both as free histones and nucleosomes (Figure 4C) (Chen et al., 2001a).
Figure 4.
AtNSI Has Acetyltransferase Activity in Vitro.
(A) AtNSI acetylates histones. Purified AtNSI, Drosophila p300, or FLAG-HMK peptide (vector control) (300 ng each) was incubated with 1 μg of calf thymus histones and 3H-acetyl-CoA. 3H-acetate incorporation was determined by filter binding assays.
(B) AtNSI has autoacetylation activity. AtNSI (300 ng) was incubated with 3H-acetyl-CoA in the presence or absence of 1 μg of calf thymus histones. Incorporated counts were determined by filter binding assays.
(C) AtNSI acetylates histones H3 and H2A in vitro. Purified AtNSI or Drosophila p300 (300 ng each) was incubated with 3H-acetyl-CoA in the presence (+) or absence (−) of 4 μg of purified Drosophila histones. Reaction products were resolved on 15% SDS-PAGE gels and analyzed by autoradiography. The lanes at right show overexposures of lanes AtNSI (+) histones and p300 (+) histones, showing resolution of all four histones.
(D) AtNSI does not acetylate NSP in vitro. AtNSI (300 ng) was incubated with 3H-acetyl-CoA and 1 μg of either purified CLCV NSP or calf thymus histones. Incorporated counts were determined by filter binding assays.
(E) AtNSI acetylates CP in vitro. His6:AtNSI (300 ng) was incubated with 3H-acetyl-CoA and 1 μg of calf thymus histones, GST:TrAP, GST:Ren, GST, His6:CP, or His6:Rep. 3H-acetate incorporation was determined by trichloroacetic acid precipitation.
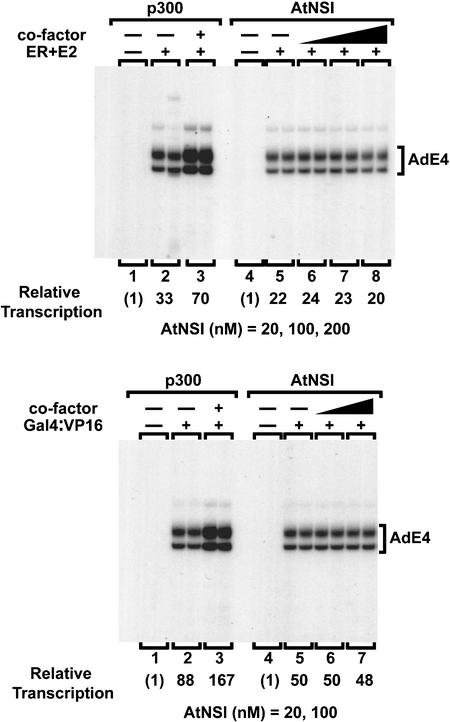
We tested the ability of AtNSI to enhance transcription mediated by the estrogen receptor bound to its ligand or by a Gal4:VP16 fusion protein using an in vitro chromatin assembly and transcription system. As shown in Figure 5, p300 enhanced transcriptional activation by the estrogen receptor or by Gal4:VP16 by approximately twofold, as reported previously (Kraus et al., 1999). By contrast, AtNSI, tested over a range of concentrations, did not increase the activity of these sequence-specific transcriptional activators.
Figure 5.
AtNSI Does Not Act as a Transcriptional Coactivator in Vitro.
Plasmid pERE, containing four copies of the estrogen response element, or pGIE0, containing five GAL4 binding sites upstream of the adenovirus E4 core promoter (AdE4), was assembled into chromatin with the estrogen receptor (ERα) bound to 17-β-estradiol (E2) or a GAL4:VP16 fusion, respectively (Kraus et al., 1999), after which purified p300 or AtNSI (20, 100, and 200 nM) was added as indicated. RNA transcripts were analyzed by primer extension, and duplicate reactions were quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
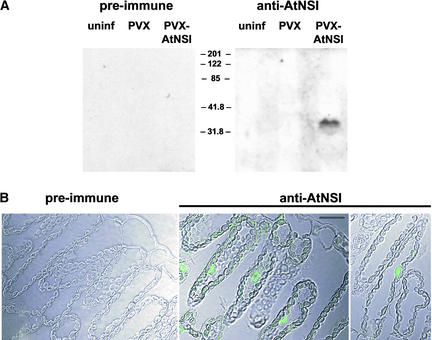
AtNSI Accumulates in the Nuclei of Plant Cells
Acetyltransferases have been reported to be active in both the nuclear and cytoplasmic compartments. Thus, to determine the potential role of AtNSI in NSP function in vivo, we investigated the subcellular localization of AtNSI in plant cells. Using rabbit polyclonal antiserum specific for AtNSI, we could not detect AtNSI in immunoblots of protein extracts from Arabidopsis. As an alternative approach, AtNSI was expressed in Nicotiana benthamiana plants by infecting them with a Potato virus X (PVX) vector that was engineered to express AtNSI from a duplicated copy of the viral coat protein promoter (PVX-AtNSI). As shown in Figure 6A, AtNSI antiserum specifically detected AtNSI in extracts from PVX-AtNSI systemically infected leaves migrating at ∼35 kD, similar to the size of AtNSI expressed in E. coli. This protein was not present in systemically infected leaves from wild-type PVX-infected plants, nor was it detected by preimmune serum in extracts from PVX- or PVX-AtNSI–infected plants.
Figure 6.
AtNSI Is a Nuclear Protein.
(A) AtNSI is detected only in PVX-AtNSI–infected plants. Immunoblot of extracts from uninfected N. benthamiana plants (uninf) or plants infected with either wild-type PVX or PVX-AtNSI. Membranes were incubated with preimmune or AtNSI antiserum (anti-AtNSI).
(B) Indirect immunofluorescence staining of methacrylate-embedded sections of symptomatic systemic leaves from N. benthamiana plants infected with PVX-AtNSI. Sections were incubated with preimmune or AtNSI antiserum, as indicated, followed by AlexaFluor 488–conjugated goat anti-rabbit antibody, and imaged by confocal microscopy. Each immunofluorescence image is superimposed on the Nomarski image. Bar = 20 μm.
To localize AtNSI in plant cells, symptomatic leaves from N. benthamiana plants infected with PVX-AtNSI or wild-type PVX were fixed and embedded in methacrylate, and tissue sections were examined by indirect immunofluorescence staining and confocal laser-scanning microscopy. AtNSI was localized specifically to the nuclei of mesophyll and epidermal cells in PVX-AtNSI–infected plants (Figure 6B). AtNSI was not detected when sections from PVX-AtNSI–infected plants were incubated with preimmune serum (Figure 6B), nor was it found when anti-AtNSI antiserum was used to stain sections from wild-type PVX-infected plants (data not shown). This localization of AtNSI to plant cell nuclei, in the context of the nuclear localization of NSP and its function as a nuclear shuttle protein (Pascal et al., 1994; Sanderfoot et al., 1996), suggests that AtNSI and protein acetylation function in the nuclear activity of NSP to modulate NSP shuttling, its binding to the viral genome, and/or other nuclear aspects of geminivirus replication.
CP, and Not NSP, Is the in Vitro Target of AtNSI Acetylation
To further address the potential role of AtNSI in the nuclear aspects of virus replication and movement, we tested the five geminivirus-encoded proteins that are targeted to the nucleus—NSP, CP, the viral transcriptional transactivator TrAP, the replication initiation protein Rep, and the replication enhancer Ren—as potential substrates for acetylation by AtNSI. NSP was a potential substrate for acetylation given its interaction with AtNSI. However, CP also was considered a likely target given its role in sequestering progeny genomes away from the replication pool so that they are available for nuclear export by NSP (Qin et al., 1998).
CLCV NSP was not acetylated by AtNSI. When AtNSI was incubated with purified NSP in vitro in the presence of 3H-acetyl CoA (acetyl-CoA), there was no increase in the level of 3H-acetate incorporation above that found for AtNSI autoacetylation (Figure 4D). SDS-PAGE analysis of proteins labeled in vitro confirmed that all of the labeled acetate was incorporated into AtNSI, with none being detected in NSP (data not shown). Similarly, CLCV TrAP, Rep, and Ren were not acetylated by AtNSI in vitro (Figure 4E). However, AtNSI did acetylate CLCV CP in vitro, with 3H-acetate being incorporated to ∼75% of the levels found using calf thymus histones as a substrate (Figure 4E). Affinity-purified AtNSI containing an N-terminal HIS6 tag was used in this last set of studies. The autoacetylation activity of this His6:AtNSI was reproducibly lower than that of thrombin-cleaved AtNSI prepared from the GST:AtNSI fusion protein (compare Figure 4E with Figures 4A, 4B, and 4D). Correlated with this, the thrombin-cleaved AtNSI preparations contained, in addition to the full-length AtNSI, truncated AtNSI consisting of the C-terminal ∼80% of the protein. This finding suggests that the folded structure of intact AtNSI may be important in regulating its activity.
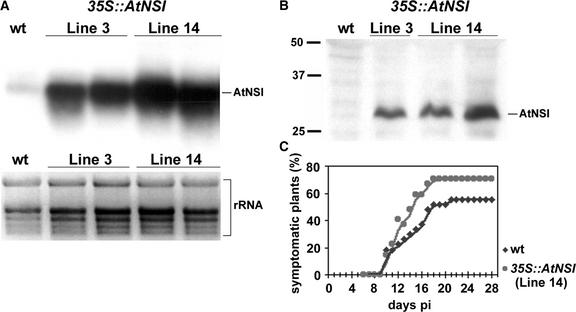
To demonstrate the relevance of AtNSI acetylation of viral CP to virus infectivity, we examined the ability of CLCV to infect transgenic Arabidopsis lines that overexpressed AtNSI from a 35S::AtNSI transgene. As shown in Figures 7A and 7B, Arabidopsis 35S::AtNSI T3 lines accumulated higher levels of AtNSI transcript and protein than did wild-type plants. When the AtNSI-overexpressing transgenic Arabidopsis plants were inoculated with CLCV, the efficiency of virus infection was increased compared with that in wild-type Arabidopsis (Figure 7C). In duplicate experiments, the efficiency of CLCV infection on transgenic 35S::AtNSI Arabidopsis was 1.2- and 1.3-fold higher that it was on wild-type plants.
Figure 7.
Overexpression of AtNSI Enhances CLCV Infection Efficiency in Arabidopsis.
(A) and (B) RNA gel blot (A) and immunoblot (B) analyses of extracts from wild-type Arabidopsis (wt) or transgenic Arabidopsis lines expressing a 35S::AtNSI transgene. In (A), equal amounts of total RNA were loaded for each sample and hybridized with a gene-specific riboprobe. rRNA was used as an internal loading control. RNA from siblings for two different T3 lines are shown. In (B), the immunoblot was incubated with AtNSI antiserum. Samples are as marked in (A). Siblings for T3 line 14 are shown. AtNSI at the expected mobility of ∼30 kD is marked.
(C) Infectivity assay of CLCV on wild-type (wt) Arabidopsis plants or transgenic Arabidopsis T3 line 14 that overexpresses AtNSI (35S::AtNSI). Values shown are percentages of plants that display systemic disease symptoms at different days after inoculation (pi).
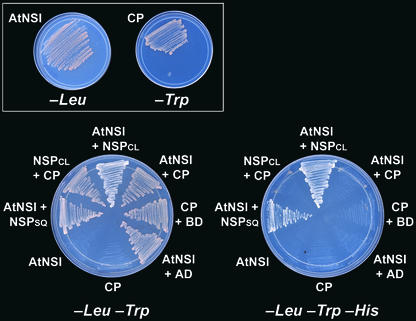
Given its ability to acetylate the viral CP, we investigated whether AtNSI could interact directly with CLCV CP. We further examined the general significance of AtNSI binding to CLCV NSP by testing whether AtNSI also could interact with NSP encoded by SqLCV. Although CP encoded by CLCV, SqLCV, and the other New World bipartite geminiviruses are essentially identical in sequence, CLCV NSP and SqLCV NSP are only 63% identical in sequence (∼90% similar), similar to the divergence of NSP among the other New World bipartite geminiviruses. AtNSI did not interact with CLCV CP when tested in a yeast two-hybrid assay. However, using this same assay, AtNSI did interact with NSP encoded by both SqLCV and CLCV, as shown in Figure 8.
Figure 8.
AtNSI Interacts with Both CLCV and SqLCV NSP but Not with Viral CP.
Yeast two-hybrid assays testing the ability of AtNSI to interact with NSP of SqLCV (NSPSQ) and CLCV (NSPCL) and with CLCV CP. Yeast coexpressing proteins as indicated were grown on SC −Leu −Trp medium (left plate), and interaction was tested by selection on SC −Leu −Trp −His (right plate). Negative controls included AtNSI fused to the GAL4 binding domain, expressed alone (AtNSI) or with the GAL4 activation domain (AtNSI + AD), and CLCV CP fused to the GAL4 activation domain, expressed alone (CP) or with the GAL4 binding domain (CP + BD). The inset shows the viability of yeast expressing AtNSI or CP alone.
DISCUSSION
To propagate bipartite geminivirus infection within the plant requires the cooperative interaction of NSP and MP to move the viral genome from its site of replication in the nucleus through the cytoplasm and across the cell wall. The interaction of NSP with replicated viral genomes and with MP, as well as the availability of these movement proteins, must be regulated to ensure that progeny genomes are directed to and across the cell wall and that this does not occur before transcription and replication of the incoming viral DNA. Previous studies of NSP have shown that its ability to export viral progeny ssDNA genomes from the nucleus is dependent on the activity of the CP to bind these newly replicated genomes and sequester them away from the replication pool (Sanderfoot et al., 1996; Qin et al., 1998). Our identification of AtNSI, based on its specific interaction with NSP, and our demonstration that it is an acetyltransferase that can acetylate CLCV CP in vitro now suggest that protein acetylation is important in regulating these nuclear events to facilitate virus movement.
Our data support the conclusions that AtNSI and NSP interact directly and that this interaction is specific, physiologically relevant, and important for CLCV infection. AtNSI and CLCV NSP were shown to interact both in vitro, in the absence of other proteins, and in vivo, using three independent assays: the classic yeast two-hybrid system, a GST pull-down assay, and coimmunoprecipitation from Sf21 insect cells. In addition, AtNSI, when expressed from a PVX vector, accumulated in the nuclei of infected N. benthamiana cells. Thus, AtNSI is a nuclear protein in plant cells, consistent with a role in the nuclear aspects of NSP function. The finding that CLCV infectivity was enhanced in transgenic Arabidopsis plants that overexpressed AtNSI suggests the importance of this interaction in vivo for CLCV infection. The extent of this enhancement (∼30%) is in good agreement with published studies showing that in the absence of CP, bipartite geminivirus infection of natural host plants is delayed and the efficiency may be decreased somewhat (∼20% lower levels at most in the case of SqLCV) (Ingham et al., 1995, and references cited therein). Also consistent with our results, preliminary studies show that the efficiency of CLCV infection is decreased in a T-DNA–mutated line that does not express AtNSI (R. McGarry and S. Lazarowitz, unpublished data). That AtNSI, which appears to be highly conserved in plants, interacted with NSP encoded by both CLCV and SqLCV further suggests its general importance in bipartite geminivirus infection.
The pattern of AtNSI transcript expression also correlated with the pattern of CLCV infection and spread in Arabidopsis. We detected the highest levels of AtNSI transcript in young cauline leaves, rosette leaves, and stems. Consistent with this finding, CLCV infection of Arabidopsis produces disease symptoms of downward leaf curl and severe stunting, killing the plant before bolting, and virus does not move to the roots (Hill et al., 1998). Our AtNSI antiserum did not detect AtNSI in wild-type Arabidopsis plants using leaf tissue prints, immunoblots of total plant extracts, or indirect immunofluorescence staining of methacrylate-embedded sections of rosette leaves or isolated protoplasts. This finding suggests that AtNSI accumulates to low levels in the plant and/or its expression is restricted to specific cell types, a suggestion also supported by preliminary in situ hybridization studies (Y.D. Barron and S.G. Lazarowitz, unpublished data). SqLCV infection in squash and pumpkin is restricted to the phloem (Lazarowitz et al., 1998), and geminivirus replication in general is restricted to cells in S-phase (Nagar et al., 1995). If AtNSI expression was limited similarly, it would be difficult to detect it by the methods used. We are currently investigating the developmental and tissue-specific expression of AtNSI to address these issues.
Since the identification of GCN5, CBP, and p300 as histone acetyltransferases, a large number of novel eukaryotic acetylases, most of which are linked to transcriptional regulation, have been identified, and the majority of these fall into two categories based on structural similarity to GCN5 (GNAT) or to MOZ, yeast Ybf2/Sas3 and Sas2, and TIP60 (MYST) (Chen et al., 2001b). The acetyltransferase domain of AtNSI is predicted to be related to that of the GNAT family of nuclear histone acetyltransferases, and our studies show that AtNSI can acetylate free histones in vitro. Based on its primary amino acid sequence and phylogenetic analyses, this region of AtNSI is related only distantly to known members of the GNAT family. Its acetyltransferase domain was more similar to that of a few known prokaryotic acetyltransferases such as E. coli RimI (Yoshikawa et al., 1987) and putative acetyltransferase domains within hypothetical proteins from Synechocystis, Porophyra, and Prochlorococcus species than to that of eukaryotic histone acetyltransferases such as yeast GCN5 and its homologs from Drosophila and Arabidopsis (Smith et al., 1998; Stockinger et al., 2001). Beyond this, except for having an acetyl-CoA binding site, AtNSI was not related to the MYST family or other characterized nuclear histone acetyltransferases, such as the transcriptional coactivators p300/CBP, SRC-1, and ACTR.
AtNSI has structural features and properties that distinguish it from all characterized histone acetyltransferases. Many histone acetyltransferases, including p300/CBP, TAFII250, and GNAT family members such as GCN5 and PCAF, possess an ∼110–amino acid module termed the bromodomain. Although the function of this domain has yet to be resolved, structural and mutational studies suggest that during transcriptional activation the bromodomain binds specific acetylated Lys residues in the N-terminal tails of histones H3 and H4, indicating a role in protein–protein interactions to anchor histone acetyltransferases and other factors onto active chromatin (Dhalluin et al., 1999; Jacobson et al., 2000; Owen et al., 2000). AtNSI does not contain a bromodomain. MYST family members also lack a bromodomain; however, they are characterized by a conserved ∼270–amino acid domain that encompasses an atypical C2HC zinc finger motif that in HBO1 has been shown to be essential for interactions with both the replication protein MCM2 and the origin-recognition protein ORC1 (Burke et al., 2001). p300 and CBP interact with a wide range of cellular and mammalian virus sequence-specific transcription factors to “bridge” these to the basal transcription machinery and thereby integrate diverse signaling pathways to coordinately regulate eukaryotic gene expression, and these interactions are mediated by three Cys/His (C/H)-rich regions, together with N-terminal and KIX interactive domains (reviewed by Chen et al., 2001b). AtNSI does not contain any putative zinc finger or C/H-rich regions; rather, it contains only a single Cys (residue 124) found upstream of the acetyltransferase domain.
Perhaps most striking, AtNSI strongly acetylated purified histone H3 and, to a lesser extent, histone H2A, but only weakly acetylated histone H4. This finding is in contrast to all characterized transcriptional regulators with demonstrated histone acetyltransferase activity, which, with the exception of ATF-2, have been reported to predominantly acetylate free H3 and H4 in vitro (Chen et al., 2001b, and references cited therein). ATF-2, a sequence-specific transcription factor, acetylates both free and nucleosome-bound H2B and H4 in vitro (Kawasaki et al., 2000). Consistent with these distinct structural and enzymatic features of AtNSI, AtNSI did not act as a coactivator when tested with two distinct transcriptional activators in vitro. We conclude that AtNSI is not a transcriptional coactivator and suggest that histones may not be the relevant in vivo targets of AtNSI.
Beyond their key role in chromatin remodeling during eukaryotic transcriptional activation and elongation, nuclear histone acetyltransferases have been implicated in DNA replication, DNA recombination, and DNA repair and apoptosis (Iizuka and Stillman, 1999; Ikura et al., 2000; McMurry and Krangel, 2000; Burke et al., 2001). p300/CBP and PCAF, in particular, through their action on nonhistone target proteins, are emerging as key integrators in stabilizing or destabilizing protein complexes to modulate transcription and a potential range of biological processes, including cell proliferation and differentiation. In this context, it is not surprising that cellular kinases have been reported to regulate the acetyltransferase activities of CBP and GCN5 and that several cellular and viral proteins recently were found to modulate and possibly retarget the acetyltransferase activities of p300/CBP and PCAF (reviewed by Hottiger and Nabel, 2000).
All DNA viruses control the availability of viral proteins through the temporal regulation of viral gene expression, in which “early” proteins required for replication and transcriptional regulation of viral gene expression are synthesized before the onset of DNA replication, and “late” proteins needed to package and transport progeny genomes are synthesized after the initiation of DNA replication. This tactic is one factor in geminivirus movement in that transcription of both NSP and CP is regulated by the early viral transactivator TrAP so that both are synthesized following the onset of DNA replication (Sunter and Bisaro, 1992). Coupled to this temporal regulation, those eukaryotic DNA viruses that require host cell DNA synthesizing enzymes for their replication must induce cells to enter S-phase or maintain cells in an undifferentiated state. Thus, viruses such as SV40, the oncogenic human papilloma virus HPV-16, and adenoviruses encode early proteins that bind and inactivate the two key players in inducing cell cycle arrest, Rb (and the related p107 and p130 proteins) and p53 (Flint and Shenk, 1997). Given that p300 and CBP are coactivators of p53 and play a central role in regulating processes that include cell proliferation and differentiation (Goodman and Smolik, 2000; Chen et al., 2001b), it is not surprising that some of these same virus-encoded early proteins also appear to target and modulate the activities of p300 and CBP.
Adenovirus E1A, long known to bind Rb to lead to the release of E2F family transcription factors and the induction of S-phase, has been reported to recruit p300 into a ternary complex with Rb that leads to increased acetylation of Rb, although the functional consequences of this remain to be defined (Chan et al., 2001). SV40 large T, which targets both Rb and p53, and HPV-16 E6, which inactivates p53, bind both p300 and CBP to competitively disrupt their interactions with p53 and other cellular targets (Eckner et al., 1996; Patel et al., 1999). E1A also competitively disrupts the interactions of CPB and p53, but its binding to CBP and p300 can have additional consequences, such as repressing transcriptional signaling by displacing PCAF and P/CIP. E1A also can repress p300- and PCAF-dependent transcription by directly inhibiting their acetylation of histones and the p300 acetylation of p53 to interfere with coactivator recruitment, or by stimulating the p300 acetylation of c-Jun to destabilize its promoter binding, although this may be promoter specific (Yang et al., 1996; Chakravarti et al., 1999; Hamamori et al., 1999; Vries et al., 2001). CBP, in addition to functioning as a transcriptional coactivator for many cellular transcription factors, also can act as a coactivator for viral transcriptional activators such as E1A, large T, E6, and EBV Zta; and E1A and Zta, as well as the cellular basic domain/Leu zipper proteins Nf-E2 and C/EPBα, have been reported to stimulate the nucleosomal acetyltransferase activity of CBP (Fax et al., 2000; Chen et al., 2001a).
How might the interaction of NSP and AtNSI affect virus movement and replication? Our finding that AtNSI acetylated CLCV CP in vitro suggests that this activity is important for NSP to displace CP and bind progeny genomes to export them from the nucleus. Upon infection, the geminivirus ssDNA genome is converted by host nuclear enzymes into double-stranded DNA minichromosomes, which are templates for transcription and for rolling-circle replication to generate progeny ssDNA genomes (Pilartz and Jeske, 1992; Bisaro, 1996). Genetic epistasis and biochemical studies have shown that viral CP, independent of its role in encapsidating progeny virus particles, aids NSP in the nuclear phase of virus movement by binding replicated progeny genomes to sequester them away from the replication pool, thereby making these viral ssDNAs available for NSP binding and export (Qin et al., 1998). It has been proposed that prior to the accumulation of high levels of CP sufficient to assemble capsids, NSP would cooperatively bind to the newly synthesized progeny genomes to displace CP and export the viral ssDNA to the cytoplasm. This model predicts that there are at least two pools of CP in the nucleus.
In this context, and given our findings that NSP is not acetylated by AtNSI and that AtNSI does not stably interact with CP, we propose that NSP, bound to newly replicated viral ssDNA, recruits AtNSI to a ternary complex with the genome-bound CP to acetylate CP, which would disrupt CP ssDNA binding. This would allow NSP to displace CP as NSP cooperatively binds the viral genome to export it from the nucleus. This model predicts that overexpression of AtNSI should not affect CP binding to viral ssDNA per se because, in the absence of viral ssDNA and NSP, AtNSI and CP will not interact stably. However, high levels of AtNSI would accelerate the release of CP from the ssDNA, and thus export of the CLCV genome and virus movement, because it would increase the rate at which NSP recruits AtNSI into a ternary complex to acetylate CP bound to the viral genome. Consistent with this, we found that the efficiency of CLCV infection was enhanced in transgenic Arabidopsis plants that overexpress AtNSI. This model does not preclude additional potential roles for AtNSI and histone acetylation in replication of the viral minichromosome.
The question remains as to what the function of AtNSI is in plant cells because it does not appear to be a transcriptional coactivator. That AtNSI is novel, with homologs identified to date only among divergent plant species, suggests that if it plays a role in chromosome replication, this likely would involve aspects that are unique to this process in plant cells. AtNSI also could play a role in regulating plant cell growth and differentiation, acetylating proteins that induce cellular differentiation similar to targets of p300 or TIP60. Several geminiviruses, including Tomato golden mosaic virus (TGMV) and Wheat dwarf virus (WDV), have been reported to infect differentiated plant cells, and TGMV Rep and WDV Rep and/or the transcriptional regulator RepA have been reported to interact with plant proteins related to mammalian Rb in yeast two-hybrid assays or when coexpressed in insect cells (Xie et al., 1995; Collin et al., 1996; Kong et al., 2000). These findings have led to the suggestion that Rep or RepA, like the early proteins encoded by mammalian DNA viruses, may act to induce plant cells to enter S phase. In contrast to geminiviruses such as TGMV, SqLCV infection is limited to the phloem, with SqLCV infecting and replicating in undifferentiated phloem cells, and differences in viral Rep do not appear to account for this phloem limitation (Lazarowitz et al., 1998; Kong et al., 2000). Our preliminary studies indicate that high concentrations of NSP can, in the absence of DNA, inhibit the AtNSI-catalyzed acetylation of histones in vitro. Thus, as proposed for MCM2–HBO1 and ORC2–HBO1 interactions, we suggest that an additional function of NSP binding to AtNSI may be to downregulate AtNSI acetyltransferase activity to maintain infected phloem or mesophyll cells in a dedifferentiated state. Investigation of Arabidopsis AtNSI mutants will examine this proposed role of AtNSI in growth and differentiation.
METHODS
Yeast Two-Hybrid Screens
Yeast two-hybrid screens in Saccharomyces cerevisiae strain Y190 (MATa ade2-101 gal4Δ gal80Δ his3-200 leu2-3,112 trp1-901 ura3-52 LYS::GAL1-HIS3 URA3::GAL1-lacZ) used the GAL4 binding domain plasmid pGBT9 and Arabidopsis thaliana cDNA libraries from either etiolated seedlings or mature leaves and roots cloned in the GAL4 activation domain plasmid pACT (ABRC, Columbus, OH) (Durfee et al., 1993; Kim et al., 1997). The NSP coding sequence, amplified by PCR from the CLCV B component (Hill et al., 1998) with unique EcoRI and BamHI restriction sites, was cloned directionally into pGBT9 (Clontech, Palo Alto, CA) to create pGBT9:NSP. Y190 cells were transformed sequentially with pGBT9:NSP and pACT cDNA libraries. Transformants were selected on synthetic complete (SC) medium that lacked Trp, Leu, and His (SC −Trp −Leu −His) and contained 25 mM 3-aminotriazole (Sigma) and were assayed for β-galactosidase activity. Clones were recovered in Escherichia coli JA226 by selection on M9 −Leu plates. Those that contained inserts were retransformed into Y190 with pGBT9:NSP to verify the interaction, and the inserts were sequenced. Interactions were verified further by cloning the 900-nucleotide AtNSI XhoI insert into pGAD424 (Clontech) and retesting with pGBT9:NSP.
The third screen in S. cerevisiae strain PJ694-a (MATa gal4 gal80 his3-Δ200 leu2-3,112 trp1-Δ901 LYS2::GAL1-HIS3 ade2::GAL2-ADE2 met::GAL7-lacZ) used the GAL4 binding domain plasmid pBI880 and an Arabidopsis cDNA library constructed from aerial tissues harvested at four developmental stages and cloned in the GAL4 activation domain plasmid pBI771 (Samach et al., 1999). The screens were as described above, except that (1) NSP was PCR-amplified to introduce unique SalI and NotI restriction sites, (2) interacting clones were selected by growth on SC −Trp −Leu −His plus 5 mM 3-aminotriazole or on supplemented minimal medium without adenine, and (3) plasmids were recovered in E. coli DH5α and identified by SalI and NotI restriction analysis.
To test the interaction of AtNSI with SqLCV NSP and CLCV CP, AtNSI and CP were amplified by PCR with SalI and NotI sites and cloned directionally into pBI880 and pBI771, respectively. These constructs, or pGBT9:AtNSI with pACT:NSPSQ, were cotransformed into PJ694-a cells and plated on SC −Trp −Leu −His with and without 5 mM 3-aminotriazole.
Genomic Library Screen
An Arabidopsis genomic library in λ-GEM11 (J.T. Mulligan and R.W. Davis, Department of Biochemistry, Stanford University, CA) was screened with the γ-32P-dCTP–labeled 900-nucleotide AtNSI cDNA using standard protocols (Ausubel et al., 1999). Positive clones were screened by restriction enzyme and sequence analyses.
Cloning of Full-Length AtNSI cDNA
Total RNA from aerial tissues of 8-day-old Arabidopsis plants (ecotype Columbia) grown at 22°C under a 16-h-day/8-h-night cycle was isolated with TRIzol (Gibco BRL). AtNSI 5′ and 3′ ends were amplified from the reverse-transcribed template by touchdown PCR using GeneRacer (Invitrogen, Carlsbad, CA) primers with appropriate reverse gene-specific primers and Takara ExTaq polymerase (Panvera, Madison, WI). Rapid amplification of cDNA ends products were cloned into pCR4-TOPO (Invitrogen), recovered in One Shot TOP10 E. coli (Invitrogen), and sequenced using universal T7 and M13 primers. To construct the full-length AtNSI cDNA, overlapping 5′ and 3′ AtNSI fragments were amplified by PCR with Pwo polymerase (Roche, Indianapolis, IN) using primers that introduced unique XbaI and XhoI sites at the 5′ and 3′ ends of the cDNA and that used the unique HindIII site in the AtNSI coding sequence. The XbaI-HindIII and HindIII-XhoI products were cloned into XbaI- and XhoI-digested pBluescript SK+ (Stratagene).
Semiquantitative Reverse Transcriptase–Mediated PCR
Total RNA was isolated using TRIzol (Gibco BRL) from Arabidopsis (ecotype Columbia) tissue harvested at ∼40 days after germination. To amplify a 438-nucleotide AtNSI cDNA fragment and a 302-nucleotide 18S cDNA fragment, appropriate gene-specific primers were designed to distinguish transcripts from genomic DNA. AtNSI cDNA and 18S cDNA (internal control) were reverse transcribed from 500 ng of RNA (Ausubel et al., 1999) using gene-specific primers and Omniscript (Qiagen, Valencia, CA) and were amplified separately by PCR with Taq DNA polymerase (Promega) and the appropriate gene-specific primer sets. The number of cycles for amplification of AtNSI and 18S RNAs was optimized such that each was in the linear range. Twenty microliters of each reaction product was analyzed and quantified on ethidium bromide stained agarose gels using a GelDoc (Bio-Rad).
Protein Expression
To construct a translational fusion of GST and AtNSI (pGST:AtNSI), the 900-nucleotide XhoI insert from pACT:AtNSI was ligated (Klenow-blunted) into the SmaI site of pAR(ΔRI)[59/60] (Blanar and Rutter, 1992), which contains the glutathione S-transferase (GST) gene from Schistosoma japonicum, a thrombin recognition site, the FLAG epitope tag, and a heart muscle kinase (HMK) site. E. coli BL21(DE3) expressing pGST:AtNSI or pAR(ΔRI)[59/60] was induced with 0.5 mM isopropyl-d-thiogalactoside for 3 h at 28°C and purified by standard affinity chromatography on glutathione-Sepharose 4B resin (Amersham Pharmacia Biotech). N-terminal HIS6 fusions of AtNSI (pHis6:AtNSI), CLCV CP (pHis6:CP), and Rep (pHis6:Rep) were constructed in pET24a (Novagen, Madison, WI). The AtNSI coding sequence was amplified by PCR to introduce NdeI and BglII sites. The CP and Rep coding regions were cloned as BamHI-XhoI fragments from subclones in pAR(ΔRI)[59/60]. To construct pGST:Ren and pGST:TrAP, the Ren and TrAP coding regions were amplified by PCR from the CLCV genome and subcloned in pAR(ΔRI)[59/60] and the entire GST fusion was recloned into NdeI- and XhoI-digested pET24a. These constructs were expressed in E. coli BL21(DE3), induced with 1 mM isopropyl-d-thiogalactoside, and purified by standard affinity chromatography on Talon (Clontech) or glutathione-Sepharose 4B resin.
Preparation of AtNSI Antiserum
AtNSI was cleaved from GST:AtNSI bound to glutathione-Sepharose 4B by thrombin digestion (30 units/mL) (Calbiochem) for 3 h at 25°C in 50 mM Tris, pH 8.4, 150 mM NaCl, and 0.5 mM CaCl2 and purified further on and eluted from preparative 12% SDS-PAGE gels (Pascal et al., 1994). Rabbit polyclonal antiserum was prepared at the University of Illinois Immunological Resource Center. Ammonium precipitation, storage, and use of antibodies have been described (Sanderfoot and Lazarowitz, 1995).
GST Pull-Down Assays
NSP, cloned as a transcriptional fusion to the T7 promoter in pGEM7Zf(−), was labeled with 35S-Met (10.2 mCi/mL; DuPont–New England Nuclear) by coupled transcription and translation in vitro using the TNT Coupled Reticulocyte Lysate System (Promega). Ten microliters of 35S-labeled NSP was incubated with 5 μg of GST or GST:AtNSI bound to glutathione-Sepharose 4B resin in binding buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM EDTA, 8% glycerol, and 0.02% Nonidet P-40) for 2 h at 4°C. Protein bound to the resin was washed twice in binding buffer and twice in 50 mM Tris, pH 7.5, 250 mM NaCl, and 1 mM EDTA by pelleting (5200g for 2 min), eluted from the resin in 3× SDS-PAGE loading buffer (Ausubel et al., 1999), and analyzed by SDS-PAGE and autoradiography.
In Vitro Histone Acetyltransferase and Transcription Assays
AtNSI or the C-terminal FLAG-HMK peptide was cleaved from GST by digestion with thrombin, diluted 1:5 in storage buffer B (Ogryzko et al., 1996), and stored at −80°C. Protein concentrations were determined by Bradford assay (Bradford, 1976). For acetyltransferase assays, 300 ng of thrombin-cleaved AtNSI or FLAG-HMK peptide (negative control) was incubated alone or in the presence of 1 μg of the following substrates: calf thymus histones (Sigma), NSP (as a GST:NSP fusion or purified by thrombin cleavage), GST:Ren, GST:TrAP, GST (negative control), His6:CP, and His6:Rep (Ogryzko et al., 1996). The purity and integrity of each recombinant protein was assessed by SDS-PAGE. Reaction products were spotted directly on, or trichloroacetic acid–precipitated and collected on, GF/A or P81 filters (Whatman), and 3H-acetate incorporation was determined by scintillation counting. Results from studies using biotinylated thrombin (Novagen), which was removed with streptavidin-agarose (Novagen), were identical to those described. For gel assays, 4 μg of histones purified from Drosophila melanogaster embryos was used. Reaction products were resolved on 15% SDS-PAGE gels and detected by autoradiography using EN3HANCE (DuPont–New England Nuclear). Sf9-expressed Drosophila p300 and Drosophila histones were purified and in vitro transcription assays using His6:AtNSI and p300 were performed as described (Cheung et al., 2002).
PVX and Transgenic Expression of AtNSI
The AtNSI coding sequence flanked by SalI sites was amplified by PCR and cloned into the unique SalI site downstream of the duplicated PVX coat protein promoter in the binary vector pQUIET to construct pPVX-AtNSI. pQUIET is a derivative of the binary vector pCB302-3 that contains the PVX insert from pP2C2S (Baulcombe et al., 1995) with a modified multiple cloning site (A.J. Bogdanove and G.B. Martin, unpublished data). Agrobacterium tumefaciens strain GV3101 containing the disarmed Ti plasmid pMP90 (Koncz and Schell, 1986) was grown at 28°C as described (Hill et al., 1998) and concentrated 10-fold. Nicotiana benthamiana plants grown under a 16-h-day/8-h-night (23/18°C) cycle were inoculated at the six- to seven-leaf stage with ∼150 μL of A. tumefaciens containing pQUIET or pPVX-AtNSI (Huang et al., 1988). Proteins extracted from systemically infected leaves were resolved on 12% SDS-PAGE gels and analyzed on immunoblots (Pascal et al., 1994) using enhanced chemiluminescence (Amersham Pharmacia Biotech).
To generate transgenic Arabidopsis lines that overexpressed AtNSI, the PCR-amplified AtNSI coding sequence was cloned as a transcriptional fusion to the 35S promoter in pCambia pC1302 (CAMBIA, Canberra, Australia), and Arabidopsis (ecotype Columbia) was transformed by floral dip using A. tumefaciens GV3101 (Clough and Bent, 1998). Hygromycin-resistant T3 lines were generated and agroinoculated with CLCV as described previously (Hill et al., 1998). Protein extracts from systemically infected leaves were analyzed as described above. Total RNA, extracted from systemically infected leaves using TRIzol (Gibco BRL), was analyzed on RNA gel blots using a gene-specific riboprobe as described previously (Pascal et al., 1994). Equal loading of RNA samples was confirmed by ethidium bromide staining to detect rRNA.
DNA Sequence Analyses
Symptomatic systemic (sink) leaves from PXV- or PVX-AtNSI–infected N. benthamiana were fixed in 4% paraformaldehyde and embedded in 4:1 n-butyl:methyl methacrylate (Baskin et al., 1992) with no DTT in the dehydration. Transverse 1-μm sections were cut and adhered to poly-l-Lys–coated slides (Sigma), and indirect immunofluorescence staining was performed as described (Sanderfoot and Lazarowitz, 1995) using goat anti-rabbit IgG conjugated to AlexaFluor 488 (Molecular Probes, Eugene, OR) as a secondary antibody. Slides were mounted with Prolong (Molecular Probes) and examined with a Fluoview confocal laser-scanning microscope (IX 70; Olympus, Tokyo, Japan).
DNA sequence analyses were performed at the University of Illinois Biotechnology Center (Urbana) or the Cornell University BioResource Center on an ABI Prism automated sequencer (Applied Biosystems, Foster City, CA). Genomic sequence was generated using custom oligonucleotide primers that annealed to the AtNSI cDNA sequence. The contiguous genomic sequence was assembled with the Sequencher 3.0 software package (Gene Codes, Ann Arbor, MI). Databank searches were performed through the National Center for Biotechnology Information site (http://www.ncbi.nlm.nih.gov) using the Basic Local Alignment Search Tool (BLAST).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The AtNSI full-length cDNA sequence has been deposited in GenBank with accession number AY281105. BAC T12O21 on chromosome 1 of the Arabidopsis genome sequence has GenBank accession number AC074309. The accession numbers for the sequences shown in Figure 3 are as follows: soybean (AW397089), tomato (BG128955), sorghum (BF58402), and rice (AUO57725).
Acknowledgments
We are grateful to Kari Peter for her exhaustive screening of Arabidopsis cDNA libraries and to Adam Bogdanove and Greg Martin for providing the pQUIET plasmid. As always, we thank the members of our laboratory for lively and provocative discussions. This work was supported by National Science Foundation Career Advancement Award MCB-9707580 and National Science Foundation Grants MCB-9417664 and MCB-9982622 to S.G.L., a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and National Institutes of Health Grant DK58110 to W.L.K., a Natural Sciences and Engineering Research Council of Canada postdoctoral fellowship to J.E.H., a Susan G. Komen Breast Cancer Foundation postdoctoral fellowship to E.C., a Natural Sciences and Engineering Research Council of Canada predoctoral fellowship to R.C.M., U.S. Department of Agriculture National Needs and Cornell University predoctoral fellowships to Y.D.B, and a Portuguese Government PRAXIS XXI FCT predoctoral fellowship to M.F.C.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012120.
References
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1999). Short Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Bannister, A.J., Miska, E.A., Gorlich, D., and Kouzarides, T. (2000). Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr. Biol. 10, 467–470. [DOI] [PubMed] [Google Scholar]
- Barlev, N.A., Liu, L., Chehab, N.H., Mansfield, K., Harris, K.G., Halazonetis, T.D., and Berger, S.L. (2001). Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8, 1243–1254. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., Busby, C.H., Fowke, L.C., Sammut, M., and Gubler, F. (1992). Improvements in immunostaining samples embedded in methacrylate: Localization of microtubules and other antigens throughout developing organs in plants of diverse taxa. Planta 187, 405–413. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C., Chapman, S., and Cruz, S.S. (1995). Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 7, 1045–1053. [DOI] [PubMed] [Google Scholar]
- Bisaro, D.M. (1996). Geminivirus replication. In DNA Replication in Eukaryotic Cells, M. DePamphilis, ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 833–854.
- Blanar, M.A., and Rutter, W.J. (1992). Interaction cloning: Identification of a helix-loop-helix zipper protein that interacts with c-fos. Science 256, 1014–1018. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brown, J.W.S., Smith, P., and Simpson, C.G. (1996). Arabidopsis consensus intron sequences. Plant Mol. Biol. 32, 531–535. [DOI] [PubMed] [Google Scholar]
- Burke, T.W., Cook, J.G., Asano, M., and Nevins, J.R. (2001). Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 276, 15397–15408. [DOI] [PubMed] [Google Scholar]
- Chakravarti, D., Ogryzko, V., Kao, H.Y., Nash, A., Chen, H.W., Nakatani, Y., and Evans, R.M. (1999). A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96, 393–403. [DOI] [PubMed] [Google Scholar]
- Chan, H.M., Krstic-Demonacos, M., Smith, L., Demonacos, C., and La Thangue, N.B. (2001). Acetylation control of the retinoblastoma tumour suppressor protein. Nat. Cell Biol. 3, 667–674. [DOI] [PubMed] [Google Scholar]
- Chen, C.-J., Deng, Z., Kim, A.Y., Blobel, G.A., and Liberman, P.M. (2001. a). Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol. 21, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.W., Lin, R.J., Xie, W., Wilpitz, D., and Evans, R.M. (1999). Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Chen, H.W., Tini, M., and Evans, R.M. (2001. b). HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13, 218–224. [DOI] [PubMed] [Google Scholar]
- Cheung, E., Zarifyan, A.S., and Kraus, W.L. (2002). Histone H1 represses estrogen receptor α transcriptional activity by selectively inhibiting receptor-mediated transcription initiation. Mol. Cell. Biol. 22, 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, P., Allis, C.D., and Sassone-Corsi, P. (2000). Signaling to chromatin through histone modifications. Cell 103, 263–271. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Collin, S., Fernandez-Lobato, M., Gooding, P.S., Mullineaux, P.M., and Fenoll, C. (1996). The two nonstructural proteins from wheat dwarf virus involved in viral gene expression and replication are retinoblastoma-binding proteins. Virology 219, 324–329. [DOI] [PubMed] [Google Scholar]
- Dhalluin, C., Carlson, J.E., Zeng, L., He, C., Aggarwal, A.K., and Zhou, M.M. (1999). Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Durfee, T., Becherer, K., Chen, P.L., Yeh, S.H., Yang, Y.Z., Kilburn, A.E., Lee, W.H., and Elledge, S.J. (1993). The retinoblastoma protein associates with the protein phosphatase type-1 catalytic subunit. Genes Dev. 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Eckner, R., Ludlow, J.W., Lill, N.L., Oldread, E., Arany, Z., Modjtahedi, N., DeCaprio, J.A., Livingston, D.M., and Morgan, J.A. (1996). Association of p300 and CBP with simian virus 40 large T antigen. Mol. Cell. Biol. 16, 3454–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fax, P., Lipinski, K.S., Esche, H., and Brockmann, D. (2000). cAMP-independent activation of the adenovirus type 12 E2 promoter correlates with the recruitment of CREB-1/ATF-1, E1A(12S), and CBP to the E2-CRE. J. Biol. Chem. 275, 8911–8920. [DOI] [PubMed] [Google Scholar]
- Flint, J., and Shenk, T. (1997). Viral transactivating proteins. Annu. Rev. Genet. 31, 177–212. [DOI] [PubMed] [Google Scholar]
- Goodman, R.H., and Smolik, S. (2000). CBP/p300 in cell growth, transformation, and development. Genes Dev. 14, 1553–1577. [PubMed] [Google Scholar]
- Hamamori, Y., Sartorelli, V., Ogryzko, V., Puri, P.L., Wu, H.Y., Wang, J.Y.J., Nakatani, Y., and Kedes, L. (1999). Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein Twist and adenoviral oncoprotein E1A. Cell 96, 405–413. [DOI] [PubMed] [Google Scholar]
- Hill, J.E., Strandberg, J.O., Hiebert, E., and Lazarowitz, S.G. (1998). Asymmetric infectivity of pseudorecombinants of cabbage leaf curl virus and squash leaf curl virus: Implications for bipartite geminivirus evolution and movement. Virology 250, 283–292. [DOI] [PubMed] [Google Scholar]
- Hottiger, M.O., and Nabel, G.J. (2000). Viral replication and the coactivators p300 and CBP. Trends Microbiol. 8, 560–565. [DOI] [PubMed] [Google Scholar]
- Huang, H.C., Schuurink, R., Denny, T.P., Atkinson, M.M., Baker, C.J., Yucel, I., Hutcheson, S.W., and Collmer, A. (1988). Molecular cloning of a Pseudomonas syringae cv syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J. Bacteriol. 170, 4748–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka, M., and Stillman, B. (1999). Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274, 23027–23034. [DOI] [PubMed] [Google Scholar]
- Ikura, T., Ogryzko, V.V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J., and Nakatani, Y. (2000). Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473. [DOI] [PubMed] [Google Scholar]
- Ingham, D.J., Pascal, E., and Lazarowitz, S.G. (1995). Both bipartite geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology 207, 191–204. [DOI] [PubMed] [Google Scholar]
- Jacobson, R.H., Ladurner, A.G., King, D.S., and Tjian, R. (2000). Structure and function of a human TAF(II)250 double bromodomain module. Science 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- Kawasaki, H., Schiltz, L., Chiu, R., Itakura, K., Taira, K., Nakatani, Y., and Yokoyama, K.K. (2000). ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature 405, 195–200. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Kong, L.J., Orozco, B.M., Roe, J.L., Nagar, S., Ou, S., Feiler, H.S., Durfee, T., Miller, A.B., Gruissem, W., Robertson, D., and Hanley-Bowdoin, L. (2000). A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19, 3485–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides, T. (2000). Acetylation: A regulatory modification to rival phosphorylation? EMBO J. 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus, W.L., Manning, E.T., and Kadonaga, J.T. (1999). Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19, 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz, S.G., Ward, B.M., Sanderfoot, A.A., and Laukaitis, C.M. (1998). Intercellular and intracellular trafficking: What we can learn from geminivirus movement. In Cellular Integration of Signalling Pathways in Plant Development, G. Morelli, F. Lo Schiavo, and N.V. Raikhel, eds (Heidelberg, Germany: Springer-Verlag), pp. 275–288.
- Martinez-Balbas, M.A., Bauer, U.M., Nielsen, S.J., Brehm, A., and Kouzarides, T. (2000). Regulation of E2F1 activity by acetylation. EMBO J. 19, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry, M.T., and Krangel, M.S. (2000). A role for histone acetylation in the developmental regulation of V(D)J recombination. Science 287, 495–498.10642553 [Google Scholar]
- Nagar, S., Pedersen, T.J., Carrick, K.M., Hanley-Bowdoin, L., and Robertson, D. (1995). A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry, A.O., Lucas, W.J., and Gilbertson, R.L. (1994). Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 76, 925–932. [DOI] [PubMed] [Google Scholar]
- Ogryzko, V.V., Schiltz, R.L., Russanova, V., Howard, B.H., and Nakatani, Y. (1996). The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Owen, D.J., Ornaghi, P., Yang, J.C., Lowe, N., Evans, P.R., Ballario, P., Neuhaus, D., Filetici, P., and Travers, A.A. (2000). The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 19, 6141–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal, E., Sanderfoot, A.A., Ward, B.M., Medville, R., Turgeon, R., and Lazarowitz, S.G. (1994). The geminivirus BR1 movement protein binds single-stranded DNA and localizes to the cell nucleus. Plant Cell 6, 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, D., Huang, S.M., Baglia, L.A., and McCance, D.J. (1999). The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 18, 5061–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilartz, M., and Jeske, H. (1992). Abutilon mosaic virus double-stranded DNA is packed into minichromosomes. Virology 189, 800–802. [DOI] [PubMed] [Google Scholar]
- Qin, S.W., Ward, B.M., and Lazarowitz, S.G. (1998). The bipartite geminivirus coat protein aids BR1 function in viral movement by affecting the accumulation of viral single-stranded DNA. J. Virol. 72, 9247–9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, H., Gregory, P.D., and Horz, W. (2001). A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell 7, 529–538. [DOI] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Ingham, D.J., and Lazarowitz, S.G. (1996). A viral movement protein as a nuclear shuttle: The geminivirus BR1 movement protein contains domains essential for interaction with BL1 and nuclear localization. Plant Physiol. 110, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., and Lazarowitz, S.G. (1995). Cooperation in viral movement: The geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell 7, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli, V., Puri, P.L., Hamamori, Y., Ogryzko, V., Chung, G., Nakatani, Y., Wang, J.Y.J., and Kedes, L. (1999). Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4, 725–734. [DOI] [PubMed] [Google Scholar]
- Smith, E.R., Belote, J.M., Schiltz, R.L., Yang, X.J., Moore, P.A., Berger, S.L., Nakatani, Y., and Allis, C.D. (1998). Cloning of Drosophila GCN5: Conserved features among metazoan GCN5 family members. Nucleic Acids Res. 26, 2948–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou, E., Katrakili, N., and Talianidis, I. (2000). Acetylation regulates transcription factor activity at multiple levels. Mol. Cell 5, 745–751. [DOI] [PubMed] [Google Scholar]
- Spilianakis, C., Papamatheakis, J., and Kretsovali, A. (2000). Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol. Cell. Biol. 20, 8489–8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Mao, Y.P., Regier, M.K., Triezenberg, S.J., and Thomashow, M.F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29, 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter, G., and Bisaro, D.M. (1992). Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4, 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vries, R.G.J., Prudenziati, M., Zwartjes, C., Verlaan, M., Kalkhoven, E., and Zantema, A. (2001). A specific lysine in c-Jun is required for transcriptional repression by E1A and is acetylated by p300. EMBO J. 20, 6095–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, B.M., and Lazarowitz, S.G. (1999). Nuclear export in plants: Use of geminivirus movement proteins for a cell-based export assay. Plant Cell 11, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, B.M., Medville, R., Lazarowitz, S.G., and Turgeon, R. (1997). The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 71, 3726–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Xie, Q., Suarezlopez, P., and Gutierrez, C. (1995). Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: Requirement for efficient viral DNA replication. EMBO J. 14, 4073–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.J., Ogryzko, V.V., Nishikawa, J., Howard, B.H., and Nakatani, Y. (1996). A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382, 319–324. [DOI] [PubMed] [Google Scholar]
- Yoshikawa, A., Isono, S., Sheback, A., and Isono, K. (1987). Cloning and nucleotide sequencing of the genes RimI and RimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet. 209, 481–488. [DOI] [PubMed] [Google Scholar]