Abstract
The plant mitochondrial genome is complex in structure, owing to a high degree of recombination activity that subdivides the genome and increases genetic variation. The replication activity of various portions of the mitochondrial genome appears to be nonuniform, providing the plant with an ability to modulate its mitochondrial genotype during development. These and other interesting features of the plant mitochondrial genome suggest that adaptive changes have occurred in DNA maintenance and transmission that will provide insight into unique aspects of plant mitochondrial biology and mitochondrial-chloroplast coevolution. A search in the Arabidopsis genome for genes involved in the regulation of mitochondrial DNA metabolism revealed a region of chromosome III that is unusually rich in genes for mitochondrial DNA and RNA maintenance. An apparently similar genetic linkage was observed in the rice genome. Several of the genes identified within the chromosome III interval appear to target the plastid or to be targeted dually to the mitochondria and the plastid, suggesting that the process of endosymbiosis likely is accompanied by an intimate coevolution of these two organelles for their genome maintenance functions.
INTRODUCTION
The plant mitochondrial genome is characterized by unusual structural complexity. A multipartite genome structure derives from both high-frequency recombination within defined repeated sequences in the genome and low-frequency ectopic recombination events (Fauron et al., 1995). Evidence of both types of activity is extensive in many higher plants (Mackenzie and McIntosh, 1999). Although plant mitochondrial genomes map as circular molecules, they appear to be predominantly linear in vivo (Bendich and Smith, 1990; Backert et al., 1997; Backert and Börner, 2000; Oldenburg and Bendich, 2001), and the role of recombination in their replication is not yet known. Although a rolling-circle replication mechanism has been speculated based on electron microscopic analyses of DNA structures (Backert and Börner, 2000), virtually none of the mitochondrial DNA metabolism apparatus has been well defined genetically or biochemically in plants.
One particularly intriguing behavior observed in plant mitochondria is a dynamic system of genome copy number modulation. With a multipartite genome structure, the plant mitochondrion contains a highly redundant gene assemblage organized within subgenomic DNA molecules. The copy number of at least some of these subgenomic molecules appears to be regulated independently (reviewed by Mackenzie and McIntosh, 1999). The phenomenon termed substoichiometric shifting refers to a process that permits dramatic copy number suppression of particular subgenomic DNA molecules to nearly undetectable levels during plant development. This process, first discovered in maize (Small et al., 1987), appears to be widespread in plants and may constitute a means of maintaining mitochondrial genetic variation in a silenced but retrievable conformation (Small et al., 1989; Janska et al., 1998). Substoichiometric shifting of specific portions of the genome appears to engage replicative and/or recombinational mechanisms that are uncharacterized to date.
An argument has been made for the endosymbiotic origin of mitochondria from a common ancestor of the rickettsial subdivision of the α-proteobacteria (Andersson et al., 1998; Gray et al., 1999; Emelyanov, 2001). Evidence suggests that during eukaryotic evolution, the protomitochondrial form relinquished much of its genetic complement to the nucleus by a process of interorganellar gene transfer. More recently, these events apparently occurred via processed RNA intermediates that must acquire, after integration to the nuclear genome, a promoter and targeting presequence for nuclear function (reviewed by Brennicke et al., 1993; Martin and Herrmann, 1998; Adams et al., 1999; Palmer et al., 2000). The additional components likely are acquired by intergenic recombination (Kadowaki et al., 1996).
The identification of nuclear genes that encode mitochondrial proteins has allowed more detailed investigation of their origins and likely evolutionary paths. Although a relatively small number of genes have been studied in detail, several examples exist of proteins capable of dual targeting to mitochondrial and chloroplast compartments (Small et al., 1998; Peeters and Small, 2001) and of interorganellar gene substitution. In the latter cases, nuclear genes of plastid origin have been found that now encode mitochondrial (Adams et al., 2002) or cytosolic (Krepinsky et al., 2001) proteins. Similarly, nuclear genes of mitochondrial origin have been found that encode cytosolic (Mireau et al., 1996) and plastidic (Gallois et al., 2001) proteins, and mitochondrially encoded genes of plastid origin also exist (Joyce and Gray, 1989).
We undertook this study to locate putative nuclear genes that encode mitochondrial DNA maintenance and transmission functions within the Arabidopsis genome. Our efforts resulted in the discovery of an unexpectedly large number of mitochondrial DNA and RNA metabolism genes within a single genomic interval, with a smaller number clustered in a second region. Here, we report the genes identified and their genomic associations in two plant species. Analysis of these genes revealed what appears to be a highly integrated mitochondrion-plastid association for genome maintenance functions.
RESULTS
Identification of Mitochondrial DNA and RNA Metabolism Loci on Chromosome III in Arabidopsis
An extensive survey of the Arabidopsis genome for genes that might be involved in mitochondrial genome maintenance functions revealed the presence of a number of genes on chromosome III that appeared to encode mitochondrial proteins based on their prokaryotic sequence homology and targeting capacity. Our comparisons with known rickettsial and yeast mitochondrial DNA and RNA metabolism proteins suggested that several genes in this interval were involved in similar functions. A list of the genes identified is provided in Table 1.
Table 1.
Putative Mitochondrial Genes within the Gene-Rich Region of Chromosome III
| Locus | Location (Mbp) |
Annotation | TargetP | MitoProt | Predotar | Confirmed Localization |
ESTs or cDNA |
Homology and Conserved Features |
Rice Homology and P Value |
|---|---|---|---|---|---|---|---|---|---|
| DNA metabolism loci
|
|||||||||
| At3g10140 | 3.134 | Putative RecA | 0.861 | 0.9384 | 0.998 | AY072877 | DMC1, Saccharomyces cerevisiae | Chromosome 1, 7.7e-55a | |
| At3g10270 | 3.17 | Putative DNA gyrase subunit B |
0.801 | 0.9997 | 0.997 | 1 | GyrB, Nostoc punctiforme | Chromosome 1, 2.5e-133a | |
| At3g10690 | 3.34 | Putative DNA gyrase subunit A |
ct 0.975/mt 0.879 | 0.99 | p 0.998/mt 0.967 | mtb (J.L. Heazlewood, A. Gout, J.M. Whelan, D.A. Day, and A.H. Millar, unpublished data) | 3 | GyrA, Nostoc punctiforme | Chromosome 3, 2.6e-150a |
| At3g18580 | 6.397 | Hypothetical protein | 0.717 | 0.9948 | 0.544 | mtc (Kang and Hamasaki, 2002) |
2 | Single-stranded DNA binding protein, Escherichia coli |
Chromosome 1, 5e-38a |
| At3g20390 | 7.11 | Translational inhibitor, putative |
p 0.497 | 0.795 | mt 0.617 | mtc (Oxelmark et al., 2000) |
AY066547 | MMF1p-mitochondrial DNA maintenance |
Chromosome 7, 6.2e-26a |
| At3g20540 | 7.168 | DNA polymerase, putative |
0.741 | 0.8028 | 0.776 | mt/ctd (this report) | AY195962 | γ-like DNA polymerase, Pol1-like | Chromosome 8/4, 5.6e-207a/7.7e-178 |
| At3g24340 | 8.831 | Hypothetical protein | 0.338 | 0.2168 | 0.77 | 1 | RAD26 DNA repair and recombination, helicase |
Chromosome 3, 7.4e-135a | |
| Mismatch repair loci
|
|||||||||
| At3g14890 | 5.008 | Putative DNA nick sensor |
ct 0.576/mt 0.269 | 0.8207 | ct 0.922 | mtb (L. McIntosh, unpublished data) |
AF453835 | 3′ DNA phosphatase | Chromosome 1, 6.2e-60a |
| At3g18630 | 6.413 | Putative uracil-DNA glycosylase |
ct 0.928 | 0.3255 | mt 0.933 | mtc (Kang and Hamasaki, 2002) |
3 | UNG1, human | Chromosome 4, 2.1e-70a |
| At3g24320 | 8.823 | Putative mismatch protein |
0.857 | 0.961 | 0.943 | mtd (Abdelnoor et al., 2003) |
AY191303 | MSH1, yeast | Chromosome 4, 1e-221a |
| At3g26690 | 9.804 | MutT-like protein | 0.79 | 0.5745 | 0.151 | 3 | bis(5′nucleosyl) tetraphosphatase | Chromosome 4, 6.2e-12a | |
| At3g46940 | 17.60 | dUTP pyrophosphatase-like |
ct 0.305 | None | ct 0.985 | mtc (Kang and Hamasaki, 2002) |
2 | DUT1, yeast, dUTPase | Chromosome 3, 0.000025a |
| At3g50880 | 19.22 | Putative DNA 3-methyladenine glycosylase |
ct 0.843 | 0.1253 | None | mtc (Kang and Hamasaki, 2002) |
1 | Yeast MAG, human MPG | Chromosome 1, 1.1e-66a |
| At3g51690 | 19.49 | DNA helicase homolog PIF1 |
ct 0.253 | 0.017 | ct 0.188 | mtc (Kang and Hamasaki, 2002) |
2 | DNA helicase PIF1, yeast | Chromosome 10, 6.3e-53 |
| At3g51880 | 19.56 | HMG2-like protein | mt 0.121 | 0.0851 | ct 0.716 | mtc (Kang and Hamasaki, 2002) |
10 | Yeast ABF2, human TFAM | Chromosome 8, 0.00075a |
| RNA metabolism loci
|
|||||||||
| At3g01410 | 1.536 | RNAse H | 0.651 | 0.938 | 0.962 | 4 | Chromosome 8, 2.6e-33a | ||
| At3g09210 | 2.825 | Unknown protein | 0.536 | 0.571 | 0.583 | 2 | NusG, transcription anti-termination | Chromosome 3, 7.3e-45a | |
| At3g18090 | 6.195 | DNA-directed RNA polymerase II |
None | None | None | mtb (L. McIntosh, unpublished data) |
2 | DNA-directed RNA polymerase II, second largest chain |
Chromosome 4, 3.3e-312a |
| At3g22310 | 7.887 | Putative RNA helicase | 0.771 | 0.9552 | 0.982 | mtb (J.L. Heazlewood, A. Gout, J.M. Whelan, D.A. Day, and A.H. Millar, unpublished data) | 7 | DEAD-box family helicases | Chromosome 4, 2.5e-116a |
| At3g22330 | 7.892 | Putative RNA helicase | 0.635 | 0.9406 | 0.943 | mtb (Millar et al., 2001) |
3 | RRP3/DBP1, S. cerevisiae | Chromosome 4, 1.6e-127a |
| At3g23780 | 8.568 | DNA-directed RNA polymerase, putative |
0.42 | 0.253 | 0.92 | mtb (L. McIntosh, unpublished data) |
2 | DNA-directed RNA polymerase, Arabidopsis, yeast |
Chromosome 4, 1.8e-294a |
| At3g23830 | 8.607 | Gly-rich RNA binding protein |
0.504 | 0.8331 | ct 0.920 | mtb (Vermel et al., 2002) |
6 | RNP-T, Arabidopsis; NSR-1, S. cerevisiae |
Chromosome 1, 6.4e-18a |
| At3g25430 | 9.22 | Hypothetical protein | 0.464 | 0.9284 | 0.982 | AY062664 | Poly(A)-specific ribonuclease | Chromosome 8, 3.2e-69a | |
| Other relevant loci
|
|||||||||
| At3g05780 | 1.714 | LON Ser-type protease | mt 0.587 | 0.9110 | None | mtb (J.L. Heazlewood, A. Gout, J.M. Whelan, D.A. Day, and A.H. Millar, unpublished data) | 0 | Mitochondrial DNA binding affinity in yeast |
Chromosome 7, 2.8e-228a |
| At3g05790 | 1.720 | LON Ser-type protease | ct 0.589 | 0.8992 | ct 0.852 | 0 | Mitochondrial DNA binding affinity in yeast |
Chromosome 7, 2.5e-265a | |
| At3g07200 | 2.291 | RING Zn finger protein, putative |
0.84 | 0.87 | 0.98 | 1 | Zinc finger, 3HC4 | Chromosome 1, 1.5e-14a | |
| At3g10010 | 3.111 | DML2 | None | None | None | 1 | Nth nuclease homolog | Chromosome 1, 3.2e-64a | |
| At3g10110 | 3.116 | TIM22-3 | –e | – | – | 1 | Mitochondrial import, inner membrane translocator | Chromosome 3, 0.002a | |
| At3g10810 | 3.38 | Ring Zn finger, putative | 0.819 | 0.935 | 0.790 | 2 | Zinc finger, C3HC4 type | Chromosome 1, 1.1e-31a | |
| At3g15000 | 5.050 | Expressed protein | ct 0.914 | 0.990 | mt 0.989 | mtb (Kruft et al., 2001) |
AF428427 | Similar to DAG protein, Antirrhinum |
Chromosome 9/6, 3e-21a |
| At3g20000 | 6..967 | TOM40 | – | – | – | mtb (Kruft et al., 2001) |
11 | Membrane import protein | Chromosome 3/1, 3.8e-49a |
| At3g23990 | 8.668 | Hsp60 | mt 0.843 | 0.9897 | 0.999 | mtb (Kruft et al., 2001) |
9 | Influences mitochondrial DNA stability in yeast |
Chromosome 10, 4.5e-150a |
| At3g25100 | 9.144 | Cdc45-like | 0.087 | 0.131 | 0.998 | 0 | CDC45 minichromosome, DNA replication |
Chromosome 11, 1.7e-199a | |
| At3g26480 | 9.687 | WD-repeat protein | mt 0.457 | 0.736 | 0.805 | 2 | DIP2, CRN1, meiosis | Chromosome 12, 7.4e-58a | |
| At3g27070 | 9.982 | TOM-20-1 | – | – | – | mtb (Werhahn et al., 2001) |
2 | Mitochondrial import receptor | Chromosome 1, 4.6e-10a |
| At3g27080 | 9.984 | TOM-20-3 | – | – | – | mtb (Werhahn et al., 2001) |
8 | Mitochondrial import receptor | Chromosome 1, 7.5e-22a |
| At3g27360 | 10.129 | Histone H3 | ct 0.765 | 0.992 | mt 0.995 | mtb (Vermel et al., 2002) |
AY077654 | Binds single-stranded mitochondrial DNA |
Chromosome 1/4/5/ 6/11, 1.8e-55a |
| At3g58610 | 21.98 | Ketol acid reductoisomerase |
ct 0.980 | 0.963 | mt 0.995 | mtb (J.L. Heazlewood, A. Gout, J.M. Whelan, D.A. Day, and A.H. Millar, unpublished data) | 71 | ILV5, yeast | Chromosome 1, 2.5e-209a |
ct, chloroplast; mt, mitochondria; None, no significant targeting value; p, plastic.
Reciprocal best hit.
Localization confirmed by proteomics data.
A homologous protein is mitochondrial in other species.
Localization confirmed by experimental data.
This protein does not have a target peptide.
In addition, a number of genes that encode mitochondrially targeted proteins of yet unknown function were present (data not shown). Conservatively, we have identified >50 genes within a 6-Mbp interval, both annotated and unannotated for function, that appear to encode proteins with mitochondrial targeting capacity. This estimate does not include the >100 genes predicted to encode plastid proteins within the same interval and several that appear to be of rickettsial origin by homology but with no clear targeting features annotated (data not shown). The association of several genes from this interval with organellar DNA and RNA maintenance suggests that at least some of the genes that lack functional annotation also might participate in related functions or their regulation.
We also surveyed the Arabidopsis genome for plant equivalents of proteins involved in eukaryotic mitochondrial DNA maintenance functions on the basis of published sequences in yeast and human (Kang and Hamasaki, 2002; MITOP database at http://mips.gsf.de/services/genomes). Nearly all of the plant equivalents for mitochondrial DNA repair are uncharacterized. Several of the identified candidates were located on chromosome III (Table 1). A complete list of the putative Arabidopsis homologs can be found in the supplemental data online (http://psiweb.unl.edu/mackenzie/table1.html).
The chromosome III interval that encompasses the identified mitochondrial genes can be estimated to extend as much as 26 Mbp, although mitochondrial gene density appeared to be greatest within the 6.5- to 9.7-Mbp interval, with a second, smaller cluster at 17.0 to 22.0 Mbp on the chromosome III map. Another statistically significant cluster was found on chromosome V. To determine whether the apparent clustering of related organellar genes in Arabidopsis was a conserved tendency in higher plants, we investigated the genomic locations of the homologous sequences in rice. Although in some cases it was not possible to identify a corresponding gene candidate from the annotated rice sequences currently available, a surprisingly large proportion of identified homologous genes were located on either chromosome I (37%) or chromosome IV (23%) in the rice genome (Table 1). This result suggests that linkage of genes for mitochondrial DNA and RNA metabolism exists within the rice genome.
The Organization of Rickettsia-Homologous Genes for DNA Metabolism in the Arabidopsis Genome
Because Rickettsia americana has been postulated to represent one of the closest living relatives of present-day mitochondria (Andersson et al., 1998), we surveyed the Rickettsia genome for genes involved in DNA maintenance and RNA metabolism functions. Of 43 genes identified, 25 (58%) homologous sequences were identified in the Arabidopsis genome, as shown in the supplemental data online (http://psiweb.unl.edu/mackenzie/table2a.html and http://psiweb.unl.edu/mackenzie/table2b.html). Of the genes in Arabidopsis predicted to be analogous to Rickettsia DNA metabolism loci, 14 (56%) were located on chromosome III, and all but 3 encoded organelle-targeting proteins. For those Rickettsia homologs located elsewhere in the genome, seven (64%) were predicted to encode proteins with ambiguous organelle-targeting discrimination values. Remarkably, of all the Rickettsia-homologous loci predicted to encode organelle-targeting proteins, half appeared to be dual targeting or plastid targeting (see supplemental data online). Together, these observations suggest an intimate coevolution of mitochondrial and plastid DNA maintenance functions during the evolution of higher plants and a striking trend toward the clustering of organellar DNA maintenance components.
We were surprised to find that a relatively large proportion of the Rickettsia-homologous loci (19%) encoded proteins with ambiguous or undetected organelle-targeting presequences. Some of these genes appeared to encode proteins that function within the nucleus, but others may represent organellar proteins targeted by a distinct mechanism or genes misannotated for translation start site.
Organellar DNA and RNA Metabolism Loci Appear to Be Clustered in Two Genomic Intervals in Arabidopsis
A permutation test for the consolidation of organellar DNA and RNA maintenance genes was constructed by comparing the observed clustering with the pattern of distribution that would be expected as a result of random rearrangement and duplication of Arabidopsis organellar genes. A total of 791 genes predicted to target mitochondria were identified using the Predotar program to survey the Arabidopsis genome database (http://www.inra.fr/Internet/Produits/Predotar/Arabidopsis_mit.html). From within this list, genes were classified by whether or not they had a putative DNA maintenance function. The chromosome location of each gene also was determined. From chromosome III loci (Table 1), only those involved in DNA and RNA metabolism were included. In addition to the 23 chromosome III loci indicated in Table 1, 3 additional loci homologous with rickettsial DNA and RNA metabolism genes but lacking apparent organelle-targeting capacity were included. Because gene density varies between chromosomes, the number of organelle-targeting genes present on each chromosome was held fixed. Then, 10,000 permutation samples were obtained by permuting the locations of the organellar genes.
Table 3.
Evidence of Protein Targeting of Presequence Exchanges in Arabidopsis
| Locus | Putative Protein Function (Predicted Localization) |
Presequence Homology with |
Putative Protein Function | Predicted Localization |
Confirmed Localization | Presequence Alignment (identical/positive [%]) |
|---|---|---|---|---|---|---|
| At3g20540 | DNA polymerase (mt) | At1g50840 | DNA polymerase | ct | cta (this report) | 61/64 |
| At3g14530 | GGPS3 | ct | ctb (Okada et al., 2000) | 34/52 | ||
| At3g32040 | GGPS, putative | ct | 37/59 | |||
| At2g47270 | Unknown protein | ct | 30/53 | |||
| At3g18420 | Unknown protein | ct | 36/53 | |||
| At3g22310 | RNA helicase (mt) | At3g22330 | RNA helicase | mt | mtb (Millar et al., 2001) | 66/83 |
| At3g22300 | 40S ribosomal protein S10 | mt | mta (Wischmann and Schuster, 1995) |
51/68 | ||
| At3g27620 | Alternative oxidase 1c | mt | mta (Saisho et al., 1997) | 50/66 | ||
| At1g60950 | Ferredoxin precursor | mt | ctb (Peltier et al., 2000) | 45/64 | ||
| At1g10960 | Ferredoxin precursor | ct | ctb (Peltier et al., 2000) | |||
| At5g38430 | Rubisco small subunit 1b | ct | ctb (Peltier et al., 2000) | |||
| At3g23830 | RNA binding protein (mt) | At4g13850 | AtGRP2-like | mt | mtb (Kruft et al., 2001) | 72/81 |
| At5g47320 | 40S ribosomal protein S19 | mt | mta (Sanchez et al., 1996) | 66/72 | ||
| At5g49210 | Unknown protein | mt | 58/67 | |||
| At5g61030 | RNA binding-like | mt | J.L. Heazlewood, A. Gout, J.M. Whelan, D.A. Day, and A.H. Millar, unpublished data b |
36/53 | ||
| At5g66660 | At14a-like | ct | 35/48 |
ct, chloroplast; mt, mitochondria; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
Localization confirmed by experimental data.
Localization confirmed by proteomics data.
A standardized gene consolidation score was obtained for each chromosome along with the maximum consolidation score. A consolidation score, Zc, for chromosome c was defined as Zc = |nc − nc|/sc, where nc is the number of unique organellar genome maintenance genes present on chromosome c, nc is the average number of unique organellar genome maintenance genes present on chromosome c across the 10,000 permutation samples, and sc is the standard deviation of the number of unique organellar genome maintenance genes present on chromosome c across the 10,000 permutation samples. The maximum consolidation score, Zmax, across the five chromosomes was defined as Zmax = maxc|Zc|. P values then were estimated as the proportion of times that the permutation sample consolidation scores were greater than or equal to the observed consolidation score. Both the overall gene consolidation test and the chromosome III–specific consolidation test supported the hypothesis of gene clustering (P < 0.0001). These data are shown in Table 2. In addition, a smaller degree of gene clustering on chromosome V was indicated as well (P < 0.005).
Table 2.
Statistical Test of Gene Clustering for Genes Involved in Organellar DNA and RNA Maintenance Functions
| Unique DNA and RNA Maintenance Genes
|
||||||
|---|---|---|---|---|---|---|
| Chromosome | Total Number of Genes |
Observed Number (nc) |
Mean Number (nc) |
Standard Deviation (sc) |
Consolidation Score (Zc) |
Estimated P Value |
| I | 219 | 13 | 17.5 | 2.9 | −1.6 | 0.0862 |
| II | 180 | 9 | 14.9 | 2.8 | −2.1 | 0.0364 |
| III | 148 | 26 | 12.9 | 2.7 | 4.9 | 0.0000 |
| IV | 173 | 7 | 14.5 | 2.8 | −2.7 | 0.0093 |
| V | 71 | 13 | 6.6 | 2.2 | 2.9 | 0.0000 |
| Maximum consolidation score (Zmax) | 4.9 | 0.0000 | ||||
Patterns of Genomic Duplication Suggest Selection for the Present-Day Arrangement and Interorganellar Substitutions
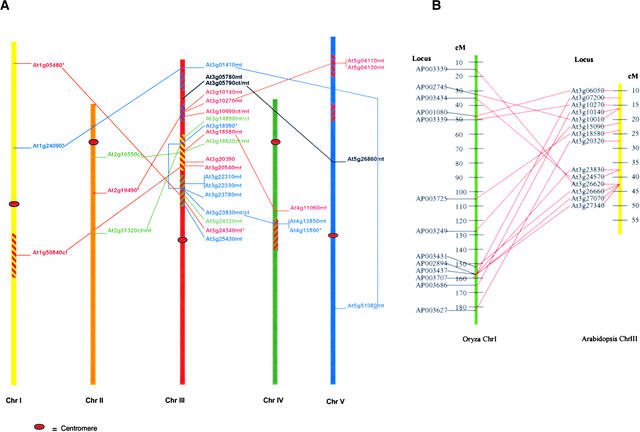
As is common for many regions of the Arabidopsis genome (Blanc et al., 2000), extensive duplication has occurred for the region of interest on chromosome III. In some cases, duplication has involved large intervals, whereas in other instances, only one or a few genes were involved. The duplication pattern of the chromosome III interval is shown in Figure 1. The extent of duplication implies that the chromosome III genomic interval has undergone significant rearrangement during the evolution of the Arabidopsis genome. This extent of rearrangement and duplication likely precludes the conservation of any ancient gene clusters that might have resulted during the process of endosymbiosis. Likewise, the gene order of the linked loci on Arabidopsis chromosome III is rearranged dramatically relative to the homologous loci on rice chromosome I (Figure 1B). No evidence of recombination suppression was detected within the chromosome III interval (data not shown).
Figure 1.
Arabidopsis Gene Duplications Involving the Organelle Gene-Rich Interval of Chromosome III.
(A) Extensive interchromosomal and intrachromosomal duplications involving genes on chromosome III. Members of larger gene families were excluded from the figure. Genes involved in DNA metabolism (red), RNA metabolism (blue), and mismatch repair (green) are designated. Predicted targeting is indicated by m (mitochondrial) or cp (chloroplast), and asterisks indicate ambiguous target predictions. Duplicated genomic regions, as described by Blanc et al. (2000), are indicated with like-colored stripes.
(B) Rice chromosome I homologous gene alignment with Arabidopsis chromosome III. Organellar genes with sequence homology were identified and aligned on rice chromosome I and Arabidopsis chromosome III to assess the conservation of gene order. cM, centimorgan.
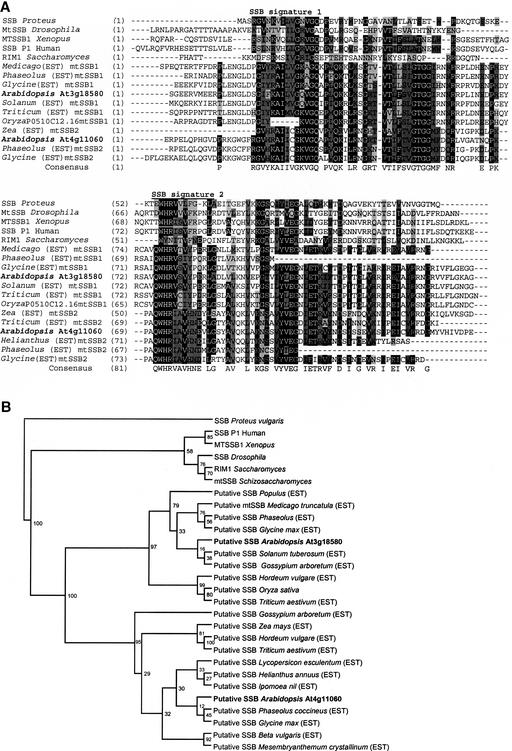
Analysis of duplicated loci indicates that functional divergence has occurred by two routes. In some cases, gene duplication may have resulted in functional divergence of the gene. This appears to be the case for a mitochondrial single-stranded DNA binding (SSB) protein gene, as shown in Figure 2. A mitochondrial SSB (mtSSB) locus present in higher plants apparently was duplicated early during plant evolution, resulting in two diverged copies in all plant species surveyed. In Arabidopsis, one copy of the gene resides within the chromosome III interval and another resides on chromosome IV. The pattern of sequence diversification after gene duplication results in two gene clusters (Figure 2B) and suggests that mtSSBs may have evolved for an additional plant-specific function. All identified plant mtSSB genes identified were predicted to encode mitochondrially targeted proteins (data not shown).
Figure 2.
Sequence Comparison of Two Arabidopsis Genes Predicted to Encode SSB-Like Proteins.
(A) Alignment of SSB-like sequences. The N-terminal portions of the sequences, which encode the mitochondrion-targeting presequence, were truncated before alignment using CLUSTAL X. Identical residues are shown by reverse contrast and similar residues with shading, with dark gray indicating a block of similar and light gray weakly similar amino acid residues. In the interest of space, only a portion of the identified genes are shown.
(B) Phylogenetic tree developed from SSB-like sequences reveals two plant forms of the gene. The phylogenetic tree was constructed using the NJ method (Saitou and Nei, 1987) based on a distance matrix computed using the PROTDIST program in the PHYLIP software package version 3.6(α3) (Felsenstein, 2002). Estimated statistical confidence was generated from 100 bootstrapped data sets using the SEQBOOT program, and the consensus tree was generated using the CONSENSE program in the PHYLIP package (Felsenstein, 2002). EST sequences were derived from BLAST searches. Two distinct ESTs were identified for nearly every plant species surveyed, providing evidence of SSB gene duplication.
A second fate of duplications appears to involve expanded protein-targeting capacity. In several cases, gene duplications result in proteins predicted to target mitochondria, plastids, or both organelles dually (Figure 1). These data, combined with the observed targeting features of the identified Rickettsia homologs (see supplemental data online [http://psiweb.unl.edu/mackenzie/table2a.html and http://psiweb.unl.edu/mackenzie/table2b.html]), suggest that the interchange of mitochondrial and plastid components for DNA maintenance has been extensive. We generally used three computer programs designed to predict protein targeting within the plant cell (see Methods). It is not possible to conclude definitively a protein's targeting capacity by computer prediction programs. However, good correspondence generally is found between computer-assisted predictions and in vivo observations for those proteins for which correspondence is seen among prediction programs (I. Small, personal communication; A. Lyznik, A. Elo, and S. Mackenzie, unpublished data).
Evidence of Mitochondrial and Plastid Coevolution for DNA Metabolism Functions
Our observations suggest that the duplication of genes involved in organellar DNA and RNA maintenance may have facilitated functional specialization and organellar coevolution. Given the prediction of both mitochondrial and plastid targeting of gene products within the chromosome III interval, we investigated the fate of targeting presequence information in the evolution of genes that are involved in mitochondrial DNA and RNA metabolism. Other groups have shown that the transfer of organellar genes to the nucleus must be accompanied by the acquisition of mitochondrial or plastid protein-targeting information to allow the gene to become functional (reviewed by Martin and Herrmann, 1998). This targeting information apparently can be acquired via nuclear intergenic recombination.
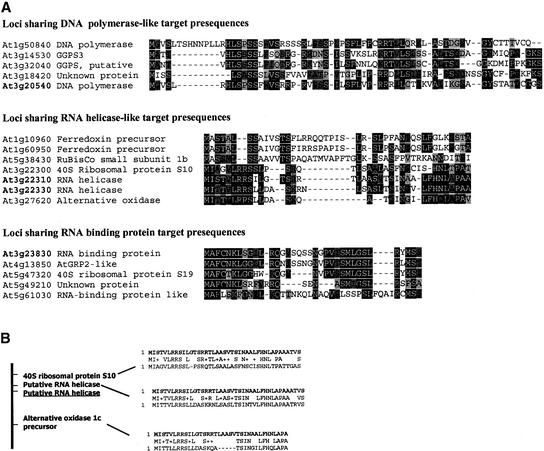
Amino acid sequence homology searches revealed that, in several cases, some genes within the chromosome III interval share protein presequence identity with functionally unrelated organelle-targeting proteins, as shown in Table 3. Putative presequences were searched using BLAST (Basic Local Alignment Search Tool) against the Arabidopsis database. The short amino acid sequences used in these searches resulted in relatively high e-values. To identify homologous presequences, the following criteria were used. Evidence of presequence exchange involved e-values of <1.0 for two sequences located at the immediate N termini of proteins, demonstrating organelle-targeting capacity. Several homologous sequences with significant similarity and low e-values were excluded by their localization other than at the N terminus. For this reason, it is possible that we missed those examples of presequence exchange that involve exon shuffling.
Analysis of Arabidopsis mitochondrial proteome data (J.L. Heazlewood, A. Gout, J.M. Whelan, D.A. Day, and A.H. Millar, unpublished data; L. McIntosh, personal communication), as well as studies by others, confirmed that several of the identified genes postulated to have shared mitochondrion-targeting presequences encode mitochondrially localized proteins (Tables 1 and 3). From this observation, we assume that intergenic recombination is the predominant mechanism involved in the acquisition of organelle-targeting capacity for genes in this region. In some cases, the intergenic exchanges occurred within the chromosome III interval, as shown in Figure 3B. The Arabidopsis nuclear rps10 gene (At3g22300; Figure 3) represents an independent and recent gene transfer event from the mitochondrion during Brassicaceae evolution. The gene remains within the mitochondrion in several plant species (Adams et al., 2000). The rps10 gene apparently acquired its targeting presequence from an adjacent RNA helicase locus on chromosome III (Figure 3).
Figure 3.
Evidence of Targeting of Presequence Exchange during the Evolution of Mitochondrial Genes in the Chromosome III Organellar Gene-Rich Interval in Arabidopsis.
(A) Amino acid sequence alignment of the Arabidopsis proteins sharing common N-terminal amino acid sequence with the DNA polymerase-like protein (At3g20540), the RNA helicase-like protein (At3g22310), or the RNA binding protein (At3g28830). Identical residues are shown by reverse contrast and similar residues with shading, with dark gray indicating a block of similar and light gray weakly similar amino acid residues.
(B) The predicted mitochondrial presequence from the RNA helicase (At3g22310; underlined) was used in a BLAST search to identify homologous sequences in the database. The three most significant hits were obtained for mitochondrial presequences of three neighboring loci within the same chromosomal interval, with amino acid identities ranging from 50 to 66%.
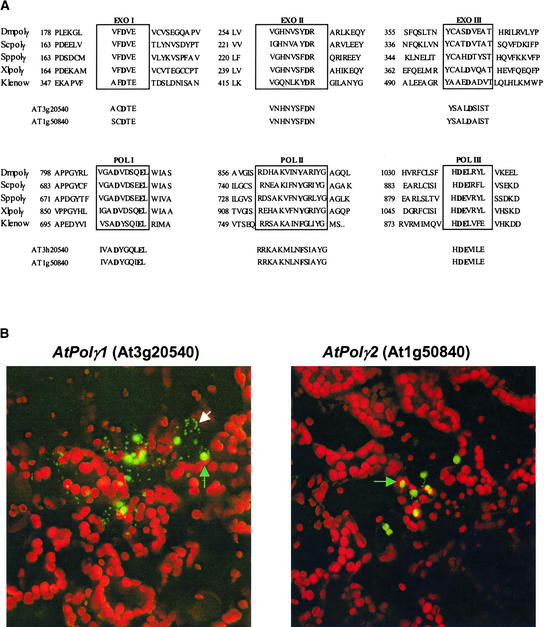
In several instances, gene duplications have resulted in mitochondrial, plastid, and dual targeting forms of a nearly identical protein (Figure 1) (A. Lyznik, A. Elo, and S. Mackenzie, unpublished data). In the case of the DNA polymerase I–homologous gene identified on chromosome III (At3g20540), designated AtPolγ1, and its duplicate on chromosome I (At1g50840), designated AtPolγ2, both genes contain features similar to those of the γ-type polymerase found in mitochondria from other species (Lewis et al., 1996; Lecrenier et al., 1997), as shown in Figure 4A. Sequence alignments that compare the Arabidopsis genes with the DNA polymerase genes from Rickettsia (34% amino acid identity) and cyanobacteria (36% amino acid identity) suggest an equidistant genetic relationship to either putative progenitor form (data not shown).
Figure 4.
Organellar Targeting of a Putative γ-Like DNA Polymerase.
(A) Amino acid sequence alignment reveals conserved motifs within exonuclease and polymerase domains of the Pol1-like DNA polymerase in both chromosome I and III versions of the Arabidopsis genes. The alignment presented here incorporates data taken directly from Lewis et al. (1996) for comparison.
(B) Arabidopsis leaf cell bombardment was used to test for organellar targeting of the putative DNA polymerase I–like gene products encoded on chromosome III (AtPolγ1; At3g20540) and chromosome I (AtPolγ2; At1g50840). Plastids are shown red as a result of autofluorescence. Enhanced GFP was used as the reporter gene. Although the product from AtPolγ2 appeared to target plastids, the product from AtPolγ1 targeted both plastids and mitochondria (the white arrow indicates mitochondria, and the green arrows indicate plastids).
The two loci share 80% DNA sequence identity and 66% amino acid identity, diverging most within the 5′ region of the genes (data not shown). The chromosome III–encoded protein was predicted to be targeted to mitochondria (MitoProt value of 0.80) or dual targeted (Predotar values of mt [mitochondria] 0.78 and cp [chloroplast] 0.64; TargetP values of mt 0.74 and cp 0.59). The chromosome I version was predicted to target to either mitochondria (MitoProt value of 0.90) or chloroplast (Predotar value of 0.96, TargetP value of 0.93). We investigated the in vivo targeting properties of both genes by developing gene constructs that fused the predicted targeting presequence information from either locus (763 bp for chromosome III or 752 bp for chromosome I) to the enhanced GREEN FLUORESCENT PROTEIN (GFP) reporter gene. The gene constructs were used for particle bombardment of Arabidopsis leaf tissues.
Confocal analysis suggested that the product from the chromosome I AtPolγ2 (At1g50840) gene was targeted to the plastid, whereas the chromosome III AtPolγ1 (At3g20540) gene product was dual targeted to the mitochondria and the plastid (Figure 4B). Dual targeting of at least one gene product would be consistent with biochemical data for similar activities of mitochondrial and plastid DNA polymerases reported by Sakai (2001) and Kimura et al. (2002). Whether the two enzyme forms can reciprocally compensate for function is being investigated.
DISCUSSION
Our investigations revealed a nuclear genomic interval in Arabidopsis that contains a large number of genes involved in DNA and RNA metabolism functions. Several of these genes have never been characterized in higher plants. A large number of unidentified genes predicted to encode mitochondrial proteins also were found in this interval. For example, two C3HC4 zinc RING finger proteins were identified with compelling discrimination values predicting mitochondrial targeting (At3g07200 and At3g10810; Table 1). This RING finger motif has been associated functionally with E3 ubiquitin protein ligase activity in a significant number of proteins (Joazeiro et al., 1999; Freemont, 2000). Interestingly, a process involving ubiquitination has been described to influence mitochondrial inheritance in yeast (Fisk and Yaffe, 1999) and the maternal transmission of mitochondria in mammals (Sutovsky et al., 1999, 2000). An intriguing possibility is that the identified Arabidopsis proteins might represent components of the maternal mitochondrial inheritance apparatus of higher plants.
We suspect that more intensive mining of this genomic interval will provide valuable insights into several similarly undetailed aspects of higher plant organelle biology. Some of the loci listed in Table 1 do not appear, by functional annotation, to be involved in aspects of genome maintenance functions. However, it has become increasingly clear that nucleus-encoded mitochondrial proteins can serve dual functions (Bateman et al., 2002), and mutation of such proteins can result in the destabilization of the mitochondrial genome in yeast (Suzuki et al., 1994). These observations in yeast and other species were considered in our analysis.
Until recently, the genetic clustering of genes for related functions was considered unusual in higher plant genomes. With the exception of disease resistance gene clusters (Richter and Ronald, 2000; Hulbert et al., 2001) and other gene families arising by duplication, genes associated with the same pathway or developmental process more often appeared to be unlinked. In fact, we were unable to detect any apparent genomic association pattern among most nuclear genes involved in mitochondrial processes for electron transport, energy transduction, biosynthetic pathways, and protein translation in Arabidopsis.
However, more extensive whole-genome quantitative trait loci analysis in maize has suggested that particular functional gene clusters may exist (Khavkin and Coe, 1997). Several loci involved in the regulation of maize development have been shown to link to quantitative trait loci associated with related developmental processes. Likewise, clustering of classes of housekeeping genes is evident in the human genome (Lercher et al., 2002). In Arabidopsis, terpenoid synthase genes are clustered and associated with genes that encode consecutive steps in terpenoid biosynthesis (Aubourg et al., 2002). In the case of genes associated with mitochondrial DNA maintenance, genetic linkage is observed not only in the higher plant genome but apparently also within the genome of Drosophila melanogaster (Lefai et al., 2000). Five D. melanogaster loci are found in close association, and their clustering is assumed to be a vestige of their cotranslocation from the mitochondrion to the nuclear genome.
What accounts for the presence of a large number of mitochondrial genes of related function within a single plant genomic interval? The observed linkage could result from the preferential integration of genes transferred from the mitochondrion to this region of the nuclear genome, either as gene blocks or as individual genes. Perhaps subsequent gene evolution, including the acquisition of regulatory and target presequence information, was facilitated by the presence of like genes in physical proximity, as appears to be the case for rps10. Most mitochondrial genes successfully transferred and expressed in the nucleus in recent evolutionary time appear to have been transferred individually via RNA intermediates (Martin and Herrmann, 1998). However, it is likely that many of the genes involved in organellar genome maintenance were transferred before the advent of organelle RNA editing; therefore, they may have integrated as multigene segments. The presence on Arabidopsis chromosome II of a 620-kb segment representing the entire mitochondrial genome with additional duplications (Stupar et al., 2001) demonstrates that large multigene DNA transfers are feasible (reviewed by Henze and Martin, 2001).
Several observations in our study suggest that regardless of whether the linkage association on chromosome III is a consequence of the original integration pattern, the present-day gene linkages likely were maintained by selection. Organellar genes found on chromosome III do not appear to have been integrated to the nuclear genome simultaneously. The DNA and RNA metabolism genes identified on chromosome III appeared to be derived from both cyanobacterial and rickettsial origins. No evidence was found for conserved gene order between the Rickettsia or Reclinomonas genomes and the genes found in the Arabidopsis interval (data not shown). This is not surprising, given the degree of genome reduction that has occurred within Rickettsia (Lang et al., 1999; Gray et al., 2001). Gene order also differed dramatically between genes linked on Arabidopsis chromosome III and their homologs on rice chromosome I. These details, and the remarkable degree of genome rearrangement that has occurred during the evolution of the Arabidopsis genome (Blanc et al., 2000), make it difficult to envision the conservation of gene linkages established early in the endosymbiotic process without their selection.
Although the selective advantage for retaining a significant number of organellar DNA and RNA metabolism loci within a single linkage group is not clear, the observed genome synteny for this interval between two diverse plant species is intriguing. We have likely underestimated the degree of Arabidopsis chromosome III/rice chromosome I synteny, because the assembly of the rice genomic data is still in progress. Comparisons between rice and Arabidopsis by other groups have revealed similar colinearity between the two genomes (Liu et al 2001).
Unlike the many other functions required by plant mitochondria, genome maintenance and transmission is one in which maternal and paternal nuclear contributions likely differ. In most plant species, mitochondria are strictly maternally inherited, and exceptions to this rule are rare (Mogensen, 1996). It is possible that gene imprinting might influence the mitochondrial genome replication and transmission process within the gametes during fertilization to ensure nuclear-mitochondrial compatibility. Coordinated gene regulation via epigenetic mechanisms or trans-acting regulators is thought to be facilitated by physical linkage of the coregulated genes (Boutanaev et al., 2002; Oliver et al., 2002; Spellman and Rubin, 2002). At least one gene that has been shown to be epigenetically regulated by hypermethylation, the SUPERMAN locus (Jacobsen and Meyerowitz, 1997), is known to reside within the chromosome III interval. As one test of this hypothesis, we are currently investigating whether the expression of organellar DNA maintenance loci on chromosome III is predominantly maternal during ovule and/or embryo development.
The role of nuclear gene duplication in gene function is of particular interest to studies of plant organellar biology, because plant mitochondrial genomes display many novel functions and genomic features. One means by which new traits originate is the evolution of novel gene functions in sequences that arise by duplication (Mitchell-Olds and Clauss, 2002). The case of mtSSB, with observed divergence of two different forms in higher plants, is intriguing because it allows for the possibility that one form of the SSB might now play a novel role. This hypothesis is consistent with what has been observed for certain fungi (Tomáška et al., 2001). In Candida species in which the mitochondrial genome has linearized, a divergent mtSSB now appears to function as a telomere binding protein (Nosek et al., 1995, 1998). With the apparent linear conformation of the plant mitochondrial genome, that could be the situation in plants as well.
The chromosome III interval encompassed both cyanobacteria-related and Rickettsia-related sequences, with gene products predicted to target mitochondria, plastids, or both. Such intimate coevolution of mitochondria and plastids also is evident for organellar RNA polymerases (Hedtke et al., 2000), aminoacyl tRNA synthases (Small et al., 1998; Peeters et al., 2000), and ribosomal proteins (Adams et al., 2002). As with these previously reported cases, several original components of DNA and RNA metabolism reported here apparently have been supplanted by interorganellar protein substitutions. Indeed, it appears from our analysis that these two subcellular organelles may share a surprisingly large proportion of their DNA and RNA metabolism apparatus. From these observations, we conclude that beyond the acquisition of important energy metabolic capacity, the endosymbiotic process has provided important elements of genomic plasticity to higher plant genomes.
METHODS
Bioinformatics
The search for N termini as well as whole amino acid sequence similarities was performed using BLAST (Basic Local Alignment Search Tool) (Altschul et al., 1990, 1997) through the GenBank and MatDB (http://mips.gsf.de/proj/thal/db/index.html) (Schoof et al., 2002) databases. Rice-homologous sequence information was obtained by BLASTX analysis of the TIGR Rice Genome Database. All sequence alignments were performed using CLUSTAL X (Thompson et al., 1997).
The transit peptide predictions were performed using Predotar version 0.5 (http://www.inra.fr/Internet/Produits/Predotar/), TargetP (Emanuelsson et al., 2000; http://www.cbs.dtu.dk/services/TargetP/), and MitoProt (Claros and Vincens 1996; http://mips.gsf.de/cgi-bin/proj/medgen/mitofilter).
Particle Bombardment for Presequence Targeting Analysis
The targeting presequence derived from the putative DNA polymerase I homolog on chromosome III (At3g20540) was amplified by PCR using the primers 17401MF (5′-GGAGATTAGCTTCACTTACTAC-3′) and 18142MR (5′-GGCCATGGCTTGTTTCACAGTTGGCGGAAC-3′). The targeting presequence from the putative DNA polymerase I homolog on chromosome I (At1g50840) was amplified by PCR using the primers 28431CF (5′-TCTCCGACTCTGTCGTCACTT-3′) and 29162CR (5′-GGCCATGGCTACCTCCGTCTGATTTCCAAC-3′). PCR products were ligated to the TOPO cloning vector (Invitrogen, Carlsbad, CA) and digested using the resulting NcoI site to release the insert. Fragments then were ligated to the pCAMBIA 1302 vector (http://www.cambia.org) within the NcoI site that resides at the start codon of the gene for enhanced GFP.
In separate experiments, 7 μg of DNA from the respective constructs was delivered into 4-week-old leaves of Arabidopsis thaliana (Columbia) using tungsten particles and the Biolistic PDS-1000/He system (Bio-Rad). Particles were bombarded into Arabidopsis leaves using 900-p.s.i. rupture discs under a vacuum of 26 inches of Hg. After the bombardment, Arabidopsis leaves were allowed to recover for 18 to 22 h on Murashige and Skoog (1962) medium plates at 22°C in 16 h of daylight. Localization of GFP expression was conducted by confocal laser scanning microscopy with Bio-Rad 1024 MRC-ES with excitation at 488 nm and two-channel measurement of emission, 522 nm (green/GFP) and 680 nm (red/chlorophyll).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank Natalya Nersesian and the Center for Biotechnology Core Microscopy Facility for assistance in tissue imaging. We also thank Michael Gray for valuable discussions and Alan Christensen for critical review of the manuscript. This work was supported by grants to S.A.M. from the National Science Foundation (MCB-0090813) and the Department of Energy (DE-FG03-00ER15075).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010009.
Footnotes
Online version contains Web-only data.
References
- Abdelnoor, R.V., Yule, R., Elo, A., Christensen, A., Meyer-Gauen, G., and Mackenzie, S. (2003). Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 100, 5968–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L., Daley, D.O., Qiu, Y.-L., Whelan, J., and Palmer, J.D. (2000). Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357. [DOI] [PubMed] [Google Scholar]
- Adams, K.L., Daley, D.O., Whelan, J., and Palmer, J.D. (2002). Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell 14, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L., Song, K., Roessler, P.G., Nugent, J.M., Doyle, J.L., Doyle, J.J., and Palmer, J.D. (1999). Intracellular gene transfer in action: Dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc. Natl. Acad. Sci. USA 96, 13863–13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, S.G.E., Zomorodipour, A., Andersson, J.O., Sicheritz-Ponten, T., Alsmark, U.C.M., Podowski, R.M., Naslund, A.K., Eriksson, A.S., Winkler, H.H., and Kurland, C.J. (1998). The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–143. [DOI] [PubMed] [Google Scholar]
- Aubourg, S., Lecharny, A., and Bohlmann, J. (2002). Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267, 730–745. [DOI] [PubMed] [Google Scholar]
- Backert, S., and Börner, T. (2000). Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 37, 304–314. [DOI] [PubMed] [Google Scholar]
- Backert, S., Nielsen, B.L., and Börner, T. (1997). The mystery of the rings: Structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 2, 477–483. [Google Scholar]
- Bateman, J.M., Perlman, P.S., and Butow, R.A. (2002). Mutational bisection of the mitochondrial DNA stability and amino acid biosynthetic functions of Ilv5p of budding yeast. Genetics 161, 1043–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich, A.J., and Smith, S.B. (1990). Moving pictures and pulsed field gel electrophoresis show linear DNA molecules from chloroplasts and mitochondria. Curr. Genet. 17, 421–425. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Barakat, A., Guyot, R., Cooke, R., and Delseny, M. (2000). Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev, A.M., Kalmykova, A.I., Sheveloyov, Y.Y., and Nuriminsky, D.I. (2002). Large clusters of co-expressed genes in the Drosophila genome. Nature 420, 666–669. [DOI] [PubMed] [Google Scholar]
- Brennicke, A., Grohmann, L., Hiesel, R., Knoop, V., and Schuster, W. (1993). The mitochondrial genome on its way to the nucleus: Different stages of gene transfer in higher plants. FEBS Lett. 325, 140–145. [DOI] [PubMed] [Google Scholar]
- Claros, M.G., and Vincens, P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emelyanov, V.V. (2001). Rickettsiaceae, rickettsia-like endosymbionts, and the origin of mitochondria. Biosci. Rep. 21, 1–17. [DOI] [PubMed] [Google Scholar]
- Fauron, C., Casper, M., Gao, Y., and Moore, B. (1995). The maize mitochondrial genome: Dynamic, yet functional. Trends Genet. 11, 228–235. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (2002). PHYLIP, Phylogeny Inference Package version 3.6a3. (Seattle: Department of Genome Sciences, University of Washington).
- Fisk, H.A., and Yaffe, M.P. (1999). A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145, 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont, P.S. (2000). Ubiquitination: RING for destruction? Curr. Biol. 10, 84–87. [DOI] [PubMed] [Google Scholar]
- Gallois, J.-L., Achard, P., Green, G., and Mache, R. (2001). The Arabidopsis chloroplast ribosomal protein L21 is encoded by a nuclear gene of mitochondrial origin. Gene 274, 179–185. [DOI] [PubMed] [Google Scholar]
- Gray, M.W., Burger, G., and Lang, B.F. (1999). Mitochondrial evolution. Science 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- Gray, M.W., Burger, G., and Lang, B.F. (2001). The origin and early evolution of mitochondria. Genome Biol. 2, REVIEWS1018.1–REVIEWS 1018.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke, B., Börner, T., and Weihe, A. (2000). One RNA polymerase serving two genomes. EMBO Rep. 1, 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze, K., and Martin, W. (2001). How do mitochondrial genes get into the nucleus? Trends Genet. 17, 383–387. [DOI] [PubMed] [Google Scholar]
- Hulbert, S.H., Webb, C.A., Smith, S.M., and Sun, Q. (2001). Resistance gene complex: Evolution and utilization. Annu. Rev. Phytopathol. 39, 285–312. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Meyerowitz, E.M. (1997). Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277, 1100–1103. [DOI] [PubMed] [Google Scholar]
- Janska, H., Sarria, R., Woloszynska, M., Arrieta-Montiel, M., and Mackenzie, S. (1998). Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10, 1163–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro, C.A.P., Wing, S.S., Huang, H.-K., Leverson, J.D., Hunter, T., and Lui, Y.-C. (1999). The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286, 309–312. [DOI] [PubMed] [Google Scholar]
- Joyce, P.B., and Gray, M.W. (1989). Chloroplast-like transfer RNA genes expressed in wheat mitochondria. Nucleic Acids Res. 17, 5461–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, K., Kubo, N., Ozawa, K., and Hirai, A. (1996). Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J. 15, 6652–6661. [PMC free article] [PubMed] [Google Scholar]
- Kang, D., and Hamasaki, N. (2002). Maintenance of mitochondrial DNA integrity: Repair and degradation. Curr. Genet. 41, 311–322. [DOI] [PubMed] [Google Scholar]
- Khavkin, E., and Coe, E. (1997). Mapped genomic locations for developmental functions and QTLs reflect concerted groups in maize (Zea mays L.). Theor. Appl. Genet. 95, 343–352. [Google Scholar]
- Kimura, S., et al. (2002). A novel DNA polymerase homologous to Escherichia coli DNA polymerase I from a higher plant, rice (Oryza sativa L.). Nucleic Acids Res. 30, 1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepinsky, K., Plaumann, M., Martin, W., and Schnarrenberger, C. (2001). Purification and cloning of chloroplast and cytosolic isoenzymes encoded in eukaryotic chromosomes. Eur. J. Biochem. 268, 2678–2686. [DOI] [PubMed] [Google Scholar]
- Kruft, V., Eubel, H., Jansch, L., Werhahn, W., and Braun, H.-P. (2001). Proteomics approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Lang, B.F., Gray, M.W., and Burger, G. (1999). Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33, 351–397. [DOI] [PubMed] [Google Scholar]
- Lecrenier, N., Van Der Bruggen, P., and Foury, F. (1997). Mitochondrial DNA polymerases from yeast to man: A new family of polymerases. Gene 185, 147–152. [DOI] [PubMed] [Google Scholar]
- Lefai, E., Fernandez-Moreno, M.A., Kaguni, L.S., and Garesse, R. (2000). The highly compact structure of the mitochondrial DNA polymerase genomic region of Drosophila melanogaster: Functional and evolutionary implications. Insect Mol. Biol. 9, 315–322. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Urrutia, A.O., and Hurst, L.D. (2002). Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31, 180–183. [DOI] [PubMed] [Google Scholar]
- Lewis, D.L., Farr, C.L., Want, Y., Lagina, A.T., III, and Kaguni, L.S. (1996). Catalytic subunit of mitochondrial DNA polymerase from Drosophila embryos. J. Biol. Chem. 271, 23389–23394. [DOI] [PubMed] [Google Scholar]
- Liu, H., Sachidanandam, R., and Stein, L. (2001). Comparative genomics between rice and Arabidopsis shows scant collinearity in gene order. Genome Res. 11, 2020–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, S., and McIntosh, L. (1999). Higher plant mitochondria. Plant Cell 11, 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., and Herrmann, R.G. (1998). Gene transfer from the organelles to the nucleus: How much, what happens, and why? Plant Physiol. 118, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.H., Sweetlove, L.J., Giege, P., and Leaver, C.J. (2001). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127, 1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Mireau, H., Lancelin, D., and Small, I.D. (1996). The same Arabidopsis gene encodes both cytosolic and mitochondrial alanyl-tRNA synthetases. Plant Cell 8, 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds, T., and Clauss, M.J. (2002). Plant evolutionary genomics. Curr. Opin. Plant Biol. 5, 74–79. [DOI] [PubMed] [Google Scholar]
- Mogensen, H.L. (1996). The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Bot. 83, 383–404. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nosek, J., Dinouel, N., Kovac, L., and Fukuhara, H. (1995). Linear mitochondrial DNAs from yeasts: Telomeres with large tandem repetitions. Mol. Gen. Genet. 247, 61–72. [DOI] [PubMed] [Google Scholar]
- Nosek, J., Tomaska, L., Fukuhara, H., Suyama, Y., and Kovac, L. (1998). Mitochondrial telomere-binding protein of a yeast Candida parapsilosis suggests an evolutionary adaptation of a nonspecific single-stranded DNA binding protein. J. Biol. Chem. 274, 8850–8857. [DOI] [PubMed] [Google Scholar]
- Okada, K., Saito, T., Nakagawa, T., Kawamukai, M., and Kamiya, Y. (2000). Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 122, 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg, D.J., and Bendich, A.J. (2001). Mitochondrial DNA from the liverwort Marchantia polymorpha: Circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J. Mol. Biol. 310, 549–562. [DOI] [PubMed] [Google Scholar]
- Oliver, B., Parisi, M., and Clark, D. (2002). Gene expression neighborhoods. J. Biol. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxelmark, E., Marchini, A., Malanchi, I., Magherini, F., Jaquet, L., Hajibagheri, M.A., Blight, K.J., Jauniaux, J.C., and Tommasino, M. (2000). Mmf1p, a novel yeast mitochondrial protein conserved throughout evolution and involved in maintenance of the mitochondrial genome. Mol. Cell. Biol. 20, 7784–7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, J.D., Adams, K.L., Cho, Y., Parkinson, C.L., Qiu, Y.L., and Song, K. (2000). Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc. Natl. Acad. Sci. USA 97, 6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N., and Small, I. (2001). Dual targeting to mitochondria and chloroplasts. Biochim. Biophys. Acta 1541, 54–63. [DOI] [PubMed] [Google Scholar]
- Peeters, N.M., Chapron, A., Giritch, A., Grandjean, O., Lancelin, D., Lhomme, T., Vivrel, A., and Small, I. (2000). Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J. Mol. Evol. 50, 413–423. [DOI] [PubMed] [Google Scholar]
- Peltier, J.-B., Friso, G., Kalume, D.E., Roepsorff, P., Nilsson, F., Adamska, I., and van Wijk, K.J. (2000). Proteomics of the chloroplast: Systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12, 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, T.E., and Ronald, P.C. (2000). The evolution of disease resistance genes. Plant Mol. Biol. 42, 195–204. [PubMed] [Google Scholar]
- Saisho, D., Nambara, E., Naito, S., Tsutsumi, N., Hirai, A., and Nakazono, M. (1997). Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol. Biol. 35, 585–596. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sakai, A. (2001). In vitro transcription/DNA synthesis using isolated organelle-nuclei: Application to the analysis of the mechanisms that regulate organelle genome function. J. Plant Res. 114, 199–211. [Google Scholar]
- Sanchez, H., Fester, T., Kloska, S., Schroder, W., and Schuster, W. (1996). Transfer of rps19 to the nucleus involves the gain of an RNP-binding motif which may functionally replace RPS13 in Arabidopsis mitochondria. EMBO J. 15, 2138–2149. [PMC free article] [PubMed] [Google Scholar]
- Schoof, H., Zaccaria, P., Gundlach, H., Lemcke, K., Rudd, S., Kolesov, G., Arnold, R., Mewes, H.W., and Mayer, K.F. (2002). MIPS Arabidopsis thaliana Database (MAtDB): An integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res. 30, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I.D., Isaac, P.G., and Leaver, C.J. (1987). Stoichiometric differences in DNA molecules containing the atpA gene suggest mechanisms for the generation of mitochondrial genome diversity in maize. EMBO J. 6, 865–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I.D., Suffolk, R., and Leaver, C.J. (1989). Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell 58, 69–76. [DOI] [PubMed] [Google Scholar]
- Small, I., Wintz, H., Akashi, K., and Mireau, H. (1998). Two birds with one stone: Genes that encode products targeted to two or more compartments. Plant Mol. Biol. 38, 265–277. [PubMed] [Google Scholar]
- Spellman, P.T., and Rubin, G.M. (2002). Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar, R.M., Lilly, J.W., Town, C.D., Cheng, Z., Kaul, S., Buell, C.R., and Jiang, J. (2001). Complex mtDNA constitutes an approximate 620 kb insertion on Arabidopsis thaliana chromosome 2: Implication of potential sequencing errors caused by large-unit repeats. Proc. Natl. Acad. Sci. USA 98, 5099–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky, P., Moreno, R.D., Ramalho-Santos, J., Dominko, T., Simerly, C., and Schatten, G. (1999). Ubiquitin tag for sperm mitochondria. Nature 402, 371–372. [DOI] [PubMed] [Google Scholar]
- Sutovsky, P., Moreno, R.D., Ramalho-Santos, J., Dominko, T., Simerly, C., and Schatten, G. (2000). Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 63, 582–590. [DOI] [PubMed] [Google Scholar]
- Suzuki, C.K., Suda, K., Wang, N., and Schatz, G. (1994). Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science 264, 273–276. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomáška, L., Nosek, J., and Kucejová, B. (2001). Mitochondrial single-stranded DNA-binding proteins: In search for new functions. Biol. Chem. 382, 179–186. [DOI] [PubMed] [Google Scholar]
- Vermel, M., Guermann, B., Delage, L., Grienenberger, J.M., Marechel Drouard, L., and Gualberto, J.M. (2002). A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc. Natl. Acad. Sci. USA 99, 5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werhahn, W., Niemeyer, A., Jansch, L., Kruft, V.V., Schmitz, U.K., and Braun, H.P. (2001). Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis: Identification of multiple forms of TOM20. Plant Physiol. 125, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischmann, C., and Schuster, W. (1995). Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: Evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett. 374, 152–156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.