Abstract
Proper control of the floral transition is critical for reproductive success in flowering plants. In Arabidopsis, FLOWERING LOCUS C (FLC) is a floral repressor upon which multiple floral regulatory pathways converge. Mutations in PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1) suppress the FLC-mediated delay of flowering as a result of the presence of FRIGIDA or of mutations in autonomous pathway genes. PIE1 is required for high levels of FLC expression in the shoot apex, but it is not required for FLC expression in roots. PIE1 is similar to ATP-dependent, chromatin-remodeling proteins of the ISWI and SWI2/SNF2 family. The role of PIE1 as an activator of FLC is consistent with the general role of ISWI and SWI2/SNF2 family genes as activators of gene expression. The pie1 mutation also causes early flowering in noninductive photoperiods independently of FLC; thus, PIE1 appears to be involved in multiple flowering pathways. PIE1 also plays a role in petal development, as revealed by the suppression of petal defects of the curly leaf mutant by the pie1 mutation.
INTRODUCTION
Flowering, the transition from vegetative to reproductive growth, is a major developmental switch in plants. Plants have evolved several pathways to regulate the timing of the floral transition and thus ensure maximum reproductive success. In Arabidopsis, four major floral promotion pathways have been identified through molecular genetic studies (for reviews, see Koornneef et al., 1998; Simpson and Dean, 2002). Two of these pathways are involved in interpreting environmental cues: the photoperiod and vernalization pathways. The photoperiod pathway promotes flowering in response to daylength. In Arabidopsis, long days induce flowering, whereas short days delay the floral transition (Koornneef et al., 1998). The vernalization pathway promotes flowering in response to the prolonged exposure to cold temperatures that occur in winter. A vernalization requirement is an adaptation that prevents plants from flowering prematurely in autumn and enables them to flower in spring (Michaels and Amasino, 2000; Simpson and Dean, 2002). The other two major floral promotion pathways, which are relatively independent of environmental cues, are the gibberellin (GA) and autonomous pathways. The autonomous pathway may coordinate flowering with the developmental state (Simpson and Dean, 2002). The GA pathway mediates the floral promotion effects of GA and is required for flowering in noninductive photoperiods (Wilson et al., 1992; Blazquez and Weigel, 2000).
Genetic analyses of naturally occurring flowering-time variation among different Arabidopsis accessions revealed that allelic variation at FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) determined the winter-annual versus summer-annual habit (Burn et al., 1993; Lee et al., 1993, 1994; Clarke and Dean, 1994; Koornneef et al., 1994). Winter-annual accessions have dominant alleles of FRI and FLC and require vernalization for rapid flowering (Michaels and Amasino, 2000), whereas many summer-annual accessions have a nonfunctional fri allele (Johanson et al., 2000), which accounts for the lack of a vernalization requirement in these accessions. FRI encodes a novel protein that increases the transcript level of FLC (Michaels and Amasino, 1999; Sheldon et al., 1999; Johanson et al., 2000). FLC, which encodes a MADS-domain transcription factor, acts as a floral repressor by negatively regulating the expression of genes that promote flowering, such as AGL20/SOC1 and FT (Lee et al., 2000; Samach et al., 2000). Genes in the autonomous pathway, such as LD, FCA, FPA, and FVE, promote flowering by repressing FLC expression; thus, mutations in the autonomous pathway genes lead to late flowering through increased FLC expression (Michaels and Amasino, 1999, 2001; Sheldon et al., 1999). Floral promotion by vernalization also is achieved in part by downregulating FLC expression (Michaels and Amasino, 1999, 2001; Sheldon et al., 1999, 2000). Thus, vernalization antagonizes the activation of FLC by FRI in winter-annual Arabidopsis accessions (reviewed by Michaels and Amasino, 2000; Simpson and Dean, 2002). Therefore, FLC is a convergence point for the regulation of flowering by FRI, the autonomous pathway, and vernalization. Studies of the FLC promoter indicate that regulatory regions exist in both the region upstream of the start site of transcription and within the first intron (Sheldon et al., 2002).
Once the vernalized state is achieved, it is maintained through mitotic cell divisions in the absence of cold (Lang, 1965). Thus, vernalization causes an epigenetic change in cold-exposed cells. The level of FLC expression reflects the vernalized state. In vernalization-requiring winter-annual types of Arabidopsis, FLC is highly expressed in nonvernalized cells of the shoot apex, but after a vernalizing cold treatment, FLC remains repressed even after plants are returned to warm growing conditions (Michaels and Amasino, 2000; Sheldon et al., 2000). In the next generation, however, the highly expressed state of FLC is restored; this resetting of the epigenetic state in the next generation is similar to genomic imprinting (Michaels and Amasino, 2000). The recent isolation of genes involved in the stable maintenance of FLC repression after vernalization (Gendall et al., 2001; Levy et al., 2002) and the resemblance of one of these gene products, VERNALIZATION2 (VRN2), to Polycomb-group proteins indicate that the repression of FLC by vernalization requires chromatin-remodeling factors.
Here, we report that PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1), a gene that encodes a member of the ISWI class of ATP-dependent, chromatin-remodeling proteins, is required for the activation of FLC by FRI and by autonomous pathway mutations. Thus, PIE1 activity is necessary to permit FLC to be expressed at levels that inhibit flowering. The role of PIE1 in flowering and FLC regulation is consistent with the general role of ISWI and SWI2/SNF2 family ATPases as transcriptional activators. PIE1 also plays a role in other developmental programs. A role in petal development is revealed by the frequent formation of extra petals in pie1 mutant flowers and by the suppression of the petal defects of curly leaf mutants by the pie1 mutation.
RESULTS
pie1 Mutations Cause Early Flowering
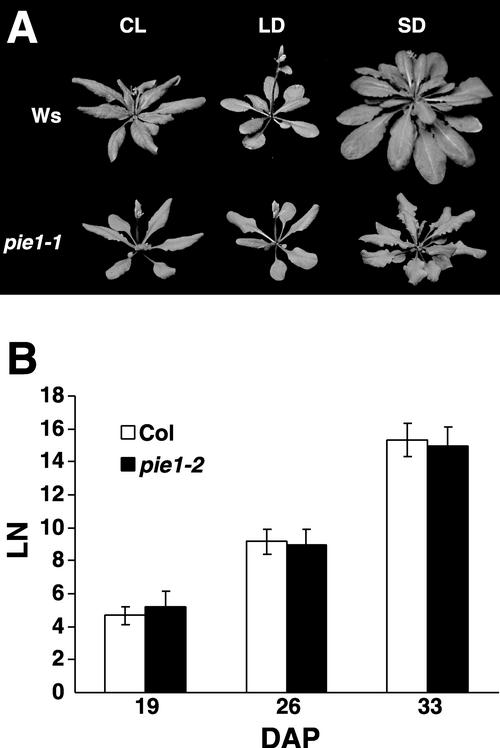
The pie1 mutant was identified in a screen for mutants that are early flowering in short days (8 h of light and 16 h of dark), conditions that are not inductive for flowering in Arabidopsis (Koornneef et al., 1998). pie1-1 mutants were found subsequently to have an early-flowering phenotype in both short days and long days (16 h of light and 8 h of dark) as well as in continuous light (Figure 1A, Table 1). However, pie1 mutants flower earlier in inductive photoperiods (long days and continuous light) than in short days; thus, a certain degree of photoperiod responsiveness is maintained. The pie1-1 mutation is recessive.
Figure 1.
Flowering and Leaf Initiation Rate of pie1-1 Compared with the Wild Type.
(A) Wild-type (Ws) and pie1-1 plants were grown under continuous light (CL), long days (LD), or short days (SD). Photographs were taken when flowering initiated and the inflorescence stem began to elongate.
(B) The number of visible leaves (LN) in wild-type (Col) or pie1-2 plants grown in short days was scored at 19, 26, and 33 days after planting (DAP). Data shown are means ± sd of 12 plants per genotype.
Table 1.
Primary Leaf Number at Flowering of pie1 Mutant Alleles in Different Photoperiodic Conditions
| Light Condition | Ws | pie1-1 | Col | pie1-2 | pie1-3 | pie1-4 |
|---|---|---|---|---|---|---|
| Continuous light | 10.67 ± 1.03 | 7.21 ± 0.80 | ND | ND | ND | ND |
| (4.00 ± 0.89) | (2.64 ± 0.63) | ND | ND | ND | ND | |
| Long days | 7.90 ± 0.32 | 5.67 ± 0.62 | 15.42 ± 1.24 | 7.38 ± 0.74 | 7.56 ± 0.53 | 7.08 ± 0.79 |
| (3.10 ± 0.32) | (2.80 ± 0.56) | (4.14 ± 0.69) | (2.71 ± 0.49) | (3.00 ± 0.71) | (2.80 ± 0.79) | |
| Short days | 35.33 ± 2.29 | 17.44 ± 1.88 | 56.00 ± 3.25 | 31.11 ± 2.98 | 30.33 ± 3.72 | 26.25 ± 3.58 |
| (9.56 ± 1.01) | (7.13 ± 0.99) | (8.50 ± 1.38) | ND | ND | ND |
Values shown are mean numbers ± sd of rosette and cauline leaves (in parentheses) at flowering. At least 12 plants were scored for each genotype and treatment. ND, not determined.
pie1-1 was identified in a T-DNA insertion population in the Wassilewskija (Ws) background. A sequence flanking a site of T-DNA insertion obtained from pie1-1 revealed a T-DNA inserted at the 3′ end of At3g12810 (MBK21.19). This T-DNA cosegregated with the early-flowering phenotype of pie1-1 in an F2 population obtained from a backcross (data not shown); thus, the pie1 phenotype appeared to be caused by this T-DNA insertion. To confirm that the loss of At3g12810 leads to the pie1 phenotype, three more T-DNA insertion alleles of At3g12810 in the Columbia (Col) background were analyzed (pie1-2, pie1-3, and pie1-4, corresponding to SALK003776, SAIL78C11, and SAIL209B1, respectively). All three Col alleles also exhibited a recessive early-flowering phenotype in short days and long days (Table 1, Figure 2B), confirming that a lesion in At3g12810 results in early flowering. The rate of leaf initiation was identical in pie1-2 and Col in short days (Figure 1B).
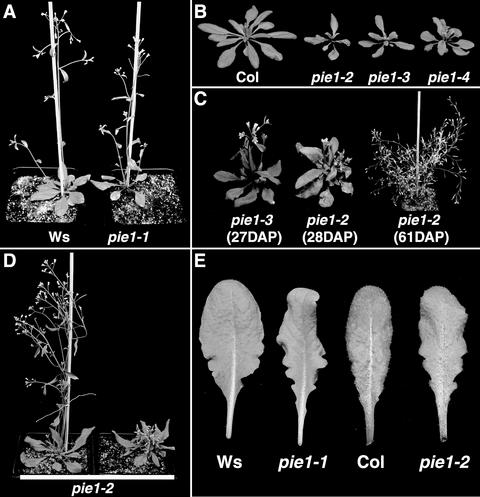
Figure 2.
pie1 Mutant Phenotype in Different Genetic Backgrounds.
(A) pie1-1 in the Ws background and wild-type Ws grown for 25 days in long days.
(B) Wild-type Col and pie1-2, pie1-3, and pie1-4 in the Col background grown in long days until flowering had initiated.
(C) The pleiotropic phenotype of Col pie1 alleles after the transition to flowering. A small fraction of pie1 mutant plants in Col showed some degree of primary inflorescence elongation (left); however, the majority of pie1 mutant plants displayed a more severe inhibition of primary inflorescence elongation and the development of numerous secondary inflorescences (middle), which led to a bushy phenotype in older plants (right). DAP, days after planting.
(D) The genetic background influences the pie1 phenotype. Shown are representative pie1-2 plants from the F2 population obtained from a cross between pie1-2 in Col and Ws. Approximately three-quarters of the pie1-2 mutant plants displayed the more normal phenotype (left), whereas approximately one-quarter of the pie1-2 mutant plants displayed the more pleiotropic phenotype (right) characteristic of Col pie1 alleles.
(E) Leaf phenotypes of pie1 mutants. Shown are the fifth rosette leaves from wild-type plants (Ws and Col) and pie1 mutants in the corresponding genetic backgrounds (pie1-1 and pie1-2, respectively) grown in short days.
Genetic Background-Dependent Phenotypes of pie1
In the Ws genetic background, pie1-1 displays, in addition to early flowering, leaves that are slightly narrower and more serrated at the base (Figure 2E) and, as discussed below, extra petals in a fraction of the flowers (see Figure 6C). After the floral transition, all three Col alleles of pie1 display the additional phenotypes of reduced fertility and reduced elongation of the primary flowering stem (bolt) accompanied by the production of numerous secondary and tertiary bolts, leading to a “bushy” phenotype (Figure 2C).
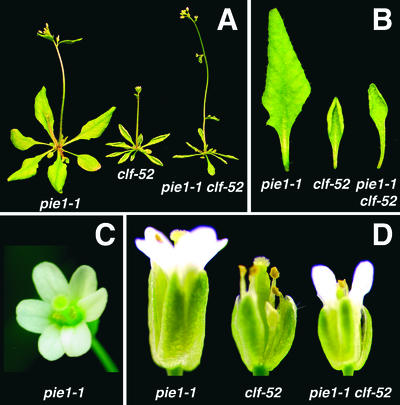
Figure 6.
Effect of pie1 on the clf Phenotype.
(A) pie1-1, clf-52, and pie1-1 clf-52 plants grown in long days to the flowering stage.
(B) Fourth rosette leaves of pie1-1, clf-52, and pie1-1 clf-52 plants grown in long days.
(C) pie1-1 flower with six petals.
(D) Flowers of pie1-1, clf-52, and pie1-1 clf-52 plants grown in long days.
There are two possibilities to account for the more severe phenotype of the pie1 alleles in Col compared with pie1-1 in Ws: pie1-1 could be a weaker allele compared with the Col alleles, or the difference in severity could result from differences in the Ws and Col genetic backgrounds. To distinguish between these possibilities, pie1-1 in Ws was crossed with Col and the phenotypic segregation in the F2 population was analyzed. Approximately one-quarter of the F2 plants (21 of 88) exhibited the early flowering of the pie1 phenotype. Approximately three-quarters of these pie1 plants (16 of 21) displayed the relatively normal growth pattern similar to pie1-1 in Ws. However, approximately one-quarter of the pie1 plants (5 of 21) exhibited the inhibition of primary bolt elongation and the bushy phenotype characteristic of Col alleles of pie1. An F2 population from the reciprocal cross of one of the Col alleles (pie1-2) with Ws also was analyzed. In this F2 population, 31 of 119 F2 plants had the pie1 phenotype, and 23 (approximately three-quarters) of these pie1 plants displayed the more normal phenotype, whereas the remaining 8 (approximately one-quarter) exhibited the inhibition of primary bolt elongation and the bushy phenotype (Figure 2D). Therefore, the difference in the Ws and Col pie1 phenotypes results from differences in the Ws and Col genetic backgrounds rather than from differences in allele strength. Furthermore, the more severe phenotype may be attributable to a single recessive locus in the Col background.
PIE1 Encodes an ISWI Family Chromatin-Remodeling Protein
The consistent early-flowering phenotype of all four insertion alleles of At3g12810 indicated that this annotated sequence encoded PIE1. Because no EST had been found for PIE1, a cDNA was cloned by reverse transcriptase–mediated PCR. Comparison of the genomic and cDNA sequences revealed that PIE1 consists of 20 exons in an 8.4-kb genomic region (Figure 3A). There were a few differences between the annotation for At3g12810 and the cDNA sequence, and the corrected amino acid sequence was deposited in GenBank. The predicted start codon noted in the GenBank accession was chosen because there were no other ATG start codons in frame that were followed by more than two codons within the 877-bp intergenic region between this predicted start codon and the adjacent gene, At3g12800 (MBK21.18). The PIE1 open reading frame is predicted to encode a 2055–amino acid protein. The T-DNA in pie1-1 is inserted at the start of the last exon, and the T-DNAs in pie1-2, pie1-3, and pie1-4 are inserted in the large fifteenth exon (Figure 3A).
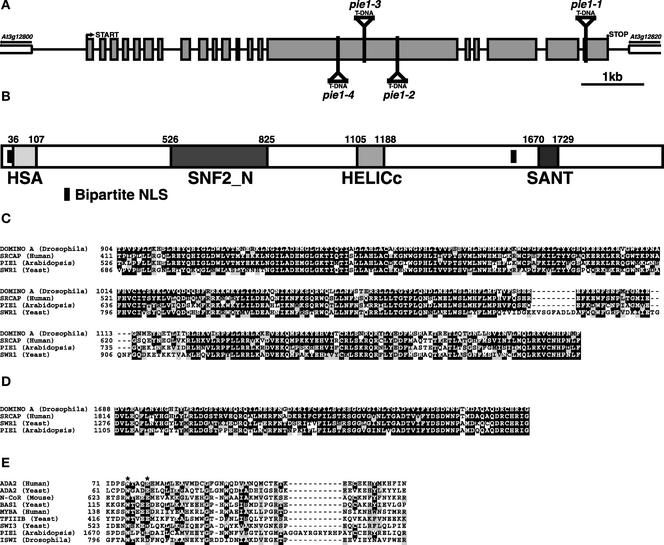
Figure 3.
PIE1 Gene and Protein Structure.
(A) Genomic arrangement of PIE1. The translation start and stop sites and the T-DNA insertion sites in pie1-1, pie1-2, pie1-3, and pie1-4 are indicated. Gray boxes indicate translated exons, and lines indicate introns or intergenic sequences.
(B) Domains of PIE1. Domains predicted by the SMART (http://smart.embl-heidelberg.de/) program and the amino acid numbers of these domains are indicated. The two putative bipartite nuclear localization signals (NLS) at the N-terminal and C-terminal regions of PIE1 are KRQKTLEAPKEPRRPKT and KKRDLIVDTDEEKTSKK, respectively.
(C) Sequence alignment of the SNF_N domain of PIE1 with Drosophila DOMINO A, human SRCAP, and yeast SWR1. Numerals indicate amino acid positions.
(D) Sequence alignment of the HELICc domain.
(E) Sequence alignment of the SANT domain. The PIE1 SANT domain was compared with the SANT domains from human ADA2, yeast ADA2, mouse N-CoR, yeast BAS1, human MYBA, yeast TFIIIB, yeast SWI3, and Drosophila ISWI. The two residues marked with stars have been shown to be important for the function of yeast ADA2 (Sterner et al., 2002).
Two domains of PIE1 are highly similar to the SNF2_N and HELICc domains of proteins belonging to the SWI2/SNF2 and ISWI class of chromatin-remodeling proteins. These types of proteins generally are involved in the transcriptional activation of target genes via chromatin remodeling, and these two domains together constitute the SWI/SNF ATPase domain that is essential for their chromatin-remodeling activity (recently reviewed by Francis and Kingston, 2001; Narlikar et al., 2002). In these two domains, PIE1 has the highest similarity with DOMINO from Drosophila (Ruhf et al., 2001), SRCAP from human (Johnston et al., 1999), and SWR1 from yeast (Jacq et al., 1997), with amino acid identities ranging from 58 to 77% (Figures 3B to 3D).
A C-terminal region of PIE1 also exhibits similarity to the SANT domain (Figures 3B and 3E). The SANT domain was found originally in SWI3, ADA2, N-CoR, TFIIIB B, and ISWI (Aasland et al., 1996). A C-terminal SANT domain is characteristic of ISWI family members (SWI2/SNF2 family members have a bromodomain in the corresponding region); thus, the domain organization of PIE1 most closely resembles that of ISWI family proteins rather than that of the SWI2/SNF2 family (Narlikar et al., 2002). Although the relatedness among SANT domains is not high, conserved and functionally important residues of the SANT domain are present in this region of PIE1. Recent studies indicate that the N-terminal half of the SANT domain is required for interaction with histone acetyltransferases or histone deacetylases and that the C-terminal half is required for the interaction with chromatin (Guenther et al., 2001; You et al., 2001; Sterner et al., 2002). It is interesting that the PIE1 SANT domain has a unique 11–amino acid linker (GGAYRGRYRHP) between the N-terminal and C-terminal halves (Figure 3E). The HSA domain, which is found in some proteins that contain the SANT domain, also is detected at the N-terminal region of PIE1 by the module-detection program SMART (http://smart.embl-heidelberg.de/; Figure 3B). Two bipartite nuclear localization signals are detected as well (Figure 3B), indicating that PIE1 likely is localized to the nucleus. Although PIE1 is similar to other SWI2/SNF2 and ISWI family proteins in the domains discussed above, other regions of PIE1 do not display any relatedness to other predicted proteins in the databases. A striking feature of PIE1 is that it exhibits a higher degree of relatedness to proteins from human, yeast, and Drosophila than to any other predicted proteins in Arabidopsis.
pie1 Suppresses FLC-Dependent Late Flowering
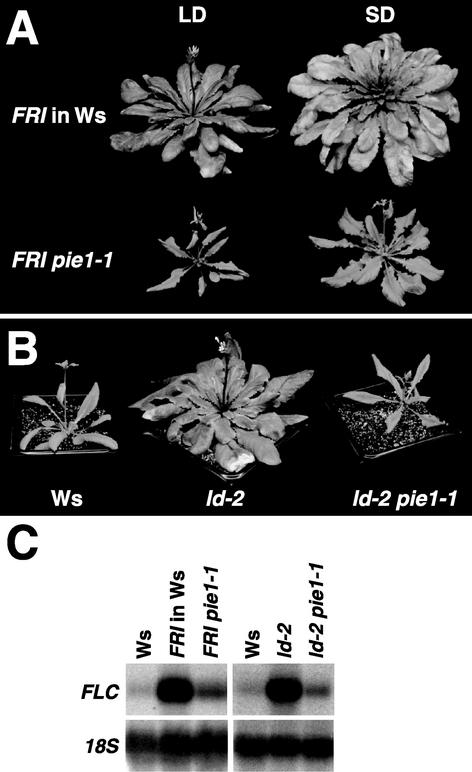
Several flowering pathways in Arabidopsis converge on the regulation of FLC expression; thus, it was of interest to evaluate the interaction of PIE1 with these pathways. Accordingly, pie1 was introduced into three genotypes that are late flowering as a result of increased FLC expression: FRI, ld, and fpa. When combined with FRI, a dominant positive regulator of FLC (Lee et al., 1993; Michaels and Amasino, 1999; Johanson et al., 2000), the late-flowering phenotype was suppressed effectively by pie1-1 and pie1-2; the FRI pie1-1 and FRI pie1-2 plants flowered at times similar to the corresponding wild-type strains Ws and Col, respectively (Figure 4A, Table 2). When pie1-1 was introduced into the autonomous pathway mutant ld-2 (Aukerman and Amasino, 1996) or fpa-6 (Schomburg et al., 2001), late flowering also was suppressed effectively and the flowering time of the double mutants was similar to that of wild-type Ws (Figure 4B, Table 3).
Figure 4.
Suppression of FLC-Dependent Late Flowering by pie1.
(A) Suppression of FRI-mediated late flowering by pie1. Representative plants of wild-type Ws and pie1-1 mutants homozygous for FRI grown in long days (LD) and short days (SD) are shown. Photographs were taken at the initiation of flowering.
(B) Suppression of ld-mediated late flowering by pie1. Representative plants of wild-type Ws, the ld-2 mutant in the Ws background, and the ld-2 pie1-1 double mutant grown in long days are shown. Photographs were taken at the initiation of flowering.
(C) Repression of FRI- or ld-mediated FLC activation by pie1. RNA was isolated from 10-day-old seedlings grown under continuous light. The blots were probed first with FLC and then reprobed with 18S ribosomal DNA (18S) as a loading control.
Table 2.
Primary Leaf Number at Flowering of FRI-Containing Lines in Long and Short Days
| Light Condition | FRI in Ws | FRI pie1-1 | Wsa | FRI in Col | FRI pie1-2 | Cola |
|---|---|---|---|---|---|---|
| Long days | 54.43 ± 9.50 | 8.56 ± 1.20 | 7.94 ± 0.94 | 72.17 ± 3.85 | 13.23 ± 1.36 | 15.42 ± 1.24 |
| (10.43 ± 1.09) | (3.56 ± 0.62) | (3.58 ± 0.61) | (9.67 ± 0.91) | ND | (4.14 ± 0.69) | |
| Short days | 89.53 ± 11.30 | 25.50 ± 4.83 | 28.60 ± 1.67 | >100 | ND | 56.00 ± 3.25 |
| (13.50 ± 1.59) | (7.07 ± 1.38) | (9.20 ± 0.45) | ND | ND | (8.50 ± 1.38) |
Values shown are mean numbers ± sd of rosette and cauline leaves (in parentheses) at flowering. At least 12 plants were scored for each genotype and treatment. ND, not determined.
Wild-type Ws and Col accessions were scored as fri-homozygous controls.
Table 3.
Primary Leaf Number at Flowering of ld- or fpa-Containing Lines in Long Days
| Light Condition | Wsa | ld-2 | ld-2 pie1-1 | fpa-6 | fpa-6 pie1-1 |
|---|---|---|---|---|---|
| Long days | 9.58 ± 1.24 | 41.36 ± 8.62 | 9.30 ± 1.26 | 52.13 ± 2.47 | 13.71 ± 1.38 |
| (3.83 ± 0.58) | (8.82 ± 0.87) | (3.78 ± 0.60) | (9.75 ± 0.71) | (4.36 ± 0.63) |
Values shown are mean numbers ± sd of rosette and cauline leaves (in parentheses) at flowering. At least 12 plants were scored for each genotype.
The wild-type Ws accession was scored as a control.
Because FRI and mutations in autonomous pathway genes cause late flowering by increasing FLC mRNA levels (Michaels and Amasino, 1999; Sheldon et al., 1999), we determined whether the pie1-mediated suppression of the FRI and ld-2 phenotypes involved an effect on FLC transcript levels. FLC transcript levels in FRI and ld-2 homozygous plants were decreased by the pie1-1 lesion (Figure 4C). Thus, PIE1 activity is required for the increased FLC expression that results from the presence of FRI or from mutations in the autonomous pathway.
PIE1 Also Regulates Flowering Time Independently of FLC
Although the largest effects of pie1 mutations on flowering time were observed in the late-flowering FRI and autonomous pathway mutant backgrounds (Tables 2 and 3), as discussed above, pie1 mutants also flower earlier than the rapid-flowering wild-type strains Col and Ws (Table 1). Because the suppression of FRI-containing lines and autonomous pathway mutants by pie1 is associated with reduced FLC expression, it was of interest to determine the fraction of the early-flowering phenotype of pie1 that is independent of the effect of the pie1 lesion on FLC expression. Accordingly, the effect of a pie1 mutation was compared with that of an flc null mutation, flc-3 (Michaels and Amasino, 1999) in both long days and short days. For the comparison with the pie1-1 allele in the Ws background, flc-3 was introduced into Ws from Col by backcrossing with Ws three times. In the F2 population resulting from the third backcross, both FLC homozygous (wild type) and flc-3 homozygous plants were scored for leaf number at flowering. flc-3 homozygous plants flowered slightly earlier than wild-type plants in both long days and short days, as reported previously for flc-3 in Col (Michaels and Amasino, 2001); however, pie1-1 plants flowered even earlier than flc-3 plants in both conditions, particularly in short days (Table 4). Confirmation that the pie1 lesion in the Col background has an FLC-independent effect on flowering time is provided by the fact that pie1-2, pie1-3, and pie1-4 mutants also flowered substantially earlier in short days (Table 1) than flc-3 in Col, which in short days flowered with 50.81 ± 2.29 rosette leaves. These results indicate that the pie1 lesion causes early flowering via FLC-independent pathways as well as by reducing FLC expression.
Table 4.
Primary Leaf Number of flc Null and pie1 at Flowering in Long and Short Days
| Light Condition | Wild Type | flc-3 | pie1-1 |
|---|---|---|---|
| Long days | 9.85 ± 1.46 | 7.63 ± 0.52 | 6.17 ± 0.58 |
| (3.15 ± 0.69) | (2.13 ± 0.35) | (3.08 ± 0.79) | |
| Short days | 41.17 ± 4.39 | 33.42 ± 3.48 | 18.07 ± 1.49 |
| (10.17 ± 0.83) | (8.25 ± 2.14) | (6.93 ± 0.62) |
Values shown are mean numbers ± sd of rosette and cauline leaves (in parentheses) at flowering. At least 12 plants were scored for each genotype and treatment. See text for the details of introgressing flc-3 into the Ws genetic background.
PIE1 Is Expressed Preferentially in the Shoot Apical Meristem and PIE1 mRNA Level Is Not Affected by Other Flowering Pathways
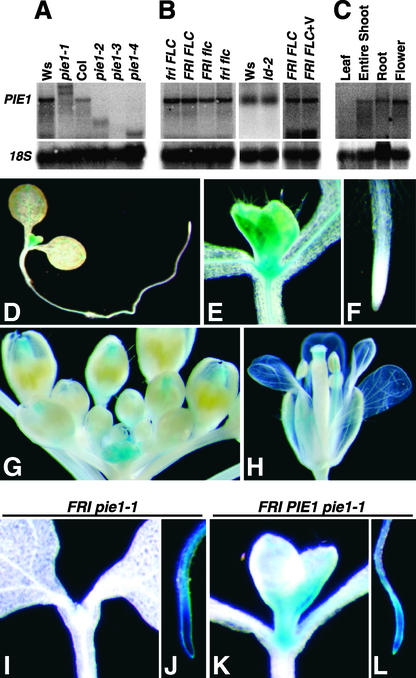
PIE1 mRNA was detectable on RNA gel blots using total RNA from whole seedlings, although the expression level was low (Figure 5A). Wild-type-size PIE1 transcript was not detected in pie1-1, but two larger bands were detected, indicating that transcription terminates at two sites in the T-DNA insertion. Smaller PIE1 transcripts were detected in pie1-2 and pie1-4, consistent with the T-DNA insertion points in the middle of the gene in these mutants. In pie1-3, no PIE1 transcript was detected, indicating that if a truncated PIE1 message is produced in pie1-3, the mRNA is unstable.
Figure 5.
PIE1 Expression Pattern and the Effect of the pie1 Mutation on the FLC Expression Pattern.
(A) PIE1 mRNA expression in wild-type and pie1 mutant alleles. RNA was isolated from 10-day-old seedlings of each genotype grown under continuous light (see [A] and [B]). The blots were probed first with PIE1 and then reprobed with 18S ribosomal DNA (18S) as a loading control (see [A], [B], and [C]).
(B) The steady state PIE1 mRNA level is not regulated by FRI, FLC, LD, or vernalization. Genotype designations for FRI and FLC lines have been described previously (Michaels and Amasino, 2001). FRI FLC+V RNA was isolated from FRI FLC seedlings that had been vernalized for 40 days and then grown under continuous light for 7 days.
(C) PIE1 mRNA expression in different tissues. Tissues were collected from adult plants grown in long days.
(D) to (H) Histochemical GUS staining of transgenic Arabidopsis containing a PIE1 promoter:GUS fusion.
(D) Seven-day-old whole seedling grown under continuous light.
(E) and (F) Magnification of the shoot apical meristem region (E) and the root tip (F) of the seedling.
(G) and (H) Inflorescence meristem region (G) and flower (H) from an adult plant grown in long days.
(I) to (L) Histochemical GUS staining of transgenic Arabidopsis containing an FLC:GUS fusion. Shown are the shoot apical meristem region ([I] and [K]) and the root tip ([J] and [L]) of representative FRI pie1-1 homozygous and FRI PIE1 pie1-1 heterozygous seedlings grown for 7 days under continuous light.
Because the late-flowering phenotype and the high level of FLC mRNA caused by the presence of FRI or autonomous pathway mutations are suppressed by the pie1 lesion or by vernalization, it is possible that the autonomous or the vernalization pathway or FRI regulates FLC by affecting PIE1 activity. However, steady state PIE1 mRNA levels were not affected by the presence or absence of FRI, LD, or FLC or by vernalization (Figure 5B). Thus, if PIE1 activity is regulated by any of these flowering pathways, this regulation does not occur at the mRNA level.
The spatial expression pattern of PIE1 was studied using a fusion of a 1.53-kb region upstream of the start codon of PIE1 to the reporter gene β-glucuronidase (GUS). In seedlings, PIE1 promoter activity was highest in the shoot apical meristem (SAM), but lower GUS activity also was detected in cotyledons, young leaves, hypocotyls, and roots, especially along the vascular tissues in these organs (Figures 5D and 5E). Unlike other genes involved in FLC-mediated flowering time regulation, such as LD (Aukerman et al., 1999), FCA (Macknight et al., 2002), FLC (Michaels and Amasino, 2000), and FPA (Schomburg et al., 2001), PIE1 promoter activity was not detected in the root tip (Figure 5F). PIE1 promoter activity was maintained after the transition to flowering in the inflorescence SAM and in floral primordia (Figure 5G). In adult flowers, the PIE1 promoter was active in sepals, in anther filaments, and at the tip of the carpel but not in papillae (Figure 5H). The spatial expression pattern revealed using this GUS fusion was consistent with the lower resolution RNA gel blot results obtained from adult tissues (Figure 5C). For example, PIE1 mRNA was barely detectable in adult leaves, but its expression was increased in shoot samples that contained the SAM along with leaves, consistent with a greater abundance of PIE1 mRNA in the SAM.
The high level of PIE1 promoter activity in the SAM, but not in the root tip, raised the question of whether FLC expression could be suppressed by the pie1 lesion in the SAM but not in the root tip. To address this issue, an FLC:GUS translational fusion construct was introduced into the FRI pie1-1 background (this FLC:GUS fusion accurately represents the native pattern of FLC expression (Michaels and Amasino, 2000). In six FRI pie1-1 lines examined, FLC expression was high in the root tip but very low or undetectable in the SAM (Figures 5I and 5J). To restore PIE1 activity to the FRI pie1-1 lines containing the FLC:GUS fusion, these lines were crossed with FRI in Ws (pie1 is a fully recessive mutation). In the resulting F1 plants, FLC expression was high in the SAM as well as in the root tip (Figures 5K and 5L). These results indicate that PIE1 is required for the high level of FLC expression in the SAM but not in the root tip. They also suggest that suppression of FLC expression in the SAM but not in the roots is sufficient to render plants early flowering and that the residual expression of FLC mRNA detected in RNA samples from entire pie1 mutant seedlings (Figures 4C and 4D) might come from FLC expression in root tips.
pie1 Frequently Makes Extra Petals and Suppresses Petal Defects in curly leaf
There was an increased frequency of extra petals in the flowers of pie1 mutants (Figure 6C). In wild-type Arabidopsis (Ws), flowers with extra petals are rare: among 71 flowers examined, 66 (93.0%) had four petals, 4 (5.6%) had three petals, and 1 (1.4%) had five petals. However, in pie1-1 mutants grown in the same conditions, approximately one-half of the flowers formed extra petals: among 95 flowers examined, 45 (47.4%) had four petals, 5 (5.3%) had three petals, and the remaining 45 (47.4%) had five or more petals. Some of the pie1 flowers with extra petals also developed extra sepals, but the frequency of extra sepal development was lower than that of extra petals. These results indicate that PIE1 plays a role in petal number regulation as well as floral induction.
Many Drosophila genes that belong to the trithorax group (trxG), including SWI2/SNF2 and ISWI family genes, were identified as suppressors of Polycomb group (PcG) genes (reviewed by Kennison, 1995). These trxG proteins generally act at the chromatin level to maintain active gene expression and to counteract repression by PcG proteins (Francis and Kingston, 2001). Because one of the Arabidopsis PcG gene mutants, curly leaf (clf), exhibits reduced petal development (Goodrich et al., 1997), whereas pie1, an ISWI gene mutant, displays extra petals, it was of interest to determine whether the petal defects in clf could be suppressed by pie1. In clf mutants, flowers display a range of abnormal phenotypes depending on the strength of the mutant allele; the most prominent of these abnormalities is that petals usually are absent or reduced severely in size. In clf mutants, leaves also are smaller and typically are curled upward compared with wild-type leaves, and the mutants flower earlier than the wild type, particularly in short days (Goodrich et al., 1997).
We isolated two clf alleles (clf-52 and clf-53) in the Ws background. clf-52 and clf-53 were identical in phenotype and displayed the typical phenotypes of clf alleles in other accessions of Arabidopsis (Goodrich et al., 1997). A double mutant between pie1-1 and clf-52 showed additive phenotypes in leaves. pie1-1 clf-52 leaves were curled upward, like clf-52 leaves, and were narrower in basal regions, like pie1-1 leaves (Figures 6A and 6B). The flowering time of pie1-1 clf-52 in long days was earlier than that of clf-52 but similar to that of pie1-1 (Figure 6A). In short days, clf-52 flowered earlier than pie1-1, and pie1-1 clf-52 flowered at times similar to the clf-52 single mutant (data not shown). The reduced sizes of flowers and individual floral organs (except petals) of pie1-1 clf-52 were similar to those of the clf-52 single mutant (Figure 6D). However, the severe defect in petal development of clf-52 was suppressed in the pie1-1 clf-52 double mutant, which displayed normal petal formation (Figure 6D).
DISCUSSION
Delayed flowering caused by the presence of FRI or by mutations in autonomous pathway genes is mediated by increased mRNA levels of the floral repressor FLC (Michaels and Amasino, 1999, 2001; Sheldon et al., 1999). Repression of these late-flowering phenotypes by vernalization results from the downregulation of FLC. Therefore, the regulation of FLC is a convergence point of flowering-time regulation by FRI, the autonomous pathway, and vernalization (recently reviewed by Simpson and Dean, 2002). Our data show that pie1 mutations can suppress FLC-mediated late flowering caused by FRI or by mutations in autonomous pathway genes. PIE1 acts upstream of FLC because PIE1 activity is required for increased FLC expression.
Interestingly, the effect of the pie1 lesion on FLC expression is restricted to the shoot apex. The expression pattern of a PIE1 reporter gene fusion indicated that, in contrast to FLC, PIE1 was not expressed in the root apex. Furthermore, the expression pattern of a FLC reporter gene fusion in a pie1 mutant revealed that the loss of PIE1 reduced FLC expression only in the shoot apex. Thus, in Arabidopsis, the level of FLC expression in the shoot apex, but not in the root apex, appears to influence flowering behavior. These results also demonstrate that root tip expression of FLC does not require PIE1. Perhaps there is another gene that provides a function similar to PIE1 in root tips to promote FLC expression, or perhaps a lack of certain negative regulators of FLC in root tips renders FLC expression independent of PIE1 or related activities.
The early-flowering phenotype of pie1 mutants was first noted in noninductive short-day photoperiods. However, pie1 mutants also flowered significantly earlier than the wild type in inductive conditions (long days and continuous light), and a degree of photoperiod sensitivity was maintained in pie1 mutants (Table 1). The early flowering of pie1 mutants in short days must occur, in part, independently of the effect of the pie1 lesion on FLC, because pie1 mutants flower much earlier in short days than do flc null mutants (Table 4).
PIE1 exhibits sequence similarity to the ATPase domains of the ISWI and SWI2/SNF2 family of chromatin-remodeling proteins. The ISWI and SWI2/SNF2 family of ATP-dependent remodeling components uses the energy of ATP hydrolysis to modify chromatin structure in a noncovalent manner. Thus, PIE1 may affect the transcriptional activation of FLC via a structural change of the chromatin in the vicinity of FLC or in the vicinity of an upstream regulator of FLC. The SWI2/SNF2 family modifiers have a bromodomain, whereas the ISWI family modifiers, such as PIE1, have a SANT domain at their C-terminal regions (Narlikar et al., 2002). The SWI2/SNF2 family can affect chromatin structure by inducing conformational changes that expose nucleosomal DNA on the surface of the histone octamer, whereas the ISWI family can affect chromatin structure by sliding the DNA around the histone octamer (Langst and Becker, 2001; Narlikar et al., 2002).
The developmental roles of several Arabidopsis proteins containing the SWI/SNF ATPase domain have been characterized. For example, SPLAYED (SYD), which encodes a SWI2/SNF2-like protein with a partial bromodomain, was isolated in a screen for LEAFY enhancers. Subsequently, SYD was found to play multiple roles in apical meristem identity and carpel and ovule development (Wagner and Meyerowitz, 2002). DDM1, another SWI2/SNF2-like protein, is required for the maintenance of cytosine methylation in the genome (Jeddeloh et al., 1999) and for the maintenance of the histone H3 methylation pattern (Gendrel et al., 2002). MOM is an SWI2/SNF2-like protein that is involved in transcriptional gene silencing but that does not affect the DNA methylation pattern (Amedeo et al., 2000). PICKLE is a CHD3 protein with two chromodomains, a PHD domain, and a SWI/SNF ATPase domain that is involved in the suppression of embryonic development during germination via the repression of an embryonic development activator, LEC1 (Ogas et al., 1999).
PIE1 is the first ISWI protein in plants for which a role in development has been identified. In Drosophila, ISWI is involved in transcriptional activation as the catalytic subunit of at least three chromatin-remodeling complexes: NURF, CHRAC, and ACF (Deuring et al., 2000; Badenhorst et al., 2002). Because mutations in PIE1 affect FLC expression, FLC-independent flowering pathways, leaf morphology, and floral organ development, PIE1 might act as a catalytic subunit in several types of chromatin-remodeling complexes with distinct developmental roles in Arabidopsis. However, one type of chromatin-remodeling complex can play many developmental roles, as shown recently for NURF (Badenhorst et al., 2002), so it is possible that there is a single complex disrupted by the pie1 lesion that is responsible for the range of phenotypes observed in the mutant.
Certain SWI2/SNF2 and ISWI family genes were first characterized genetically in Drosophila as trxG genes, which, when mutated, were suppressors of PcG genes (reviewed by Kennison, 1995). trxG proteins generally promote an open and active configuration of target chromatin, whereas PcG proteins generally favor a closed and repressed configuration. The ISWI and SWI2/SNF2 family proteins provide ATP-dependent chromatin-remodeling activity, and other types of trxG proteins are associated with chromatin-remodeling complexes (Müller and Leutz, 2001). Our observation that PIE1, an ISWI family protein, is required for the expression of FLC is consistent with a trxG-like role for PIE1.
The vernalization pathway acts to repress FLC. This repression is maintained through subsequent mitotic cell divisions in the absence of cold (Michaels and Amasino, 2000) and likely is mediated by chromatin remodeling. Recently, two genes were identified, VRN1 and VRN2, that are required for the stable maintenance of FLC repression after vernalization (Gendall et al., 2001; Levy et al., 2002). One of these genes, VRN2, encodes a protein with similarities to PcG proteins. The identities of VRN2 and PIE1 are consistent with the transcriptional regulation of FLC being subject to chromatin remodeling by opposing trxG and PcG complexes. A PIE1-containing complex would favor an active FLC conformation, whereas a VRN2-containing complex would create an inactive conformation. After vernalization, PIE1-mediated activation of FLC might be replaced by VRN2-mediated inactivation.
The pie1 clf double mutant phenotype (Figure 6) provides another example of the opposing roles of PIE1 and a PcG protein, CLF. The frequent development of extra petals in pie1 mutant flowers and the severe reduction in size or the absence of petals in clf mutant flowers indicate that PIE1 and CLF perform negative and positive regulatory functions, respectively, in Arabidopsis petal development. Perhaps PIE1 is required for the expression of a repressor of petal development, whereas CLF is required for the suppression of this repressor. It will be interesting to identify the common downstream target genes of PIE1 and CLF in petal development.
In summary, our data demonstrate that PIE1 is required for both FLC expression in the shoot apex and the activity of an FLC-independent floral repression pathway. The role of PIE1 in FLC regulation is to facilitate the increase in FLC expression caused by the presence of FRI or by autonomous pathway mutations. One possible model for the regulation of FLC by PIE1 is that signaling by FRI, the autonomous pathway, or the vernalization pathway regulates the activity of a PIE1-containing chromatin-remodeling complex, which in turn leads to the regulation of FLC transcription. It will be important to determine whether FLC is a direct target of a PIE1-containing chromatin-remodeling complex and whether FRI, the autonomous pathway, or the vernalization pathway might affect the activity of such a complex.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana pie1-1 mutant in the Wassilewskija (Ws) background was isolated from the BASTA population of the Arabidopsis Knockout Facility (http://www.biotech.wisc.edu/Arabidopsis/). pie1 T-DNA insertion lines in the Columbia (Col) background were isolated from either the SALK Collection (http://signal.salk.edu/; pie1-2, which is SALK_003776) or the Syngenta Arabidopsis Insertion Library (http://www.nadii.com/pages/collaborations/; pie1-3 and pie1-4, which are SAIL78C11 and SAIL209B1, respectively). FRI in Ws was obtained by introgressing FRI into Ws by four backcrosses from FRI in Col (Lee and Amasino, 1995). FRI pie1-1 was generated by crossing pie1-1 with FRI in Ws, and FRI pie1-2 was generated by crossing pie1-2 with FRI in Col. The following mutants are in the Ws background: ld-2 (Aukerman and Amasino, 1996), fpa-6 (Schomburg et al., 2001), and clf-52 (T-DNA insertion in the eighth intron of CLF; the designation of this clf allele as clf-52 was the recommendation of Justin Goodrich). flc-3 (Michaels and Amasino, 1999) is in the Col background. All plants were grown under ∼100 μE·m−2·s−1 cool-white fluorescent light at 22°C. For vernalization treatments, seeds were plated on agar-solidified medium containing 0.65 g/L Peter's Excel 15-5-15 fertilizer (Scotts, Maysville, OH) and germinated for 4 days under short-day conditions (8 h of light and 16 h of dark) at 22°C before being transferred to 2°C for 40 days. During cold treatment, samples were kept under short-day conditions.
T-DNA–Flanking Sequence Analyses
The sequence flanking the T-DNA of pie1-1 was obtained by thermal asymmetric interlaced PCR (Liu et al., 1995); details are described elsewhere (Schomburg et al., 2003). T-DNA borders were defined by sequencing PCR products obtained using a T-DNA border primer and a gene-specific primer. The T-DNA border primers used for each T-DNA insertion population are described on the Arabidopsis Knockout Facility World Wide Web site listed above.
RNA Gel Blot Analyses
Total RNA was isolated using TRI Reagent (Sigma) according to the manufacturer's instructions. For RNA gel blot analysis, 40 μg of total RNA was separated by denaturing formaldehyde-agarose gel electrophoresis as described by Sambrook et al. (1989). The FLC probe was a cDNA fragment lacking the conserved MADS-domain sequences. The PIE1 probe was a 6.3-kb full-length cDNA fragment. Blots also were probed with an 18S rDNA as a control for the quantity of RNA loaded.
Sequence Analyses
Genes were predicted with GenScan (Burge and Karlin, 1997). Protein sequences were analyzed with SMART (Schultz et al., 2000), PSORT (Nakai and Kanehisa, 1992), and ψ-BLAST (Altschul et al., 1997). Protein sequence alignments were generated using CLUSTAL W (Thompson et al., 1994). First-strand cDNA was synthesized using an oligo-d(T) primer and Moloney murine leukemia virus Reverse Transcriptase (Stratagene), then the PIE1 cDNA was amplified by PCR using cDNA Polymerase Mix (Clontech, Palo Alto, CA) and MBK21-2 (5′-AAGTGGGAGGTTAAGAAAATGATCATCCAC-3′) and MBK21-6 (5′-CACTCTGCAGTCACTAACCATCTTCTTCTT-3′) as primers. The PIE1 cDNA PCR product was cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA), and the sequence was determined with Big-Dye reaction mix (Amersham) using an ABI automated sequencer (Applied Biosystems, Foster City, CA).
Histochemical β-Glucuronidase Assays
The PIE1 promoter:β-glucuronidase fusion construct was generated by PCR amplification of 1.53 kb of the PIE1 5′ regulatory region using PIE1P-1 (5′-TCCCCCGGGCTAATGTTAATAATCGAACCTCCACCGCTT-3′) and PIE1P-3 (5′-GGACTAGTATGATTGCGGAAATTCGTTTTAGAAGGTTC-3′) as primers; restriction sites are shown in boldface, and sequences corresponding to the PIE1 5′ regulatory region are underlined. The resulting PCR product was digested with SmaI-SpeI and ligated to pPZP211-GUS (HindIII to EcoRI fragment of pBI121 in pPZP211) digested with PstI-XbaI (PstI was blunted with T4 DNA polymerase [New England Biolabs, Beverly, MA] after digestion), resulting in pNA179. Arabidopsis (ecotype Ws) plants were transformed with pNA179-containing Agrobacterium tumefaciens strain ABI by infiltration (Clough and Bent, 1998). Transgenic lines were selected on agar-solidified medium containing 0.65 g/L Peter's Excel 15-5-15 fertilizer and 50 μg/mL kanamycin. Staining for β-glucuronidase activity was performed as described previously (Schomburg et al., 2001).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession number for the sequence of At3g12810 is AY279398. GenBank accession numbers for the Drosophila DOMINO A, human SRCAP, and yeast SWR1 sequences shown in Figure 3C are AF076776, AF143946, and NP_010621, respectively. Accession numbers for the sequences shown in Figure 3E are as follows: human ADA2 (AAB50689), yeast ADA2 (NP_010736), mouse N-CoR (XP_109199), yeast BAS1 (P22035), human MYBA (P10243), yeast TFIIIB (NP_014359), yeast SWI3 (P32591), and Drosophila ISWI (NP_523719).
Acknowledgments
We are grateful to Fritz Schomburg for creating the T-DNA mutant population in which pie1-1 was found and for help with mutant screening, to Scott Michaels for providing the FLC:GUS construct, to Kelly Buono for the construction of FRI pie1-1, and to Scott Michaels, Mark Doyle, and Colleen Bizzell for comments on the manuscript. We acknowledge The Salk Institute Genome Analysis Laboratory and the Torrey Mesa Research Institute of Syngenta for providing knockout pools containing pie alleles. This work was supported by the College of Agricultural and Life Sciences and the Graduate School of the University of Wisconsin and by grants from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program and the National Science Foundation (0133663) to R.M.A.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012161.
References
- Aasland, R., Stewart, A.F., and Gibson, T. (1996). The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21, 87–88. [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedeo, P., Habu, Y., Afsar, K., Scheid, O.M., and Paszkowski, J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405, 203–206. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., and Amasino, R.M. (1996). Molecular genetic analysis of flowering time in Arabidopsis. Semin. Cell Dev. Biol. 7, 427–433. [Google Scholar]
- Aukerman, M.J., Lee, I., Weigel, D., and Amasino, R.M. (1999). The Arabidopsis flowering-time gene LUMINIDEPENDENS is expressed primarily in regions of cell proliferation and encodes a nuclear protein that regulates LEAFY expression. Plant J. 18, 195–203. [DOI] [PubMed] [Google Scholar]
- Badenhorst, P., Voas, M., Rebay, I., and Wu, C. (2002). Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 16, 3186–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez, M.A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. [DOI] [PubMed] [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Burn, J.E., Smyth, D.R., Peacock, W.J., and Dennis, E.S. (1993). Genes conferring late flowering in Arabidopsis thaliana. Genetica 90, 147–155. [Google Scholar]
- Clarke, J.H., and Dean, C. (1994). Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol. Gen. Genet. 242, 81–89. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Deuring, R., et al. (2000). The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Francis, N.J., and Kingston, R.E. (2001). Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 6, 409–421. [DOI] [PubMed] [Google Scholar]
- Gendall, A.R., Levy, Y.Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. [DOI] [PubMed] [Google Scholar]
- Guenther, M.G., Barak, O., and Lazar, M.A. (2001). The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21, 6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq, C., et al. (1997). The nucleotide sequence of Saccharomyces cerevisiae chromosome IV. Nature 387 (suppl.), 75.–78. [PubMed] [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Johnston, H., Kneer, J., Chackalaparampil, I., Yaciuk, P., and Chrivia, J. (1999). Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J. Biol. Chem. 274, 16370–16376. [DOI] [PubMed] [Google Scholar]
- Kennison, J.A. (1995). The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29, 289–303. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Peeters, A.J.M., and Soppe, W. (1998). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 345–370. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Blankestijn-de Vries, H., Hanhart, C., Soppe, W., and Peeters, T. (1994). The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J. 6, 911–919. [Google Scholar]
- Lang, A. (1965). Physiology of flower initiation. In Encyclopedia of Plant Physiology, W. Ruhland, ed (Berlin: Springer-Verlag), pp. 1380–1536.
- Langst, G., and Becker, P.B. (2001). ISWI induces nucleosome sliding on nicked DNA. Mol. Cell 8, 1085–1092. [DOI] [PubMed] [Google Scholar]
- Lee, H., Suh, S.-S., Park, E., Cho, E., Ahn, J.H., Kim, S.-G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., and Amasino, R.M. (1995). Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108, 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Bleecker, A., and Amasino, R. (1993). Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 237, 171–176. [DOI] [PubMed] [Google Scholar]
- Lee, I., Michaels, S.D., Masshardt, A.S., and Amasino, R.M. (1994). The late-flowering phenotype of FRIGIDA and mutations in LU-MINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 6, 903–909. [Google Scholar]
- Levy, Y.Y., Mesnage, S., Mylne, J.S., Gendall, A.R., and Dean, C. (2002). Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Duroux, M., Laurie, R., Dijkwel, P., Simpson, G., and Dean, C. (2002). Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2000). Memories of winter: Vernalization and the competence to flower. Plant Cell Environ. 23, 1145–1153. [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, C., and Leutz, A. (2001). Chromatin remodeling in development and disease. Curr. Opin. Genet. Dev. 11, 167–174. [DOI] [PubMed] [Google Scholar]
- Nakai, K., and Kanehisa, M. (1992). A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar, G.J., Fan, H.Y., and Kingston, R.E. (2002). Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhf, M.L., Braun, A., Papoulas, O., Tamkun, J.W., Randsholt, N., and Meister, M. (2001). The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development 128, 1429–1441. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schomburg, F.M., Bizzell, C.M., Lee, D.J., Zeevaart, J.A.D., and Amasino, R.M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15, 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg, F.M., Patton, D.A., Meinke, D.W., and Amasino, R.M. (2001). FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Copley, R.R., Doerks, T., Ponting, C.P., and Bork, P. (2000). SMART: A Web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Conn, A.B., Dennis, E.S., and Peacock, W.J. (2002). Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14, 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Rouse, D.T., Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (2000). The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Sterner, D.E., Wang, X., Bloom, M.H., Simon, G.M., and Berger, S.L. (2002). The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277, 8178–8186. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., and Meyerowitz, E.M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Wilson, R.N., Heckman, J.W., and Somerville, C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100, 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, A., Tong, J.K., Grozinger, C.M., and Schreiber, S.L. (2001). CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]